Introduction
Glioma is one of the most common and fatal malignant
intracranial types of cancer, accounting for ~50% of central
nervous system tumors (1). Gliomas
are classified according to the World Health Organization (WHO) and
graded from I to IV based on their histology and characteristics
(2). WHO grade Ⅳ glioblastomas
typically originate from astrocytic cells and are the most commonly
diagnosed malignant brain tumors (3). At present, the primary treatment for
glioma is surgical resection and postoperative adjuvant therapy,
including radiotherapy, chemotherapy, electric field therapy, and
immunotherapy. Despite advances in current treatment strategies, a
high-grade glioma is almost always a fatal disease with an overall
median survival of 9-10 months (4). Therefore, there is an urgent need to
understand the key factors responsible for the pathogenesis and
progression of glioma to provide a theoretical basis for inhibiting
the progression of glioma. MicroRNAs (miRNAs/miRs) are short
non-coding (nc)RNA regulatory elements, which are usually >25
nucleotides in length. Initially, they are present as pre-miRNA
processed through RNase III in the nucleus and exported to the
cytoplasm where they participate in several cellular processes
(5). During gene expression,
miRNAs can regulate the post-transcriptional levels by binding to
one or multiple specific mRNA targets, promoting gene degradation
and suppressing translation (6).
Moreover, these molecules play an important role in cancer
pathogenesis; therefore, these ncRNA molecules have been thoroughly
investigated and found to be dysregulated in several types of human
cancer including gliomas (7). With
theoretical research going deeper in analyzing the function of
miRNAs, several miRNAs have been categorized as glioma oncogenes or
suppressor genes, depending on the specific actions they perform in
the carcinogenesis of gliomas (8-10).
In addition, miRNA-controlled gene expression has arisen as a novel
potential therapeutic option for the management of glioma (11-13).
The expression levels of miR-149-3p, an evolutionarily conserved
miRNA, were reported to be altered in tumors and shown to regulate
several physiological processes, including proliferation,
apoptosis, cell cycle and metastasis, amongst other cellular
behaviors. Nonetheless, the expression levels and physiological
functions of miR-149-3p in gliomas remain to be fully
elucidated.
Chromobox 2 (CBX2), a member of the
chromodomain-containing proteins, participates in the polycomb
group (PcG) family-mediated silencing of lineage-specific genes
both in embryonic stem cells and multiple cancer types (14). In 2014, the oncogenic role of CBX2
was identified for the first time through genotranscriptomic
meta-analysis in human cancers (14). Subsequent extensive research
revealed that upregulation and amplification of CBX2 were
significantly correlated with tumor metastatic progression, cell
proliferation, and poor overall survival of patients (15-17).
However, the exact function and expression levels in glioma remain
unclear.
The present study demonstrated that miR-149-3p was
downregulated in glioma, and this downregulation was associated
with increased cell proliferation and invasion. To further study
the mechanism of its oncogene role, possible downstream target
genes were screened through bioinformatics analysis. Specifically,
it was found that miR-149-3p target specific sequences of the CBX2
mRNA. It was also found that the downregulated expression of CBX2
was associated with the inhibition of the Wnt/β-catenin pathway.
The findings of the present study demonstrated the potential role
and the underlying molecular mechanism of miR-149-3p and CBX2 in
the development of glioma and may provide novel insights for the
diagnosis and treatment of glioma.
Materials and methods
Tissues and cells
A total of 10 pairs of glioma specimens and adjacent
nontumor tissues were obtained from the Department of Neurosurgery
at Shandong Provincial Hospital Affiliated with Shandong
University. All samples were primary samples and had not received
any treatment prior to the operation. Patients involved in the
present study provided written informed consent for the use of
their tissue samples. The present study was approved by the Ethics
Committee of Shandong Provincial Hospital Affiliated with Shandong
University (approval no. SZRJJ:NO.2021-077). 293T cells and human
glioma cell lines U251 and HS683 were obtained from the Cell Bank
of Type Culture Collection of the Chinese Academy of Sciences. The
normal glial cell line SVG p12 was obtained from the American Type
Culture Collection. All cell lines used in the present study were
authenticated using Short Tandem Repeat profiling to ensure their
identity. The cells were cultured in DMEM supplemented with 10% FBS
(Biological Industries) at 37˚C with 5% CO2.
Bioinformatics analysis
Gene expression data of miRNA-149-3p and CBX2 in
glioma and normal brain tissues, accession nos. GSE90603(18) and GSE147352(19) were downloaded from Gene Expression
Omnibus (GEO). The glioma (glioblastoma multiforme and low-grade
glioma) gene expression profiles and the corresponding clinical
datasets were obtained from The Cancer Genome Atlas (TCGA)
database, Gene Expression Profiling Interactive Analysis (GEPIA)
and Chinese Glioma Genome Atlas (CGGA) databases. Target sequences
of miR-149-3p and CBX2 were predicted using TargetScan.
Expression vector construction and
cell transfection
The miR-149-3p inhibitor and its corresponding
negative control (NC) were commercially synthesized by Shanghai
GenePharma Co., Ltd. The 3'-untranslated region (UTR) of CBX2 was
synthesized and cloned into the pGL4.10 vector, to generate the
wild-type (WT) and mutant-type (MT) constructs (Shanghai GenePharma
Co., Ltd). Small interfering (si)RNAs targeting CBX2 and the
corresponding NC siRNAs were purchased from Shanghai GenePharma
Co., Ltd. Vector or siRNA transfections were performed according to
standard protocols (20) using
Lipofectamine™ 2000 (Thermo Fisher Scientific, Inc.).
Reverse transcription-quantitative
(RT-q)PCR
Total RNA from glioma tissues and cells was
extracted by TRIzol® reagent and reverse transcribed
into cDNA using the PrimeScript RT Reagent kit (Takara Bio, Inc.
Japan). qPCR was performed using the Light Roche 480 Real-time PCR
System, using a real-time quantitative PCR assay kit (Takara Bio,
Inc.). GAPDH was used as the internal standard for mRNA expression,
U6 small RNA was used as the internal reference for normalizing
miRNA expression. The sequences of the primers used were:
miR-149-3p forward, 5'-TTAGGGAGGGACGGGGG-3' and
5'CAGTGCGTGTCGTGGAGT-3' and reverse; CBX2 forward,
5'-TTCTGCCAACAGTCTGTCCC-3' and reverse, 5'-ATCCAGCCGGCAATCCTATG-3';
GAPDH forward, 5'-AGAAGGCTGGGGCTCATTTG-3' and reverse,
5'-AGGGGCCATCCACAGTCTTC-3'; and U6 forward,
5'-GCTCGCTTCGGCAGCACA-3' and reverse
5'-GAACGCTTCACGAATTTGCGTG-3'.
Cell Counting Kit-8 (CCK-8) assay
Cells in the logarithmic growth phase were seeded in
a 96-well plate at a density of 2,500 cells/well and cultured in
100 µl culture medium, with three wells per condition. The cells
were collected after 24, 48, 72 and 96 h, and 10 µl CCK-8 was added
to each well. After incubation at 37˚C for 1 h, the absorbance
values were measured at a wavelength of 450 nm using a microplate
reader (EL340, BioTek Instruments, Inc.).
Colony-formation assay
The U251 and HS683 cells were obtained at 24 h
post-transfection and seeded into 6-well plates (500 cells/well).
The culture medium was replaced every 48 h. After 2 weeks of
culture to allow for colony formation, the cells were fixed with 4%
paraformaldehyde for 15 min at room temperature and stained with
0.1% crystal violet for 30 min at room temperature. Colonies were
washed with PBS and counted.
Transwell assay
U251 and HS683 cells were obtained 24 h
post-transfection, and 200 µl of the cell suspension
(1x105 cells without FBS) was added to the upper chamber
of the Transwell insert (MilliporeSigma) and 600 µl medium
supplemented with 15% FBS was added to the lower chamber. After 24
h of incubation, the cells were fixed with 4% paraformaldehyde for
15 min at room temperature and stained using 1% crystal violet for
30 min at room temperature, and the number of cells was counted in
randomly selected fields using a light microscope. Images were
captured at a magnification of 200x.
Dual-luciferase assay
293 and U251 cells were seeded into 96-well plates
(2x104 cells/well). After 24 h, 293 and U251 cells the
miR-149-3p expression vectors were co-transfected with CBX2-WT or
CBX2-MT vectors, and the pRL-TK vector was co-transfected as the
control. After 48 h of transfection, the luciferase activity was
measured using a Dual-Luciferase® Reporter Assay System
(cat. no. E1910; Promega Corporation) according to the
manufacturer's instruction. All experiments were performed in
triplicate.
Western blotting
After transfection for 48 h, total protein was
extracted, and the protein concentration was measured using a BCA
protein assay kit (Beyotime Institute of Biotechnology). The
proteins were separated using SDS-PAGE on a 10% SDS gel and
subsequently transferred to a PVDF membrane. After blocking with 5%
non-fat milk for 1 h at room temperature, the membranes were
incubated with primary antibodies overnight at 4˚C, followed by
incubation with a horseradish peroxidase-conjugated secondary
antibody. The antibodies we used are as follows: Anti-CBX2 (cat.
no. ab235305, 1:500; Abcam), anti-c-myc (1:500; cat. no. A1309;
ABclonal Biotech Co., Ltd.), anti-cyclin D1 (1:1,000; cat. no.
A11022; ABclonal Biotech Co., Ltd.), anti-c-myc (1:500; cat. no.
A1309; ABclonal Biotech Co., Ltd.), anti-β-catenin (1:1,000; cat.
no. A19657; ABclonal Biotech Co., Ltd.) and anti-GAPDH (Abcam, cat.
no. ab9485, 1:2,500). Then, the membranes were washed three times
with TBST and incubated with ECL Plus Reagent (MilliporeSigma).
GAPDH was used as the loading control. Protein bands were
visualized using ECL Plus Reagent on an Amersham Imager 600 (GE
Healthcare).
Statistical analysis
The experiments were performed independently at
least three times and data are presented as the mean ± SEM.
Differences were compared using a one-way ANOVA followed by a
Tukey's post hoc test. Analysis was performed using GraphPad Prism
version 4.0 (GraphPad Software Inc.). P<0.05 was considered to
indicate a statistically significant difference.
Results
miR149-3p expression is downregulated
in gliomas
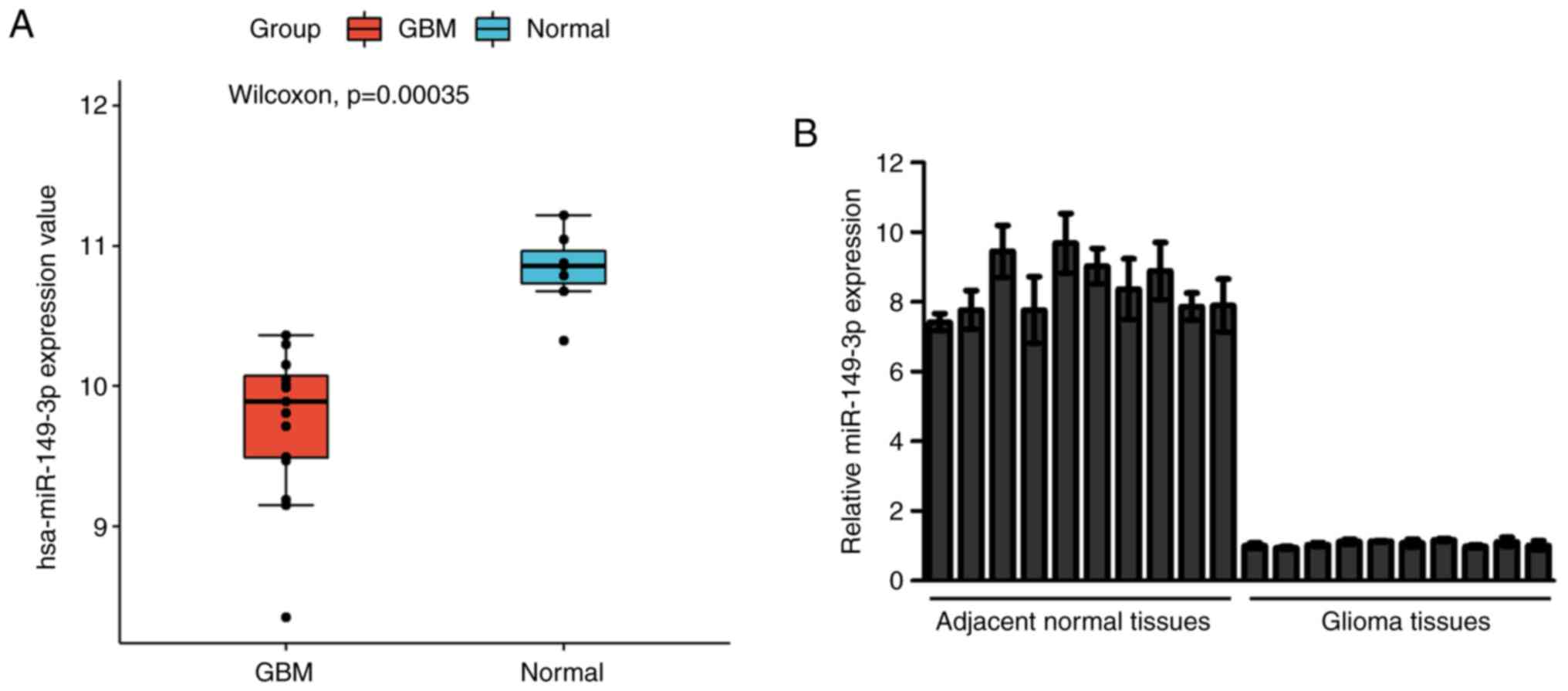
To investigate the expression of miR-149-3p in
glioblastoma the miRNAs in the GEO dataset GSE90603(18) were clustered and estimated. The
results showed that miR-149-3p expression was lower in glioblastoma
tissues compared with the normal tissues (Fig. 1A). Subsequently, the miR-149-3p
levels in 10 paired glioma tissue samples and the adjacent normal
tissue samples were measured. The RT-qPCR results indicated that
the miR-149-3p levels in glioma tissue samples were significantly
decreased compared with those in the matched paracancerous tissues
(Fig. 1B). Similarly, the levels
of miR-149-3p were higher in SVG p12 cells than in U251 and HS683
cells (Fig. S1). The
aforementioned results preliminarily showed that the expression of
miR-149-3p was downregulated in gliomas.
Downregulation of miR149-3p promotes
glioma cell proliferation and invasion
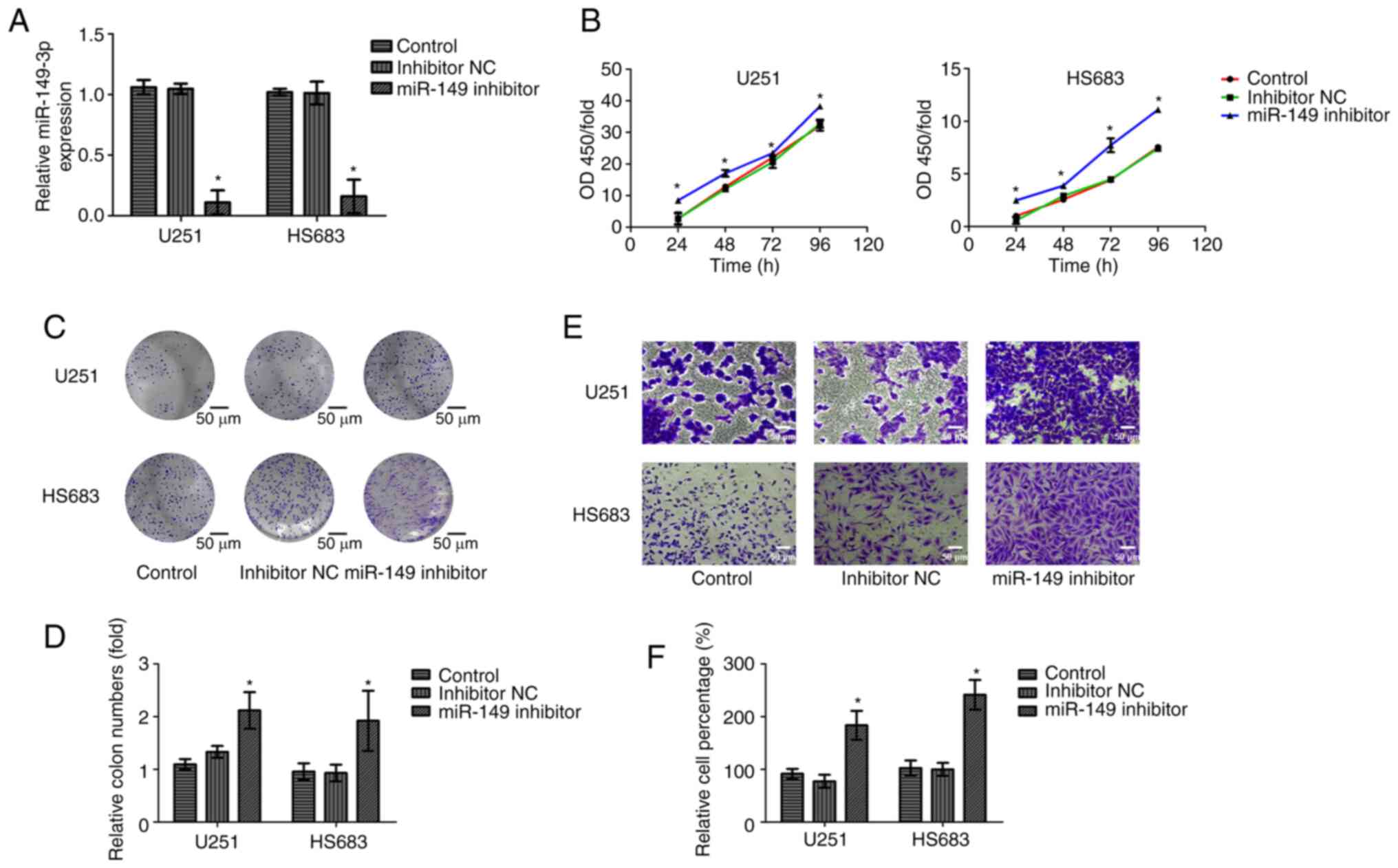
To further verify the biological function of
miR-149-3p, inhibitors of miR-149-3p were transfected into the U251
and HS683 cells. Successful transfection was confirmed using
RT-qPCR (Fig. 2A). Cell viability
and invasion of glioma cells were measured using CCK-8, colony
formation, and Transwell assays. The results indicated that
compared with those in the NC and control group, cell proliferation
and invasion were increased after miR-149-3p inhibition (Fig. 2B-F). These results reveal that
miR-149-3p may act as a tumor-suppressor gene influencing the
growth and metastasis of glioma cells.
CBX2 is overexpressed in gliomas and
is related to a poor prognosis
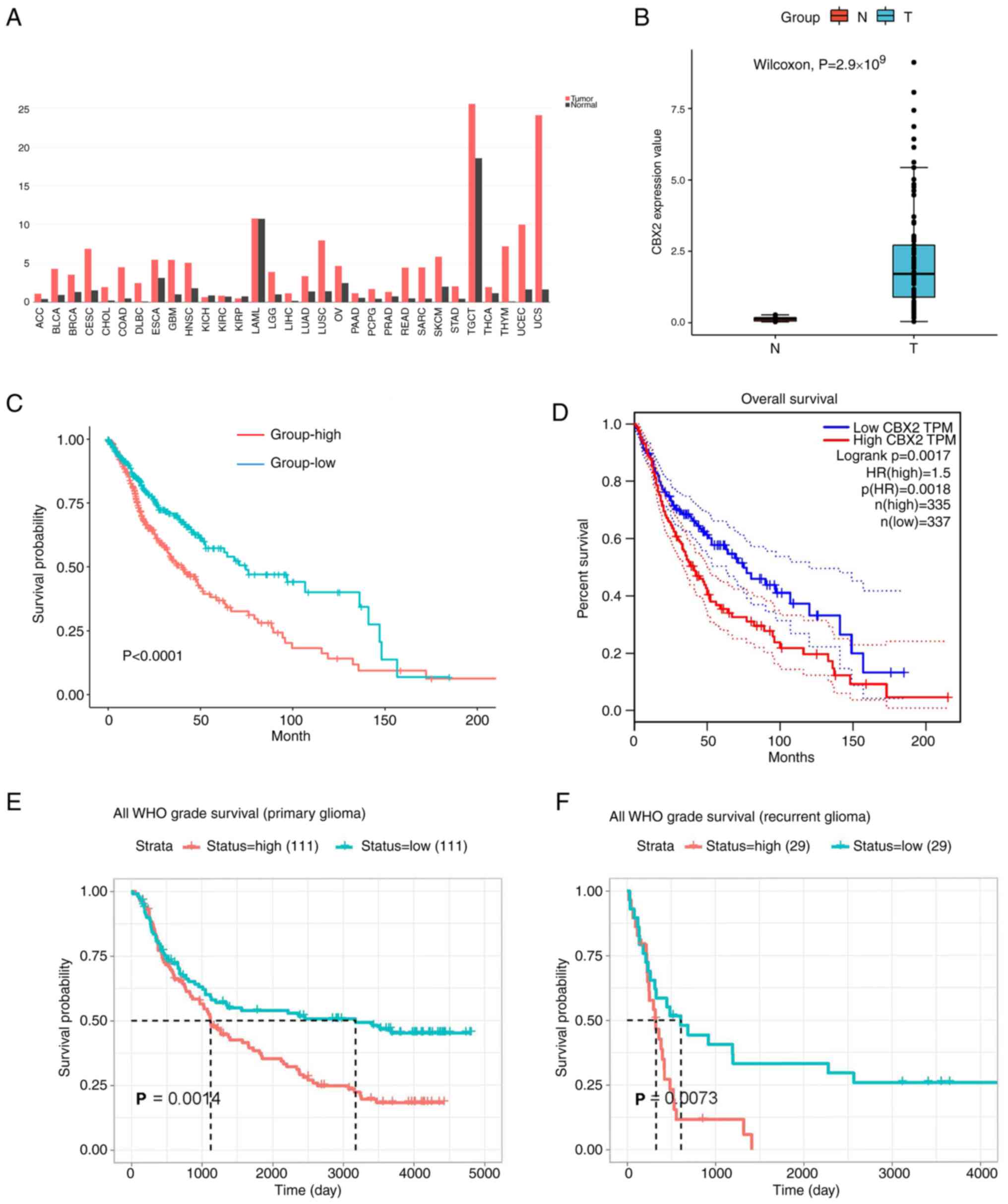
To verify the oncogenic role of CBX2, CBX2
expression in clinical samples was determined using the GEPIA
database and the results showed that CBX2 was highly expressed in
the majority of cancers including high-grade and low-grade gliomas
(Fig. 3A). To investigate CBX2
expression in glioma, the expression of CBX2 in the GEO database
array GSE147352(19) was clustered
and estimated. The results showed that CBX2 was highly expressed in
glioblastoma (Fig. 3B). To further
clarify the impact of CBX2 on the prognosis of clinical patients,
the glioma gene expression profiles and the corresponding clinical
datasets were collected from TCGA, CGGA, and GEPIA databases. The
results showed that the patients with high CBX2 expression had a
poorer prognosis (Fig. 3C-F).
These results showed that the expression of CBX2 was upregulated
and it is related to the prognosis of gliomas.
CBX2 is the target gene of
miR-149-3p
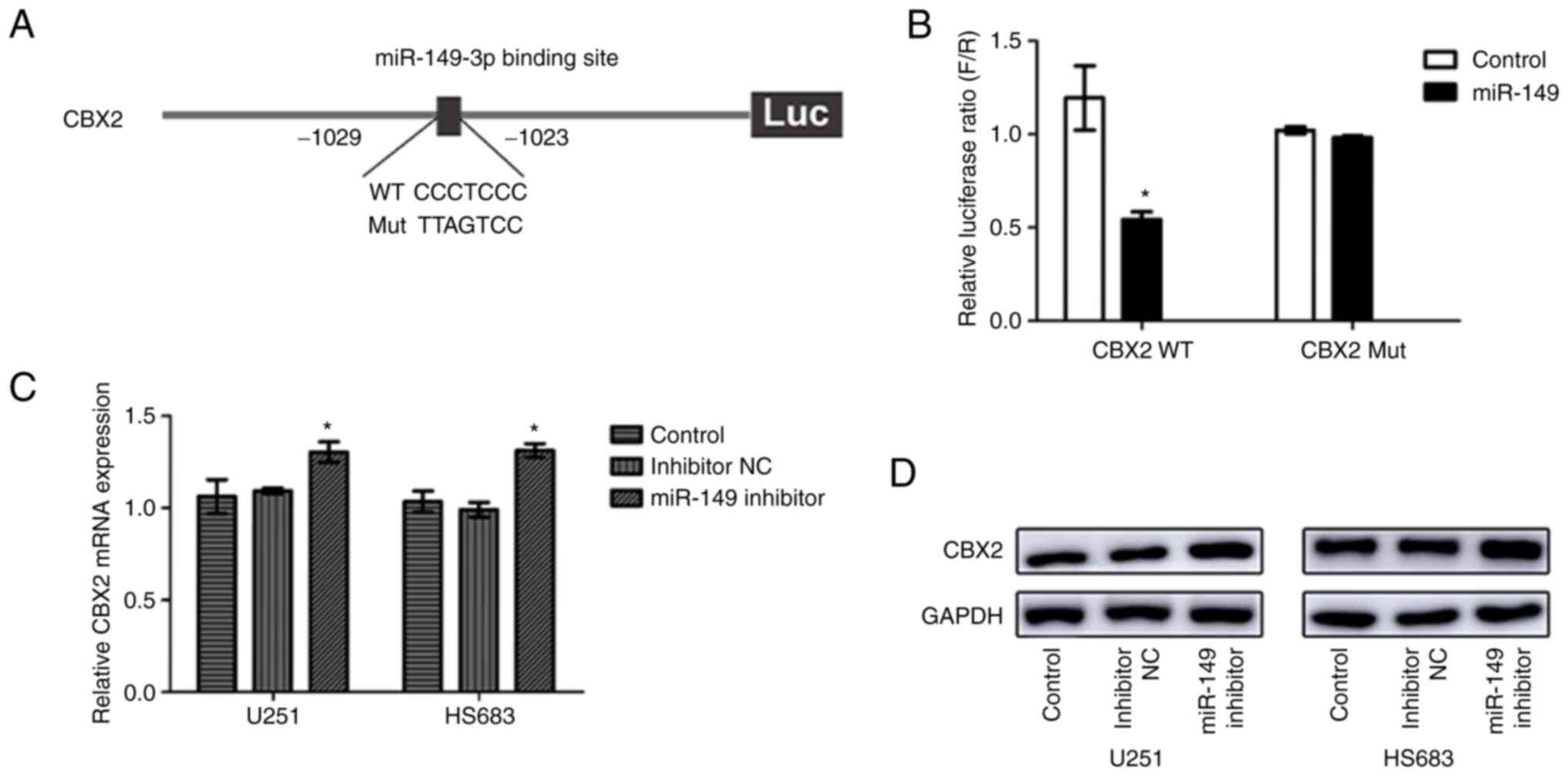
To investigate whether CBX2 expression had relevance
to miR-149-3p, bioinformatics analysis was performed to discover
the target sites in the CBX2 sequence targeted by miR-149-3p. The
results showed that miR-149-3p can bind to the 3'-UTR of CBX2 and
CBX2 may be a downstream target gene of miR-149-3p (Fig. 4A). To further verify this
hypothesis, a dual-luciferase reporter assay was performed. The
results indicated that miR-149-3p reduced CBX2 3'-UTR-WT luciferase
activity in 293T and U251 cells (Figs.
4B and S2). However,
miR-149-3p had no significant effect on the CBX2 3'-UTR-MT
luciferase activity. Moreover, downregulated miR-149-3p expression
upregulated CBX2 expression at both the mRNA and protein levels
(Fig. 4C and D). These results indicated that
miR-149-3p may bind to the 3'-UTR of CBX2 in glioblastoma
cells.
Downregulation of CBX2 suppresses
glioma cell proliferation and invasion
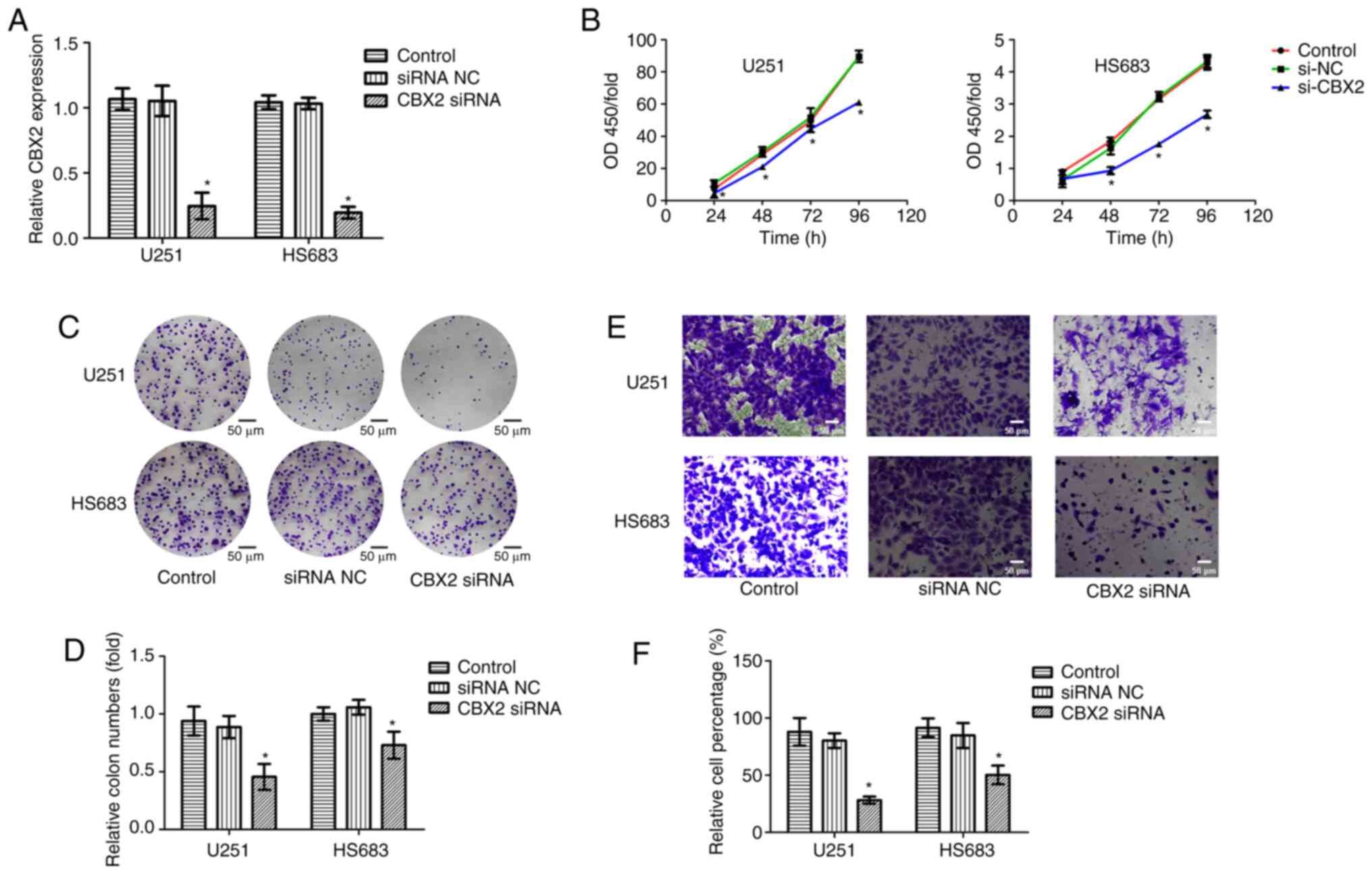
To further investigate the oncogenic role of CBX2,
CBX2 expression was knocked down in glioblastoma cell lines U251
and HS683. RT-qPCR was used to determine the CBX2 siRNA
transfection efficiency and the results indicated that the
transfection was successful (Fig.
5A). CCK-8, colony formation, and Transwell assays were
performed to measure the cell viability and invasion of glioma
cells. The results indicated that compared with that in the NC and
control groups, the cell proliferation and invasion were suppressed
after the knockdown of CBX2 expression (Fig. 5B-F). These results revealed that
CBX2 may act as an oncogene to affect the growth and metastasis of
glioma cells.
Downregulation of CBX2 suppresses the
Wnt/β-catenin pathway
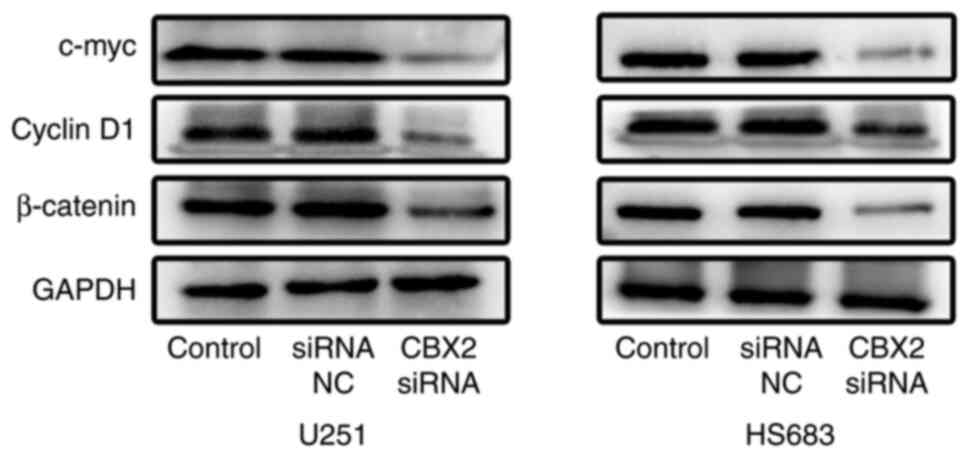
Subsequently, the effect of CBX2 on the
Wnt/β-catenin pathway was further investigated. It was found that
the upregulation of CBX2 significantly decreased the expression of
β-catenin, c-Myc, and cyclin D1 (Fig.
6). These results showed that the knockdown of CBX2 inactivated
the Wnt/β-catenin pathway in glioma cells.
Knockdown of CBX2 markedly reverses
miR-149-3p inhibitor-medicated promotion of cell proliferation and
invasion in glioma cells
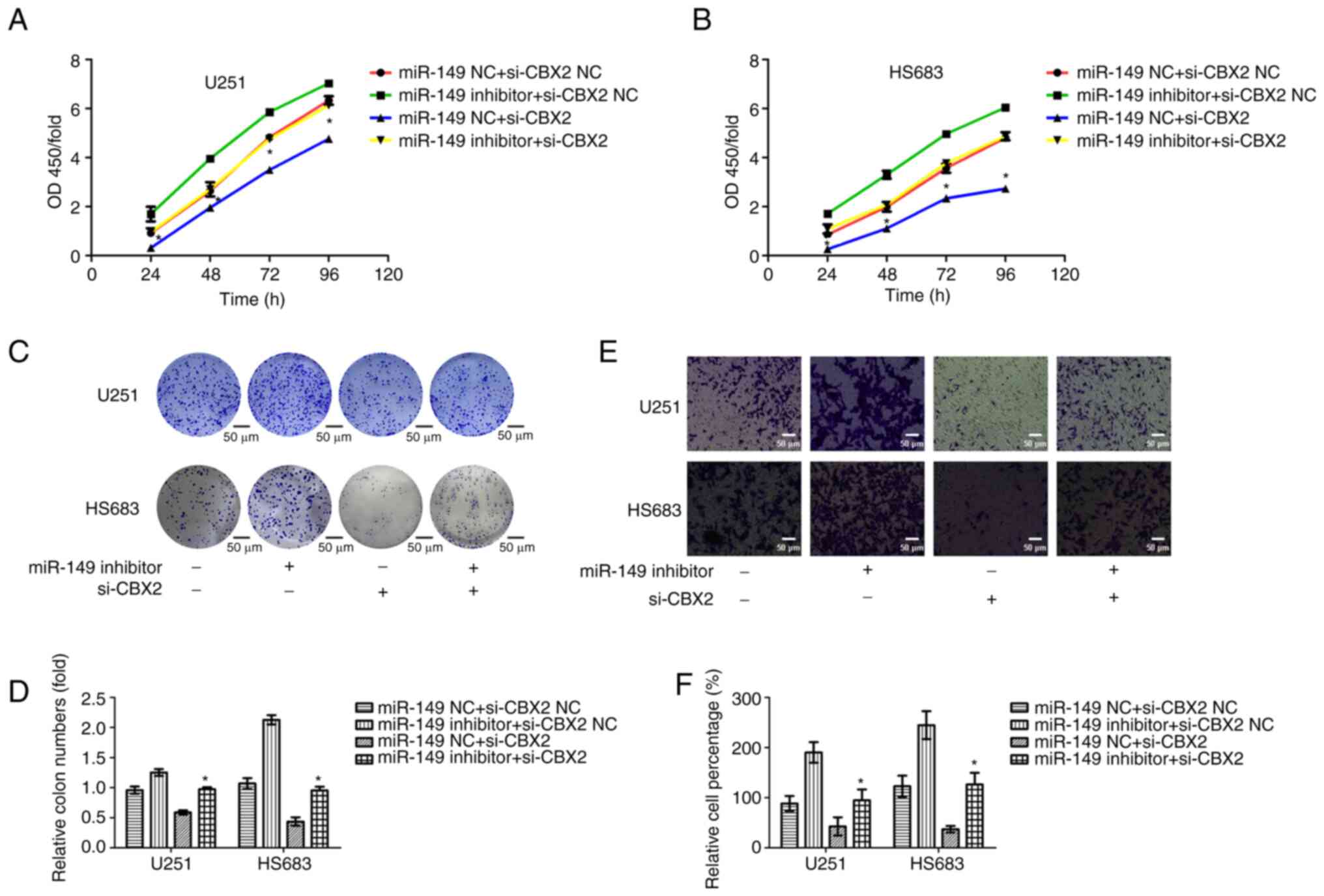
To determine whether miR-149-3p regulated cell
proliferation and invasion of glioma cells by targeting CBX2, CBX2
siRNAs, and miR-149-3p inhibitors were co-transfected into glioma
cells. Subsequently, CCK-8, colony formation, and Transwell assays
were performed to measure the cell proliferation and invasion of
glioma cells. The results indicated that the knockdown of CBX2
abrogated the promotion of cell proliferation due to miR-149-3p
inhibition. Furthermore, CBX2 siRNA markedly reversed miR-149-3p
inhibitor-mediated promotion of cell invasion in glioma cells
(Fig. 7). These results showed
that miR-149-3p promotes the proliferation and invasion of glioma
cells partly by targeting CBX2.
Discussion
The poor prognosis and high mortality rate of glioma
are a significant burden to society. Although the treatment of
glioma has made significant progress, the improvement of outcomes
for patients has been modest. Recent molecular studies on glioma
have found that several genes play a guiding role in its prognosis
and treatment; for instance, telomerase reverses transcriptase,
O-6-methylguanine-DNA methyltransferase, isocitrate dehydrogenase
1, tumor protein p53, PTEN, and epidermal growth factor receptor,
amongst others (21,22). These molecular markers were useful
to integrate findings from biological studies into clinical
practice to enhance the precision of treatment for glioma.
Accumulating evidence has shown that miRNAs can act
as oncogenes as well as suppressor genes that influence the
biological characteristics of glioma cells (23-25).
Studies performed in the past decade have found that miR-149-3p is
dysregulated in most types of cancer and it exerts its effects on
tumorigenesis and tumor progression, including proliferation,
apoptosis, cell cycle, and metastasis, amongst other cellular
behaviors. However, the expression and the influence of miR-149-3p
on cellular processes are still contested in certain types of
cancer. For example, miR-149-3p was shown to be downregulated and
suppress the proliferation and metastasis in breast cancer
(26). In gastric cancer,
miR-149-3p was also shown to be downregulated and the
downregulation of miR-149-3p inhibited tumor progression by
targeting the Wnt-1 pathway (27).
Downregulated miR-149-3p inhibited the proliferation, migration,
and invasion of renal cell carcinoma (28). However, in T-cell acute
lymphoblastic leukemia, miR-149-3p was shown to be upregulated and
promote cell proliferation and suppress apoptosis by mediating the
JunB pathway (29), and miR-149-3p
could promote Chordoma progression through the regulation of the
MAPK pathway (30). Conversely,
the expression levels and the biological functions of miR-149-3p in
glioma clinical samples and glioma cell lines remain unclear. In
the present study, it was demonstrated that miR-149-3p was
significantly downregulated in glioma tissues. Additionally, it was
also confirmed that low miR-149-3p expression enhanced the
proliferation and invasion of glioma cells.
It is well established that miRNAs play a role in
tumorigenesis by regulating various target genes in different
pathways. This study aimed to demonstrate that CBX2 is a potential
target gene of miR-149-3p through bioinformatics analysis and it
further determined their targeting relationship using
dual-luciferase reporter assay. CBX2, an epigenetics ‘reader’, is a
chromodomain-containing protein, that can recruit a protein
regulator of cytokinesis 1 to chromatin through the E3 ubiquitin
ligase activity of Ring1B via the C-terminal domain (31). Recent studies found that CBX2 is
overexpressed in several types of cancer. For example, in prostate
cancer, CBX2 was overexpressed and its depletion abrogated cell
viability, positioning CBX2 as an attractive drug target (32). Wheeler et al (33) found that CBX2 was overexpressed in
ovarian cancer which is correlated with stemness, dissemination,
chemoresistance, and worse overall survival. Moreover, several
studies showed that CBX2 is a very promising target for drug
therapy (34). Consistent with
previous studies, the present study found that CBX2 was also
upregulated in glioma. Functional studies showed that
siRNA-mediated knockdown of CBX2 decreased the cell proliferation
and invasion of glioma cells. Furthermore, it was found that CBX2
knockdown significantly decreased the expression of β-catenin,
c-Myc, and cyclin D1, indicating that CBX2 may exert an oncogenic
effect through the Wnt/β-catenin pathway. Furthermore, the rescue
experiment results indicated that the deletion of CBX2 abolished
the promotion effect of miR-149-3p inhibition on cell proliferation
and invasion in glioma cells. These results indicated that
miR-149-3p suppressed cell proliferation and invasion of glioma
cells partly by targeting CBX2. In future studies, a focus on
defining more precisely the mechanism through which miR-149-3p
inhibits the progress of glioma will be placed. Moreover, whether
exosome-derived miR-149-3p mimics have therapeutic effects on
glioma should also be explored. It is hypothesized that miR-149-3p
may be an effective biomarker and therapeutic strategy for glioma
treatment in the future.
In conclusion, the present study confirmed that the
downregulation of miR-149-3p is a common phenomenon in glioma
tissues and identified that miR-149-3p regulates cell proliferation
and invasion of glioma cells. Moreover, it was shown that CBX2 is
upregulated in gliomas, and patients with high levels of CBX2 have
poor overall survival. In addition, it was also found that the
expression of CBX2 is regulated by miR-149-3p, which may further
confirm the significance of the tumor suppressive role of
miR-149-3p in glioma. These results provide novel insights into the
mechanisms of miR-149-3p as a tumor-suppressor gene.
Supplementary Material
Expression levels of CBX2 in SVG p12,
U251 and HS683 cells. **P<0.01. CBX2, chromobox 2;
miR, microRNA.
CBX2 3'-UTR-WT and CBX2 3'-UTR-MT
activity was measured using a dual-luciferase reporter assay after
transfection with miR-149-3p inhibitors in U251 cells.
**P<0.01. UTR, untranslated region; WT, wild type;
MT, mutant type; ns, not significant; miR, microRNA.
Acknowledgements
Not applicable.
Funding
Funding: This research was funded by Shandong Provincial Natural
Science Foundation (grant no. ZR2021QH227) and Qilu Health and
Health Leading Talent Cultivation Project (grant no.
2020-2025).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YW, HP, and ZZ conceived the study. YS, JL and ZL
collected the data. YW and YS performed the experiments. YW, JL and
GW wrote the original manuscript. GW and ZZ conducted data analysis
and interpretation. HP and ZZ reviewed and edited the manuscript.
HP and ZZ confirm the authenticity of all the raw data. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Miller KD, Ostrom QT, Kruchko C, Patil N,
Tihan T, Cioffi G, Fuchs HE, Waite KA, Jemal A, Siegel RL and
Barnholtz-Sloan JS: Brain and other central nervous system tumor
statistics, 2021. CA Cancer J Clin. 71:381–406. 2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Chen R, Smith-Cohn M, Cohen AL and Colman
H: Glioma subclassifications and their clinical significance.
Neurotherapeutics. 14:284–297. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Montella L, Cuomo M, Del Gaudio N,
Buonaiuto M, Costabile D, Visconti R, Di Risi T, Vinciguerra R,
Trio F, Ferraro S, et al: Epigenetic alterations in glioblastomas:
Diagnostic, prognostic and therapeutic relevance. Int J Cancer.
153:476–488. 2023.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Kannan S, Murugan AK, Balasubramanian S,
Munirajan AK and Alzahrani AS: Gliomas: Genetic alterations,
mechanisms of metastasis, recurrence, drug resistance, and recent
trends in molecular therapeutic options. Biochem Pharmacol.
201(115090)2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S and Kim VN: The nuclear RNase
III Drosha initiates microRNA processing. Nature. 425:415–419.
2003.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Banelli B, Forlani A, Allemanni G,
Morabito A, Pistillo MP and Romani M: MicroRNA in glioblastoma: An
overview. Int J Genomics. 2017(7639084)2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Murugan AK, Munirajan AK and Alzahrani AS:
MicroRNAs: Modulators of the Ras oncogenes in oral cancer. J Cell
Physiol. 231:1424–1431. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Dai L, Liang W, Shi Z, Li X, Zhou S, Hu W,
Yang Z and Wang X: Systematic characterization and biological
functions of non-coding RNAs in glioblastoma. Cell Prolif.
56(e13375)2023.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Moore LM, Kivinen V, Liu Y, Annala M,
Cogdell D, Liu X, Liu CG, Sawaya R, Yli-Harja O, Shmulevich I, et
al: Transcriptome and small RNA deep sequencing reveals
deregulation of miRNA biogenesis in human glioma. J Pathol.
229:449–459. 2013.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Behrooz AB, Latifi-Navid H, Nezhadi A,
Świat M, Los M, Jamalpoor Z and Ghavami S: Molecular mechanisms of
microRNAs in glioblastoma pathogenesis. Biochim Biophys Acta Mol
Cell Res. 1870(119482)2023.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Anthiya S, Griveau A, Loussouarn C, Baril
P, Garnett M, Issartel JP and Garcion E: MicroRNA-based drugs for
brain tumors. Trends Cancer. 4:222–238. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yin J, Ge X, Shi Z, Yu C, Lu C, Wei Y,
Zeng A, Wang X, Yan W, Zhang J and You Y: Extracellular vesicles
derived from hypoxic glioma stem-like cells confer temozolomide
resistance on glioblastoma by delivering miR-30b-3p. Theranostics.
11:1763–1779. 2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Lei Q, Yang Y, Zhou W, Liu W, Li Y, Qi N,
Li Q, Wen Z, Ding L, Huang X, et al: MicroRNA-based therapy for
glioblastoma: Opportunities and challenges. Eur J Pharmacol.
938(175388)2023.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Clermont PL, Sun L, Crea F, Thu KL, Zhang
A, Parolia A, Lam WL and Helgason CD: Genotranscriptomic
meta-analysis of the Polycomb gene CBX2 in human cancers: Initial
evidence of an oncogenic role. Br J Cancer. 111:1663–1672.
2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Del Gaudio N, Di Costanzo A, Liu NQ, Conte
L, Dell'Aversana C, Bove G, Benedetti R, Montella L, Ciardiello F,
Carafa V, et al: CBX2 shapes chromatin accessibility promoting AML
via p38 MAPK signaling pathway. Mol Cancer. 21(125)2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bilton LJ, Warren C, Humphries RM, Kalsi
S, Waters E, Francis T, Dobrowinski W, Beltran-Alvarez P and Wade
MA: The epigenetic regulatory protein CBX2 promotes mTORC1
signalling and inhibits DREAM complex activity to drive breast
cancer cell growth. Cancers (Basel). 14(3491)2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhou H, Xiong Y, Liu Z, Hou S and Zhou T:
Expression and prognostic significance of CBX2 in colorectal
cancer: Database mining for CBX family members in malignancies and
vitro analyses. Cancer Cell Int. 21(402)2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Gulluoglu S, Tuysuz EC, Sahin M, Kuskucu
A, Kaan Yaltirik C, Ture U, Kucukkaraduman B, Akbar MW, Gure AO,
Bayrak OF and Dalan AB: . Simultaneous miRNA and mRNA transcriptome
profiling of glioblastoma samples reveals a novel set of OncomiR
candidates and their target genes. Brain Res. 1700:199–210.
2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Huang T, Yang Y, Song X, Wan X, Wu B,
Sastry N, Horbinski CM, Zeng C, Tiek D, Goenka A, et al: PRMT6
methylation of RCC1 regulates mitosis, tumorigenicity, and
radiation response of glioblastoma stem cells. Mol Cell.
81:1276–1291. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Dalby B, Cates S, Harris A, Ohki EC,
Tilkins ML, Price PJ and Ciccarone VC: Advanced transfection with
Lipofectamine 2000 reagent: primary neurons, siRNA, and
high-throughput applications. Methods. 33:95–103. 2004.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Liang A, Zhou B and Sun W: Integrated
genomic characterization of cancer genes in glioma. Cancer Cell
Int. 17(90)2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Jonsson P, Lin AL, Young RJ, DiStefano NM,
Hyman DM, Li BT, Berger MF, Zehir A, Ladanyi M, Solit DB, et al:
Genomic correlates of disease progression and treatment response in
prospectively characterized gliomas. Clin Cancer Res. 25:5537–5547.
2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zuo J, Yu H, Xie P, Liu W, Wang K and Ni
H: miR-454-3p exerts tumor-suppressive functions by down-regulation
of NFATc2 in glioblastoma. Gene. 710:233–239. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhou F, Cao W, Xu R, Zhang J, Yu T, Xu X,
Zhi T, Yin J, Cao S, Liu N, et al: MicroRNA-206 attenuates glioma
cell proliferation, migration, and invasion by blocking the
WNT/β-catenin pathway via direct targeting of Frizzled 7 mRNA. Am J
Transl Res. 11:4584–4601. 2019.PubMed/NCBI
|
|
25
|
Bian Z, Ji W, Xu B, Huo Z, Huang H, Huang
J, Jiao J, Shao J and Zhang X: Noncoding RNAs involved in the STAT3
pathway in glioma. Cancer Cell Int. 21(445)2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Chan SH, Huang WC, Chang JW, Chang KJ, Kuo
WH, Wang MY, Lin KY, Uen YH, Hou MF, Lin CM, et al: MicroRNA-149
targets GIT1 to suppress integrin signaling and breast cancer
metastasis. Oncogene. 33:4496–4507. 2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cao D, Jia Z, You L, Wu Y, Hou Z, Suo Y,
Zhang H, Wen S, Tsukamoto T, Oshima M, et al: 18β-glycyrrhetinic
acid suppresses gastric cancer by activation of miR-149-3p-Wnt-1
signaling. Oncotarget. 7:71960–71973. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Okato A, Arai T, Yamada Y, Sugawara S,
Koshizuka K, Fujimura L, Kurozumi A, Kato M, Kojima S, Naya Y, et
al: Dual strands of pre-miR-149 inhibit cancer cell migration and
invasion through targeting FOXM1 in renal cell carcinoma. Int J Mol
Sci. 18(1969)2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Fan SJ, Li HB, Cui G, Kong XL, Sun LL,
Zhao YQ, Li YH and Zhou J: miRNA-149* promotes cell proliferation
and suppresses apoptosis by mediating JunB in T-cell acute
lymphoblastic leukemia. Leuk Res. 41:62–70. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Long C, Jiang L, Wei F, Ma C, Zhou H, Yang
S, Liu X and Liu Z: Integrated miRNA-mRNA analysis revealing the
potential roles of miRNAs in chordomas. PLoS One.
8(e66676)2013.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Kaustov L, Ouyang H, Amaya M, Lemak A,
Nady N, Duan S, Wasney GA, Li Z, Vedadi M, Schapira M, et al:
Recognition and specificity determinants of the human cbx
chromodomains. J Biol Chem. 286:521–529. 2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Clermont PL, Crea F, Chiang YT, Lin D,
Zhang A, Wang JZ, Parolia A, Wu R, Xue H, Wang Y, et al:
Identification of the epigenetic reader CBX2 as a potential drug
target in advanced prostate cancer. Clin Epigenetics.
8(16)2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wheeler LJ, Watson ZL, Qamar L, Yamamoto
TM, Post MD, Berning AA, Spillman MA, Behbakht K and Bitler BG:
CBX2 identified as driver of anoikis escape and dissemination in
high grade serous ovarian cancer. Oncogenesis. 7(92)2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Jangal M, Lebeau B and Witcher M: Beyond
EZH2: Is the polycomb protein CBX2 an emerging target for
anti-cancer therapy? Expert Opin Ther Targets. 23:565–578.
2019.PubMed/NCBI View Article : Google Scholar
|