Introduction
Idiopathic pulmonary fibrosis (IPF) is a
progressive, irreversible lung disease with a poor prognosis. It
usually occurs in the adults and the 5-year survival rate is only
~20-25%, worse than most types of cancer (1,2). The
etiology of IPF remains unknown and it is often characterized by
the abnormal activation of alveolar epithelial cells and
fibroblasts, leading to the continuous accumulation of collagen and
extracellular matrix, ultimately causing structural damage to the
lung tissues and lung dysfunction (3). Currently, two drugs, nintedanib and
pirfenidone, have been approved for the treatment of IPF, which may
delay the progression of IPF to some extent (4). However, their effects remain limited
and the search for new molecules or therapeutic strategies
continues to be a research hot spots and clinical challenge.
Exosomes are a type of extracellular vesicles with a
diameter of 30-150 nm that are released by most eukaryotic cells
and circulate in the extracellular environment (5). They are able to carry various types
of cellular cargo, including nucleic acids, proteins, lipids and
metabolites, thereby playing important roles in intercellular
communication. A number of studies have indicated that exosomes
secreted from pathological lung cells and the microenvironment
exacerbate fibrosis by activating epithelial cells or fibroblasts
(6,7). However, exosomes released from the
normal lung cells, especially the widely concerned mesenchymal stem
cells (MSCs), have been found to show promising therapeutic
potential (8,9). MSCs belong to the class of
pluripotent stem cells, thus have self-renewal capabilities and are
able to differentiate into diverse types of cells (10). In recent decades, MSCs have been
highlighted for their potential use in cell therapy for various
diseases (11). MSC-derived
exosomes have gained attention due to their non-oncogenic and
immunogenic characteristics (11).
For instance, treatment with bone marrow MSC-derived exosomes was
found to inhibit the epithelial-mesenchymal transition (EMT) of
lung cells induced by silica, as well as alleviate the progression
of fibrosis in vivo (12).
Some clinical assays have shown that treatment with MSC-derived
exosomes is both safe and effective against some pulmonary
diseases, including severe COVID-19 (13,14).
In addition, exosomes derived from normal lung cells, namely human
bronchial epithelial cell, have also been found to attenuate
TGF-β-induced myofibroblast differentiation and lung epithelial
cell senescence by inhibiting TGF-β-WNT crosstalk (8).
In our previous study, a rare population of basal
cells (SOX9 positive) located at airway epithelium rugae was
isolated and identified. These stem-like cells, able to be passaged
for at least 30 doublings, were found to regenerate human lung
epithelium in patients with chronic lung diseases, as well as
rescue dysregulated pulmonary function (15). It has been reported that
MSC-derived exosomes are partly responsible for MSC-mediated
regeneration (16). Therefore,
exosomes derived from basal cells may play vital roles in the
recovery of lung function. Thus, it was hypothesized that basal
cell-derived exosomes could reverse the activation of lung
cells.
EMT is an important pathological change in the
activation process of IPF and the disruption of EMT-related
effectors can inhibit the occurrence and development of IPF
(17). In the present study, the
role of basal cell-derived exosomes on the EMT of activated lung
cells induced by TGF-β1 was investigated. RNA sequencing was used
to identify the dysregulated genes in exosome-treated lung cells.
Next, the role of one key gene, anoctamin-1 (ANO1), which encodes
the protein recognized as a Ca2+-activated chloride
channel (18), in the
TGF-β1-induced EMT of lung cells was investigated. Finally,
bleomycin-induced pulmonary fibrosis model was used to evaluate the
treatment effect of exosomes. The present study is the first to the
best of the authors' knowledge to highlight the therapeutic
potential of airway basal cell-derived exosomes on IPF. The
findings are presented in accordance with the MDAR reporting
checklist.
Materials and methods
Cells and cell culture
Primary SOX9+ airway basal cells,
accounting for ~1% of the basal cells, were isolated from patients
with IFP as reported in a previous study (15) and cultured in DMEM/F12 medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
bovine serum (FBS; Cytiva), 1% amphotericin, antibiotics and a
growth factor cocktail, as previously described (19). Human bronchial epithelial cell line
BEAS-2B (cat. no. SCSP-5067) and human embryonic fibroblasts MRC-5
(cat. no. GNHu41) were purchased from the Cell Bank of Chinese
Academy of Sciences. BEAS-2B and MRC-5 were cultured with DEME
medium (Gibco; Thermo Fisher Scientific, Inc.) and MEM medium
(Gibco; Thermo Fisher Scientific, Inc.), respectively, both of
which were supplemented with 10% FBS and 1% amphotericin. All cells
were maintained in an humidified incubator with 5% CO2
at 37˚C.
Isolation and identification of
exosomes
SOX9+ airway basal cells (within 10
passages) were cultured with exosome-free FBS for 48 h. Then, the
resulting supernatant was collected and filtered using a 0.45-µm
film to remove cell fragments. Next, Qiagen exoEasy Maxi Kit
(Qiagen GmbH) was used to extract exosomes, according to the
manufacturer's instructions. The number and size distribution of
the resulting exosomes was analyzed using NanoSight NS300 (Malvern
Instruments, Inc.). Transmission electron microscopy (TEM) was used
to observe the morphology of the exosomes. Briefly, a drop of the
diluted exosomes were loaded on a cooper mesh and fixed with 2.5%
glutaraldehyde for 5 min at room temperature. After an incubated
with 1% phosphotungstic acid for 10 min at temperature, the sample
was used for TEM imaging. The exosomal positive protein markers
[CD63 and tumor susceptibility gene 101 protein (TSG101)] and
negative marker (β-Tubulin) were analyzed using western
blotting.
Western blotting
BEAS-2B and MRC-5 were treated with 1 ng/ml TGF-β1
for 24 h before culturing with/without exosomes for an additional
24 h. Cells without any treatment were used as the control group.
Next, the cells were lysed with RIPA buffer (Beyotime Institute of
Biotechnology) and the proteins in the supernatant were quantified
using a BCA kit (Beijing Solarbio Science & Technology Co.,
Ltd.) after the lysates had been centrifuged at 12,000 x g for 5
min at 4˚C. Then, 10 µg of protein was added and separated in 10%
SDS-PAGE and the proteins in the gels were transferred onto PVDF
membranes. Subsequently, membranes were blocked with 5% fat-free
milk for 1 h at room temperature, which were then incubated with
primary antibodies at 4˚C overnight, followed by the secondary
antibody for 2 h at room temperature. Finally, the signals were
enhanced using ECL Plus Western blotting system kit (Beyotime
Institute of Biotechnology). The primary antibodies, namely ANO1
(cat. no. 14476; 1:1,000), vimentin (cat. no. 5741; 1:1,000),
E-cadherin (cat. no. 3195; 1:1,000), CD63 (cat. no. 52090;
1:1,000), TSG101 (cat. no. 72312; 1:1,000), β-Tubulin (cat. no.
2146; 1:1,000), Fibronectin (FN1; cat. no. 26836; 1:1,000),
Collagen I A1 (COL1A1; cat. no. 72026; 1:1,000), GAPDH (cat. no.
5174; 1:1,000) and secondary antibody HRP-labeled IgG (cat. no.
7074; 1:3,000), were obtained from Cell Signaling Technology, Inc.
Gray values of the blot bands were analyzed using ImageJ software
(version 1.8.0; National Institutes of Health).
RNA sequencing
After treating the cells (BEAS-2B and MRC-5) with 1
ng/ml TGF-β1 for 24 h or further treatment with 5x108
particles/ml exosomes for 24 h at 37˚C, the total RNA of the four
samples was extracted using RNeasy mini kit (Qiagen GmbH).
Integrity of total RNA was assessed using the Agilent 2100
Bioanalyzer (Agilent Technologies Inc.). Paired-end libraries were
synthesized by using the Stranded mRNA-seq Lib Prep Kit (cat. no.
RK20301; ABclonal Biotech Co., Ltd.) in accordance with the
manufacturer's protocols. Purified libraries were quantified by
Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Inc.) and
validated by Agilent 2100 bioanalyzer (Agilent Technologies Inc.)
to confirm the insert size and calculate the mole concentration.
Cluster was generated by cBot (Illumina, Inc.) with the library
diluted to 10 pM and then were sequenced on the Illumina NovaSeq
6000 (Illumina, Inc.). cDNA library construction and RNA sequencing
were performed by Shanghai Biotechnology Corporation. Nucleotide
length and the direction of sequencing were 150 bp and paired-end
respectively. The raw data was deposited in the SRA database and
accessible at https://www.ncbi.nlm.nih.gov/sra/PRJNA1051029.
Stringtie software (v1.3.3b; The Center for
Computational Biology, Johns Hopkins University) was used to count
the fragment within each gene and TMM algorithm was used for
normalization. Differential expression analysis for mRNA was
performed using the edgeR package in R (version 3.4.3; http://www.R-project.org/) (20). Fragments Per Kilobase Million
(FPKM) was used to indicate the expression level of gene.
Fold-change (FC) was calculated using the ratio of the mean FPKM of
TGF-β1 + exosome group to that of TGF-β1 group. The dysregulated
genes in TGF-β1 + exosome group were screened using the threshold
of |log2(FC)| value >1(21).
The P-value was not taken as the threshold, as the sample number in
each group was less than three.
Bioinformatical analyses
Gene Ontology (GO) and Kyoto Encyclopedia of Genes
and Genomes (KEGG) pathway analysis were conducted to determine the
biological role of the dysregulated genes via the enrich package in
R (version 3.4.3; http://www.R-project.org/) (20). Rich factor was calculated using the
ratio of
dysregulated_gene_in_this_pathway/dysregulated_gene_in_all_pathway
to all_gene_in_this_pathway/all_gene_in_all_pathway. The P-value
was used to screen the significant GO terms or KEGG pathways and
the top 30 terms or pathways were further screened by rich factor
to draw the corresponding bubble chart.
Reverse transcription-quantitative
(RT-q) PCR
The total RNA of the 5x105 cells was
extracted using a RNeasy mini kit (Qiagen GmbH) according to
manufacturer's protocols and quantified on a NanoDrop ND-1000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.). Next, 500 ng of RNA was transcribed into cDNA using a
ReverTra Ace qPCR RT kit (Toyobo Life Science) following
manufacturer's instructions. Then, 2 µl of cDNA (n=3) was mixed
with 10 µl of Master SYBR Green I mix (Roche Diagnostics), 1 µl of
primers and water to obtain a mixture of 20 µl, which was further
reacted on an ABI StepOnePlus Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The reaction was
performed at 95˚C for 5 min, followed by 40 cycles of 95˚C for 10
sec and 60˚C for 60 sec according to manufacturer's protocols. The
relative expression levels of mRNA were calculated using the
2-ΔΔCq method (22) and GAPDH was used as the internal
control. The sequences of the primers use are listed in Table I.
 | Table IPrimer sequences used for reverse
transcription-quantitative PCR. |
Table I
Primer sequences used for reverse
transcription-quantitative PCR.
| Gene | Primer sequence
(5'-3') |
|---|
| KRT81 | Forward:
GCATTGGGGCTGTGAATGTCT |
| | Reverse:
ACCCAGGGAGCTGATACCAC |
|
PCDHGA12 | Forward:
CACCGGGACTACAAAGGGC |
| | Reverse:
ATAGCGTATCTGGGTGCATCC |
| ANO1 | Forward:
CTGATGCCGAGTGCAAGTATG |
| | Reverse:
AGGGCCTCTTGTGATGGTACA |
| CXCL8 | Forward:
TTTTGCCAAGGAGTGCTAAAGA |
| | Reverse:
AACCCTCTGCACCCAGTTTTC |
| CXCL5 | Forward:
AGCTGCGTTGCGTTTGTTTAC |
| | Reverse:
TGGCGAACACTTGCAGATTAC |
| GAPDH | Forward:
GGAGCGAGATCCCTCCAAAAT |
| | Reverse:
GGCTGTTGTCATACTTCTCATGG |
Cell transfection
The overexpression vector of ANO1 (pcDNA3.1-ANO1)
and its short interfering (si)RNA (si-ANO1) were both synthetized
by Sangon Biotech Co., Ltd. Next, the overexpression vector (2.5
µg) and si-ANO1 (2 µg) were transfected into 5x105 cells
(BEAS-2B and MRC-5) using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37˚C for 48 h,
according to the manufacturer's instructions. The control vector
and the si-RNA were also transfected into 5x105 cells
(BEAS-2B and MRC-5) as negative control (NC) and si-NC,
respectively. Subsequently, cells transfected with vectors were
screened by G418 for 10 days, followed by qPCR validation. Cells
transfected with siRNAs were directly detected by qPCR to confirm
the interference efficiency. si-NC: 5'-UUCUCCGAACGUGUCACGUTT-3',
si-ANO1: 5'-CGTGTACAAAGGCCAAGTA-3'.
Transwell assay
Cells were collected and resuspended in FBS-free
medium. Then, 100 µl of cells at a density of 4x105
cells/ml were seeded into the upper chamber and 600 µl of the
complete medium was added into the lower chamber. After 24 h of
migration, the residual cells on the Inner layer of the membrane
were removed and the migrated cells were fixed with methanol for 20
min at room temperature, followed by staining with 0.1% crystal
violet for 15 min at room temperature. Finally, the migrated cells
were observed and counted under an inverted microscope.
Animals and treatments
Male C57BL/6 mice (6-8 weeks; 18-22 g) were
purchased from Shanghai Laboratory Animal Research Center. All mice
were maintained in 12-h light/dark cycle and with free access to
food and water. The temperature and relative humidity were maintain
at 22±2˚C and 50-60%, respectively. The 18 mice were randomly
divided into 3 groups (n=6): Control group, model (bleomycin
treatment) group and exosome group (bleomycin as well as exosome
treatments). Pulmonary fibrosis was induced with a single
intratracheal injection of 2 U/kg bleomycin (Nippon Kayaku Co.,
Ltd.; cat. no. H20090885) in 30 µl saline as reported (23). Mice administered with same volume
of saline were served as controls. At 10 days post bleomycin
treatment, exosomes (10x109 particles/kg) were given for
~30 min/day for 7 consecutive days using a nebulizer (Trek S
Portable Compressor Nebulizer Aerosol System; PARI GmbH). At 21
days post bleomycin treatment, all mice were anesthetized by
intraperitoneal injection of chloral hydrate (350 mg/kg), then
sacrificed by cervical dislocation. Lung tissues were collected for
protein detection and histological examination. All experimental
procedures were approved by the Animal Welfare Ethics Committee of
Shanghai Sixth People's Hospital (approval no. 2023-0374).
Histological evaluation
Pulmonary tissue samples were fixed with 4%
paraformaldehyde for 48 h at room temperature, followed by
dehydration in graded alcohol and embedding in paraffin. Embedded
tissue samples were then cut into 5 µm thick sections and stained
with hematoxylin and eosin (H&E) for 5 min or Masson's
trichrome reagents for 30 min at room temperature. Images of the
histologic sections were captured using light microscopy.
Statistical analysis
Data are presented as the mean ± standard deviation
from three independent experiments unless otherwise noted. All
statistical analyses were performed using GraphPad Prism 7.0
(Dotmatics). Statistical significance was analyzed by unpaired
two-tailed Student's t-test, and one-way ANOVA with Tukey's post
hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Airway basal cell-derived exosomes
suppress the EMT of lung cells induced by TGF-β1
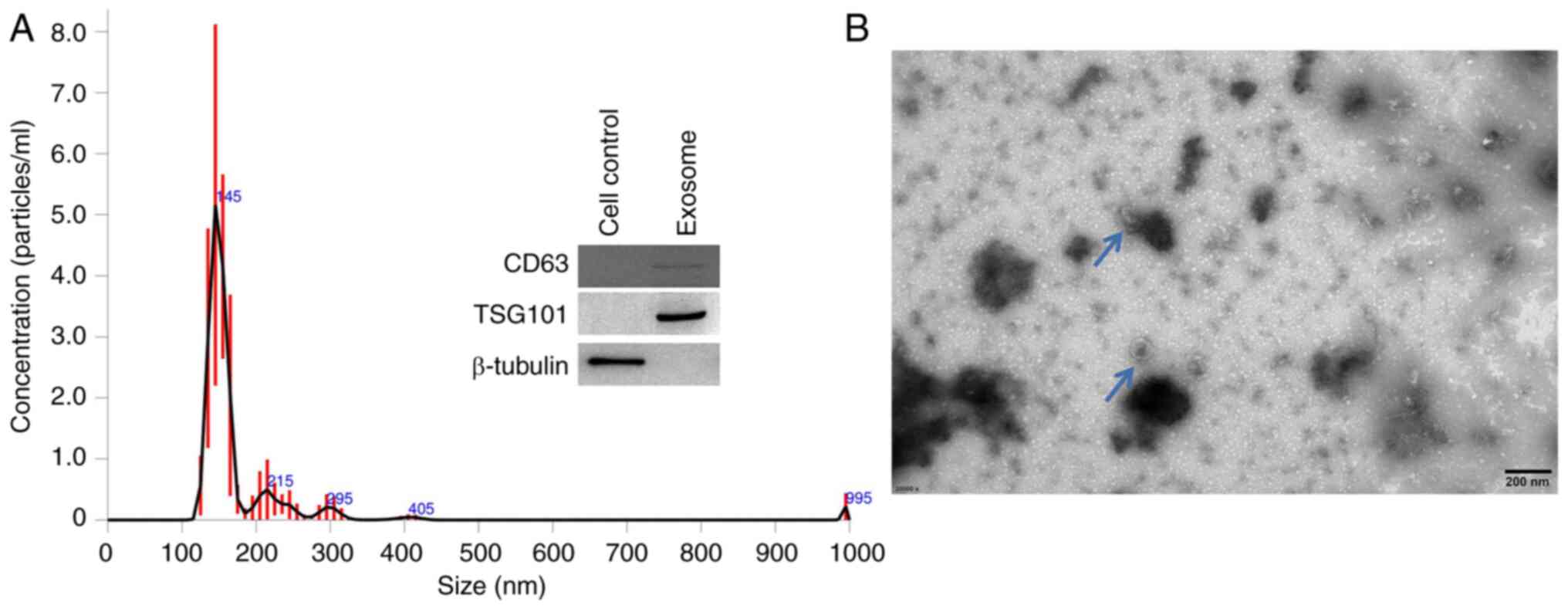
First, the exosomes were extracted from the medium
of airway basal cells and the nanoparticle tracking analysis
indicated that the majority of the particles had an average
diameter of 145 nm, with some showing a larger size (Fig. 1A). The TEM image (Fig. 1B) showed that the obtained
particles were round-like and had a size of ~100 nm. Furthermore,
exosome makers CD63 and TSG101 were both expressed in these
particles, while cellular maker β-Tubulin could not be detected
(Fig. 1A, right). These results
indicate that the extracellular vesicles were nanoparticles,
confirming that they were exosomes.
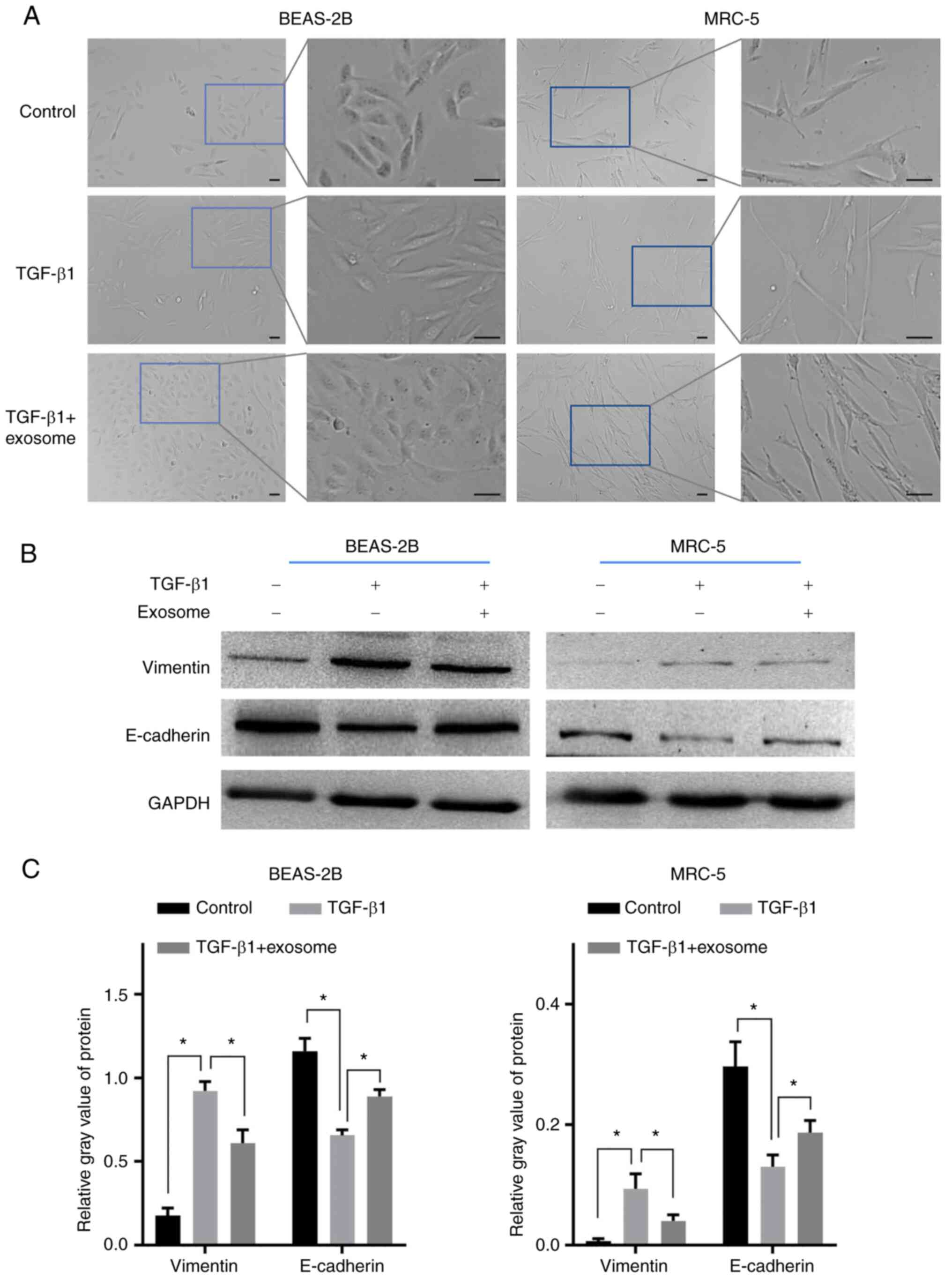
The resulting exosomes were used to reverse the EMT
of activated lung cells. As shown in Fig. 2A, most of the BEAS-2B cells were
long shuttle after 1 ng/ml TGF-β1 treatment, then became short
shuttle or polygon after further treated with exsomes. Similarly,
the morphology of the MRC-5 in TGF-β1 group became more slender
compared with the control cells and further treatment of exosomes
could reverse this morphological change. Western blot analysis
indicated that TGF-β1 stimulation resulted in the significant
upregulation of vimentin and the downregulation of E-cadherin in
the two types of cells (Fig. 2B
and C), reflecting the occurrence
of EMT. Notably, the morphological alterations of the two lung
cells induced by TGF-β1 were both reversed after further treatment
with exosomes. Consistently, the EMT of the two types of cells was
also significantly inhibited in the TGF-β1 + exosomes group
compared with the TGF-β1 group (Fig.
2B and C). These results
suggest that airway basal cell-derived exosomes could relieve the
EMT of lung epithelial cell and fibroblasts activated by
TGF-β1.
Identification of dysregulated genes
in activated lung cells treated with exosomes
To determine the underlying mechanism of the
exosomes, the cells in the TGF-β1 and TGF-β1 + exosomes groups were
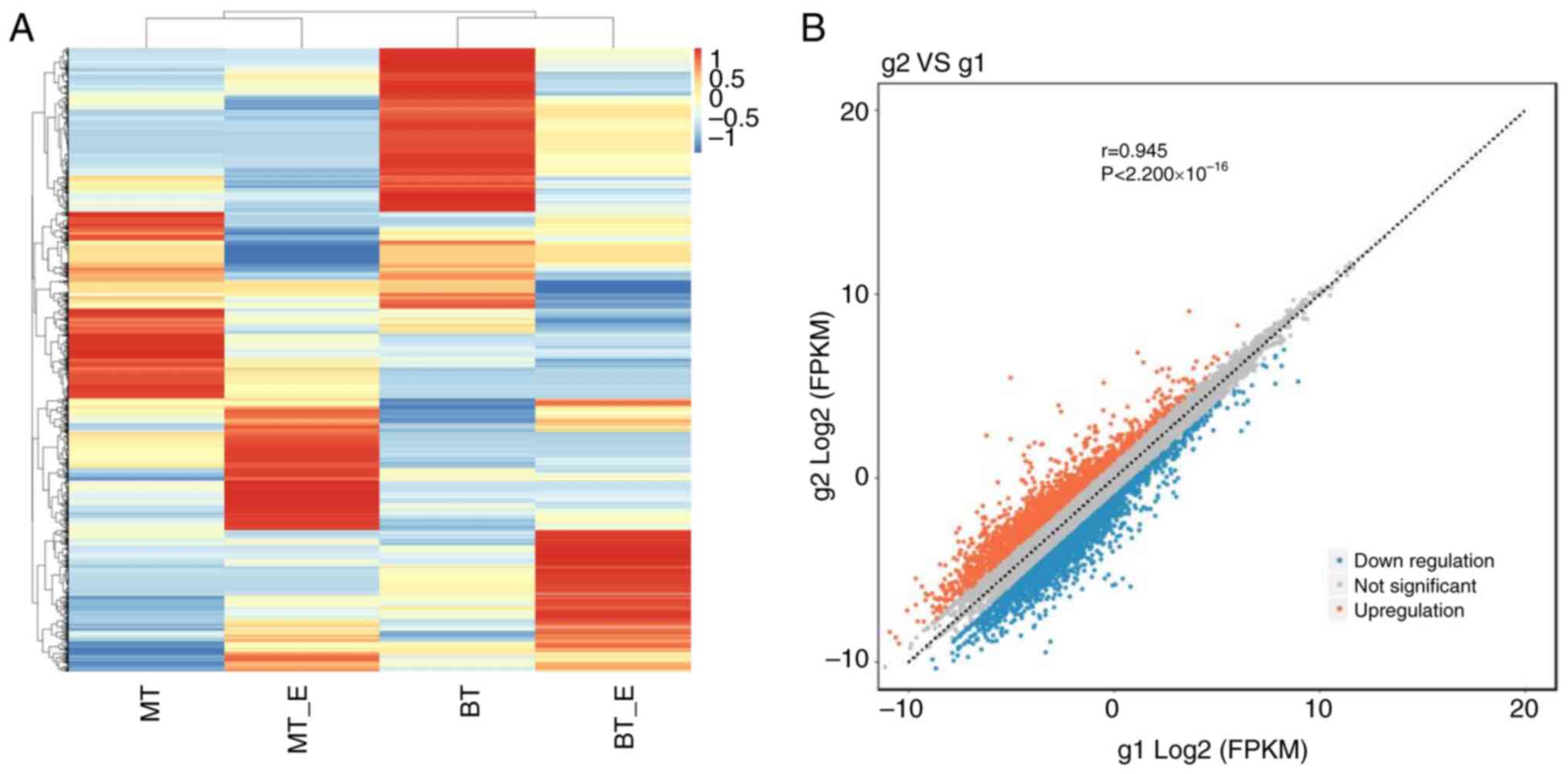
collected and analyzed by RNA sequencing. As shown in Fig. 3A, although the expression profile
of the four samples (without any parallels) varied, the fold-change
of most genes was consistent in the TGF-β1 + exosomes treated cells
compared with the corresponding cells treated with TGF-β1. A total
of 4,158 dysregulated genes, including 1,819 upregulated and 2,339
downregulated genes, were screened under the threshold of |log2FC|
value >1 (Fig. 3B). The P-value
was not taken as the threshold, as the sample number in each group
was <3.
As shown in Table
II, among the top 20 genes with the highest FC (abs) values,
AP005263.1 (unknown gene) had the highest FC (abs) value (1,442),
followed by KRT81, PCDHGA12, ANO1 and CXCL8, sharing the FC (abs)
values varying from 100-370. The P-value of the 20 genes were also
presented and were all <0.05, indicating that the FC (abs)
values were consistent in the two independent cells in the TGF-β1 +
exosomes group (compared with the cells in TGF-β1 group).
 | Table IITop 20 dysregulated genes in
exosome-treated lung cells. |
Table II
Top 20 dysregulated genes in
exosome-treated lung cells.
| | Gene id | Gene name | log2FC | log2FC abs | FC abs | P-value |
|---|
| 1 |
ENSG00000265257 |
AP005263.1 | 10.494 | 10.494 | 1442.642 |
3.54x10-9 |
| 2 |
ENSG00000205426 | KRT81 | 8.516 | 8.516 | 366.097 |
4.25x10-3 |
| 3 |
ENSG00000253159 |
PCDHGA12 | 7.164 | 7.164 | 143.379 |
8.05x10-4 |
| 4 |
ENSG00000131620 | ANO1 | -6.697 | 6.697 | 103.749 |
1.21x10-2 |
| 5 |
ENSG00000169429 | CXCL8 | 6.653 | 6.653 | 100.627 |
4.12x10-4 |
| 6 |
ENSG00000163735 | CXCL5 | 6.207 | 6.207 | 73.892 |
6.12x10-4 |
| 7 |
ENSG00000157542 | KCNJ6 | -6.135 | 6.135 | 70.294 |
2.53x10-2 |
| 8 |
ENSG00000251537 |
AC005324.2 | -5.878 | 5.878 | 58.815 |
1.43x10-2 |
| 9 |
ENSG00000242419 | PCDHGC4 | 5.855 | 5.855 | 57.871 |
3.35x10-3 |
| 10 |
ENSG00000286239 |
AC093884.1 | -5.840 | 5.840 | 57.290 |
6.71x10-3 |
| 11 |
ENSG00000260108 |
AC026464.2 | 5.791 | 5.791 | 55.373 |
4.82x10-4 |
| 12 |
ENSG00000118194 | TNNT2 | -5.788 | 5.788 | 55.251 |
5.62x10-3 |
| 13 |
ENSG00000256349 |
AP002748.5 | -5.757 | 5.757 | 54.069 |
2.24x10-3 |
| 14 |
ENSG00000149968 | MMP3 | 5.688 | 5.688 | 51.563 |
3.43x10-3 |
| 15 |
ENSG00000196611 | MMP1 | 5.674 | 5.674 | 51.051 |
3.07x10-3 |
| 16 |
ENSG00000073756 | PTGS2 | 5.640 | 5.640 | 49.874 |
3.60x10-2 |
| 17 |
ENSG00000115919 | KYNU | 5.421 | 5.421 | 42.852 |
2.06x10-2 |
| 18 |
ENSG00000105825 | TFPI2 | 5.420 | 5.420 | 42.825 |
9.16x10-3 |
| 19 |
ENSG00000123496 | IL13RA2 | 5.365 | 5.365 | 41.221 |
4.45x10-2 |
| 20 |
ENSG00000263244 |
AC087190.3 | 5.326 | 5.326 | 40.123 |
6.11x10-3 |
Bioinformatic analysis of dysregulated
genes
GO enrichment analysis indicated that the
dysregulated genes were enriched in various metabolism processes of
quinolinate, glycine and tryptophan (Fig. 4), which were regulated by HAAO,
KYNU, ACMSD and KMO (Table SI).
Some genes (WNK2, WNK3, ATP1B2 and WNK3) were involved in the
biological processes of ion import (Fig. 4, Table SI). The KEGG enrichment results
also showed that a number of the pathways were involved in the
metabolism of various amino acids and some other molecules,
including thiamine, butanoate and arachidonic acid (Fig. 5). In addition, 51 genes were
enriched in cytokine-cytokine receptor interaction and 18 genes
were involved in TGF-β signaling pathway, including downregulated
NOG, FST, GDF5, BAMBI and BMPR1B (Fig.
5; Table SII).
Another two migration-related GO terms and KEGG
pathways were also extracted, since little evidence could be found
associated with EMT in the top 30 terms and 30 pathways. As shown
in Table III, 110 of the genes
were enriched in the regulation of cell motility and 22 genes were
involved in the pathway responsible for the regulation of the actin
cytoskeleton. Next, nine overlapped genes (in red, Table III) were obtained through
comparing the genes in the two collections, including three
upregulated genes (CXCL12, ITGA2 and BDKRB1) and six downregulated
ones (FGF5, FGF22, FGF17, PDGFD, FGF1 and FGF18). These may might
be involved in the EMT process reversed by exosomes.
 | Table IIIBioinformatical analysis of genes
enriched in cell motility-related biological processes. |
Table III
Bioinformatical analysis of genes
enriched in cell motility-related biological processes.
| GO/pathway_ID |
GO_term/pathway_DES |
diff_gene_in_this_pathway | Up_genes | Down_genes | gene_UP_list | gene_DOWN_list | rich_factor | P-value |
|---|
| GO:2000145 | Regulation of cell
motility | 110 | 50 | 60 | SEMA6B, GREM1,
SERPINB3, SERPINE2, CXCL12a, TNFAIP6, ITGA2a, PAX6, JUN, DRD2, PTGS2, ACVRL1,
BDKRB1a, LBP, DNAI3,
CGA, CDH5, BMP5, VEGFA, PGF, PLXNA4, CXCL8, TNFRSF18, NRG1, CXCL10,
POSTN, CLDN3, WAS, STC1, LAMA2, CORO1A, LGR6, CD200, HGF, DUSP3,
SRCIN1, IL24, SEMA3G, SMIM22, IL1R1, NTRK3, CCL8, BMP2, CYGB,
ATP1B2, MEOX2, SPOCK2, IL1B, GPR183, EREG | S100A14, PTPRR,
ANGPTL3, EDN1, FGF22a, TACSTD2, F3, DUSP22, NEDD9,
PTGER4, HMOX1, DOCK8, MIR503, SMURF2, IL34, GPNMB, SERPINE1, TF,
ACKR3, IL12A, ADTRP, FLT4, MMP28, IGFBP3, SYNPO2, MIR221, RARRES2,
AQP1, MIR29A, PDGFDa,
SEMA3E, DCN, MGAT3, TAC4, FGF5a, ONECUT1, FLT1,
FGF17a, IL23A,
MIR27B, STK26, ANGPT1, PODN, NOG, FAM110C, CCN1, SEMA6A,
FGF18a, SULF1,
FGF1a, TPM1,
PPARGC1A, ADRA2A, PLXNC1, GAS6, DRD1, BST1, RHOJ, MSTN, CASS4 | 1.394 | 0.001 |
| hsa04810 | Regulation of actin
cytoskeleton | 22 | 7 | 15 | CXCL12a, ITGA2a, ITGB7, ITGB2, FGFR4, ARHGEF4,
BDKRB1a | CHRM4, SCIN, ITGB8,
MYH11, MYL9, FGF5a,
FGF22a, ITGA1,
FGF17a, ACTR3C,
ITGA11, PDGFDa, VAV3,
FGF1a,
FGF18a | 1.493 | 0.059 |
Overexpression of ANO1 promotes the
EMT of lung cells
Since the top 20 dysregulated genes had much higher
FC (abs) values, the first five known genes, excluding the unknown
gene AP005263.1, were selected for qPCR validation. As a result,
the log2FC values of four genes were consistent in the two lung
cells. The FC (abs) values of ANO1 in the two cells were both
higher than 4, whereas the value of the other three genes in
BEAS-2B or MRC-5 was always lower than 4 (Fig. 6A). Therefore, ANO1 was selected for
further analysis. The qPCR and western blotting results showed that
ANO1 was successfully overexpressed in the two types of cells. As
shown in Fig. 6B-D, the mRNA
expression level was enhanced by 2-3 fold and the ANO1 protein
level was also significantly increased. Further assays indicated
that the migration abilities were both significantly enhanced after
the overexpression of ANO1, along with increased levels of vimentin
and downregulated levels of E-cadherin (Fig. 6E-G). These results suggest that the
overexpression of ANO1 could induce the EMT and enhance the
migration ability of lung cells.
Knockdown of ANO1 inhibits the
TGF-β1-induced EMT of lung cells
To further confirm the role of ANO1, its expression
was evaluated in lung cells. The qPCR results indicated that the
expression level of ANO1 was reduced by ~60% (Fig. 7A). Consistently, its protein level
was also significantly decreased (Fig.
7B and C). Next, ANO1
knockdown cells were further treated with TGF-β1 for 24 h. The
Transwell results indicated that the migration ability of BEAS-2B
in the si-NC + TGF-β1 group showed a significant enhancement
compared with si-NC group, whereas the knockdown of ANO1 caused a
weakness of migration ability compared with those in the si-NC +
TGF-β1 group (Fig. 7D). The
migration ability of MRC-5 also showed a similar trend (Fig. 7D). In addition, the knockdown of
ANO1 in two lung cells also inhibited the EMT induced by TGF-β1
(Fig. 7E and F). These results suggest that the
downregulation of ANO1 induced by exosomes is an important
mechanism for the suppression of the TGF-β1-induced EMT by
exosomes.
Airway basal cell-derived exosomes
treatment ameliorates pulmonary fibrosis and inhibits the
upregulation of ANO1 induced by bleomycin
H&E and Masson's trichrome staining results
indicated that, bleomycin treatment resulted in interstitial
thickening, infiltration of inflammatory cells and collagen
deposition (indicated by arrows; Fig.
8A and B). Typical fibrosis
progression markers COL1A1 and FN1 were both significantly enhanced
in model group, compared with control group (Fig. 8C and D). In addition, a significant increase of
vimentin expression level as well as the reduction of E-cadherin
was also found following bleomycin treatment (Fig. 8C and D), which indicated the EMT of lung cells
and the progression of fibrosis. Notably, bleomycin treatment also
caused a significant upregulation of ANO1 (Fig. 8C and D).
Following exosome treatment for 7 consecutive days,
the fibrosis level was reduced, with fewer interstitial thickening
and collagen deposition were observed in exosome group (indicated
by arrows; Fig. 8A and B). Consistently, the protein levels of
fibrosis makers were both decreased and the EMT level was also
alleviated (Fig. 8C and D). Furthermore, exosome treatment also
reduced the expression level of ANO1. These results indicated that
airway basal cell-derived exosomes could ameliorate pulmonary
fibrosis and inhibited the upregulation of ANO1, basically
consistent with the in vitro results.
Discussion
In recent years, exosomes derived from MSCs and
other cells have increasingly received attention compared with the
cells themselves due to the number of advantages of exosomes in
clinical applications. First, these extracellular products can
avoid the risks associated with whole-cell transplantation, as well
as being non-oncogenic and less immunogenic compared with living
cells (24). Second, the
procedures used for the sterilization, handling and storage of
exosomes are simpler, thereby reducing manufacturing costs
(25). The present study focused
on exosomes derived from the airway basal cells and found that they
could inhibit the TGF-β1-induced EMT of lung epithelial cells and
fibroblasts and bleomycin-induced pulmonary fibrosis, providing a
potential therapeutic option for patients with IPF.
The pathological process of IPF is very complex and
involves the participation of various inflammation factors and
cytokines, among which TGF-β1 plays a predominant role. The
exposure of lung cells to TGF-β1 generally results in the
activation of cells and EMT and ECM accumulation, which is a widely
used as an in vitro model. The occurrence of EMT in damaged
epithelial cells not only causes continuous mesenchymal cell
activation and matrix remodeling, but also alterations of tissue
function once they separate from neighboring cells and migrate to
adjacent tissues (26). In
fibroblasts, EMT is regarded as an important mechanism leading to
the generation of myofibroblasts (27). Therefore, the present study focused
on reversing the EMT of lung cells, in contrast to previous
studies. As a result, it was found that the exposure of lung cells
caused the occurrence of EMT, while further treatment with exosomes
inhibited the EMT process.
Metabolic reprogramming is regarded as a hallmark of
cancer that contributes to tumorigenesis and disease progression.
It has been reported that metabolic reprogramming occurs during
TGFβ-induced EMT in IPF and non-small cell lung carcinoma (NSCLC)
(28,29). For instance, TGFβ treatment results
in an increased activity of glycolytic enzymes, thus enhancing
glycolysis metabolism, as well as the TCA cycle. Notably, extensive
metabolic changes have also been observed after exosome treatment
(29). The results of the
bioinformatical analysis indicated that the dysregulated genes were
predominantly enriched in the metabolism of amino acids, including
glycine catabolic process, as well as tryptophan metabolism.
Glycine and tryptophan are classic TCA cycle metabolites associated
with EMT-driven NSCLC progression and prognosis (30). Therefore, exosomes may inhibit the
metabolic reprogramming induced by TGF-β1 by regulating the
expression level of genes involved in the metabolism of certain key
amino acids.
Among the nine overlapped genes enriched in cell
motility-related terms and pathways, five belonged to fibroblast
growth factors (FGFs). FGFs are critical in controlling cell
proliferation, migration and differentiation through the activation
of surface receptors (31). It has
been reported that FGF1 is elevated in patients with IPF (32) and FGF18 is able to promote the
migration of lung fibroblasts (33). Therefore, the downregulation of
FGF1 and FGF18 may also be involved in the inhibition of EMT
following exosome treatment. Furthermore, 18 genes were found to be
enriched in the TGF-β signaling pathway and the dysregulation of
these genes also played important roles in the signaling
transduction of TGF-β1. Compared with the nine overlapped genes
enriched in motility-related biological processes, the top 20
dysregulated genes had a much higher fold-change, as well as a more
significant P-value. Therefore, focusing on the top 20 ones would
obtain a consistent validation result of the two types of cells. As
a result, four genes among the five tested showed a consistent
fold-change in the two cells treated with exosomes, indicating the
RNA sequencing result was reliable.
ANO1, also known as transmembrane protein 16A, is
recognized as a Ca2+-activated chloride channel and is
widely expressed in epithelial cells and smooth muscle cells
(18). In the field of fibrosis,
research on ANO1 is centered on cystic fibrosis due to its
fundamental role in regulating mucus secretion and he production of
epithelial and ciliated cells (34,35).
The role of ANO1 in the function of lung fibroblasts has also been
reported. For instance, the exposure of human lung fibroblasts to
TGF-β results in a sharp increase in the mRNA levels of ANO1 and
the inhibition of ANO1 via the inhibitor tannic acid causes the
downregulation of smooth muscle actin and fibronectin, markers of
myofibroblast differentiation (36). Additionally, the alteration of ANO1
is also able to regulate the proliferation and apoptosis of lung
fibroblasts in a mouse model of IPF (37). These studies highlight the vital
role of ANO1 in lung fibrosis. However, whether ANO1 regulates the
EMT or migration ability of lung cells is less investigated. The
present study found that the overexpression of ANO1 induced the EMT
of lung epithelial cells and fibroblasts, while the silencing of
ANO1 reversed the EMT induced by TGF-β1. Consistent with the
present study, the overexpression of ANO1 has been previously
reported to cause the EMT and stronger migration ability of various
cancer cells, including lung cancer cells (38,39).
Bleomycin-induced pulmonary fibrosis is a widely
used model, which closely mimics the pathological features of human
IPF. 9-days treatment of bleomycin initiates the fibrotic response,
resulting in the expression of pro-fibrotic factors (40), followed by the deposition of
collagen (41). The in vivo
assay of the present study indicated that exosome treatment
effectively inhibited bleomycin-induced pulmonary fibrosis,
consistent with in vitro results. It was also found that
bleomycin treatment resulted in the upregulation of ANO1 and
further exosome treatment ameliorated its upregulation. Notably,
the protein level of TGF-β1 is found to be significantly enhanced
in bleomycin-induced pulmonary fibrosis models (42,43).
The in vitro assay of the present study confirmed that
knockdown of ANO1 inhibited the TGF-β1-induced EMT of lung cells.
Therefore, the upregulation of ANO1 in pulmonary fibrosis model may
be related to the increased level of TGF-β1.
The reduction of ANO1 protein level in exosome group
indicated that exosome could inhibit the expression of ANO1, but it
is difficult to explain if this reduction is directly or indirectly
caused by the exosomes. It is reported that microRNAs (miR),
miR-142-3p (44) and miR-769-5p
(45), carried by exosomes could
directly bind with the 3'-UTR of TGF-β1 mRNA, thereby
downregulating its mRNA level. In addition, the expression level of
ANO1 could also be inhibited by miR-9(37) and miR-381(46). Therefore, the downregulation of
ANO1 in the mice model may be the synthesis results of the
molecules/miRNAs carried by exosomes and miRNAs targeting ANO1 may
be just one of the mechanisms, which also required further
investigations.
The present study has some limitations. The
screening of dysregulated genes was based on one replicate of the
two different cell samples. Herein, the addition of one replicate
for sequencing would improve the reliability of the screened genes.
Another limitation is that only the |log2FC| value of gene was
taken as the major factor for candidate gene selection, while the
functional annotation analysis was neglected. A combination of the
FC value and functional annotation analysis for gene selection
should be included in future studies. In addition, the effective
exosome contents were not detected, which also deserved further
investigation. The last limitation is that, only EMT markers were
detected, while other cytokines related to EMT were not
detected.
In the present study, airway basal cell-derived
exosomes were found to suppress the TGF-β1-induced EMT of lung
cells and bleomycin-induced pulmonary fibrosis by downregulating
ANO1. Thus, these exosomes represent a potential therapeutic option
for patients with IPF. Furthermore, the gene expression profile of
exosome-treated cells was elucidated and the corresponding
dysregulated genes provide insights into the mechanisms of
exosomes.
Supplementary Material
List of the top 30 GO terms/KEGG
pathways the dysregulated genes are enriched in.
List of the top 30 GO terms/KEGG
pathways the dysregulated genes are enriched in.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author. The raw data of RNA
sequencing was deposited in the SRA database and is accessible at
https://www.ncbi.nlm.nih.gov/sra/PRJNA1051029.
Authors' contributions
TR and SW designed the study. XG, ZL and SS
performed the experiments. XG and ZL collected the data. XG and SS
analyzed the data. XG and SW confirm the authenticity of all the
raw data. All authors contributed to the preparation of the
manuscript, and read and approved the final manuscript.
Ethics approval and consent to
participate
All experimental procedures were approved by the
Animal Welfare Ethics Committee of Shanghai Sixth People's Hospital
(approval no. 2023-0374).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Abuserewa ST, Duff R and Becker G:
Treatment of idiopathic pulmonary fibrosis. Cureus.
13(e15360)2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Glass DS, Grossfeld D, Renna HA, Agarwala
P, Spiegler P, DeLeon J and Reiss AB: Idiopathic pulmonary
fibrosis: Current and future treatment. Clin Respir J. 16:84–96.
2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Mattoo H and Pillai S: Idiopathic
pulmonary fibrosis and systemic sclerosis: Pathogenic mechanisms
and therapeutic interventions. Cell Mol Life Sci. 78:5527–5542.
2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Spagnolo P, Kropski JA, Jones MG, Lee JS,
Rossi G, Karampitsakos T, Maher TM, Tzouvelekis A and Ryerson CJ:
Idiopathic pulmonary fibrosis: Disease mechanisms and drug
development. Pharmacol Ther. 222(107798)2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Purghè B, Manfredi M, Ragnoli B, Baldanzi
G and Malerba M: Exosomes in chronic respiratory diseases. Biomed
Pharmacother. 144(112270)2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kadota T, Yoshioka Y, Fujita Y, Araya J,
Minagawa S, Hara H, Miyamoto A, Suzuki S, Fujimori S, Kohno T, et
al: Extracellular vesicles from fibroblasts induce epithelial-cell
senescence in pulmonary fibrosis. Am J Respir Cell Mol Biol.
63:623–636. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhu L, Chen Y, Chen M and Wang W:
Mechanism of miR-204-5p in exosomes derived from bronchoalveolar
lavage fluid on the progression of pulmonary fibrosis via AP1S2.
Ann Transl Med. 9(1068)2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kadota T, Fujita Y, Araya J, Watanabe N,
Fujimoto S, Kawamoto H, Minagawa S, Hara H, Ohtsuka T, Yamamoto Y,
et al: Human bronchial epithelial cell-derived extracellular
vesicle therapy for pulmonary fibrosis via inhibition of TGF-β-WNT
crosstalk. J Extracell Vesicles. 10(e12124)2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Samarelli AV, Tonelli R, Heijink I, Martin
Medina A, Marchioni A, Bruzzi G, Castaniere I, Andrisani D, Gozzi
F, Manicardi L, et al: Dissecting the Role of mesenchymal stem
cells in idiopathic pulmonary fibrosis: Cause or solution. Front
Pharmacol. 12(692551)2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Hoang DM, Pham PT, Bach TQ, Ngo ATL,
Nguyen QT, Phan TTK, Nguyen GH, Le PTT, Hoang VT, Forsyth NR, et
al: Stem cell-based therapy for human diseases. Signal Transduct
Target Ther. 7(272)2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ha DH, Kim HK, Lee J, Kwon HH, Park GH,
Yang SH, Jung JY, Choi H, Lee JH, Sung S, et al: Mesenchymal
stem/stromal cell-derived exosomes for immunomodulatory
therapeutics and skin regeneration. Cells. 9(1157)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhang E, Geng X, Shan S, Li P, Li S, Li W,
Yu M, Peng C, Wang S, Shao H and Du Z: Exosomes derived from bone
marrow mesenchymal stem cells reverse epithelial-mesenchymal
transition potentially via attenuating Wnt/β-catenin signaling to
alleviate silica-induced pulmonary fibrosis. Toxicol Mech Methods.
31:655–666. 2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Harrell CR, Miloradovic D, Sadikot R,
Fellabaum C, Markovic BS, Miloradovic D, Acovic A, Djonov V,
Arsenijevic N and Volarevic V: Molecular and cellular mechanisms
responsible for beneficial effects of mesenchymal stem cell-derived
product ‘Exo-d-MAPPS’ in attenuation of chronic airway
inflammation. Anal Cell Pathol (Amst). 2020(3153891)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Sengupta V, Sengupta S, Lazo A, Woods P,
Nolan A and Bremer N: Exosomes derived from bone marrow mesenchymal
stem cells as treatment for severe COVID-19. Stem Cells Dev.
29:747–754. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ma Q, Ma Y, Dai X, Ren T, Fu Y, Liu W, Han
Y, Wu Y, Cheng Y, Zhang T and Zuo W: Regeneration of functional
alveoli by adult human SOX9+ airway basal cell
transplantation. Protein Cell. 9:267–282. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lee C, Mitsialis SA, Aslam M, Vitali SH,
Vergadi E, Konstantinou G, Sdrimas K, Fernandez-Gonzalez A and
Kourembanas S: Exosomes mediate the cytoprotective action of
mesenchymal stromal cells on hypoxia-induced pulmonary
hypertension. Circulation. 126:2601–2611. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kropski JA and Blackwell TS: Progress in
understanding and treating idiopathic pulmonary fibrosis. Annu Rev
Med. 70:211–224. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Takayama Y, Derouiche S, Maruyama K and
Tominaga M: Emerging perspectives on pain management by modulation
of TRP channels and ANO1. Int J Mol Sci. 20(3411)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zuo W, Zhang T, Wu DZ, Guan SP, Liew AA,
Yamamoto Y, Wang X, Lim SJ, Vincent M, Lessard M, et al:
p63(+)Krt5(+) distal airway stem cells are essential for lung
regeneration. Nature. 517:616–620. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
R Core Team. R: A language and environment
for statistical computing. R Foundation for Statistical Computing,
Vienna, Austria, 2019.
|
|
21
|
Javellana M, Eckert MA, Heide J, Zawieracz
K, Weigert M, Ashley S, Stock E, Chapel D, Huang L, Yamada SD, et
al: Neoadjuvant chemotherapy induces genomic and transcriptomic
changes in ovarian cancer. Cancer Res. 82:169–176. 2022.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Liu P, Miao K, Zhang L, Mou Y, Xu Y, Xiong
W, Yu J and Wang Y: Curdione ameliorates bleomycin-induced
pulmonary fibrosis by repressing TGF-beta-induced fibroblast to
myofibroblast differentiation. Respir Res. 21(58)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Giebel B, Kordelas L and Börger V:
Clinical potential of mesenchymal stem/stromal cell-derived
extracellular vesicles. Stem Cell Investig. 4(84)2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Gimona M, Pachler K, Laner-Plamberger S,
Schallmoser K and Rohde E: Manufacturing of human extracellular
vesicle-based therapeutics for clinical use. Int J Mol Sci.
18(1190)2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Xie L and Zeng Y: Therapeutic potential of
exosomes in pulmonary fibrosis. Front Pharmacol.
11(590972)2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hu L, Ding M and He W: Emerging
therapeutic strategies for attenuating tubular EMT and kidney
fibrosis by targeting Wnt/β-catenin signaling. Front Pharmacol.
12(830340)2022.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhao H, Dennery PA and Yao H: Metabolic
reprogramming in the pathogenesis of chronic lung diseases,
including BPD, COPD, and pulmonary fibrosis. Am J Physiol Lung Cell
Mol Physiol. 314:L544–L554. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Hua W, Ten Dijke P, Kostidis S, Giera M
and Hornsveld M: TGFβ-induced metabolic reprogramming during
epithelial-to-mesenchymal transition in cancer. Cell Mol Life Sci.
77:2103–2123. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Giannos P, Kechagias KS and Gal A:
Identification of prognostic gene biomarkers in non-small cell lung
cancer progression by integrated bioinformatics analysis. Biology
(Basel). 10(1200)2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Seitz T and Hellerbrand C: Role of
fibroblast growth factor signalling in hepatic fibrosis. Liver Int.
41:1201–1215. 2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
MacKenzie B, Korfei M, Henneke I, Sibinska
Z, Tian X, Hezel S, Dilai S, Wasnick R, Schneider B, Wilhelm J, et
al: Increased FGF1-FGFRc expression in idiopathic pulmonary
fibrosis. Respir Res. 16(83)2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Joannes A, Brayer S, Besnard V,
Marchal-Somme J, Jaillet M, Mordant P, Mal H, Borie R, Crestani B
and Mailleux AA: FGF9 and FGF18 in idiopathic pulmonary fibrosis
promote survival and migration and inhibit myofibroblast
differentiation of human lung fibroblasts in vitro. Am J Physiol
Lung Cell Mol Physiol. 310:L615–L629. 2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Kunzelmann K, Ousingsawat J, Cabrita I,
Doušová T, Bähr A, Janda M, Schreiber R and Benedetto R: TMEM16A in
cystic fibrosis: Activating or inhibiting? Front Pharmacol.
10(3)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Danahay H and Gosling M: TMEM16A: An
alternative approach to restoring airway anion secretion in cystic
fibrosis? Int J Mol Sci. 21(2386)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Dulin NO, Smolyaninova LV and Orlov SN:
Control of lung myofibroblast transformation by monovalent ion
transporters. Curr Top Membr. 83:15–43. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Dai WJ, Qiu J, Sun J, Ma CL, Huang N,
Jiang Y, Zeng J, Ren BC, Li WC and Li YH: Downregulation of
microRNA-9 reduces inflammatory response and fibroblast
proliferation in mice with idiopathic pulmonary fibrosis through
the ANO1-mediated TGF-β-Smad3 pathway. J Cell Physiol.
234:2552–2565. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Chen W, Gu M, Gao C, Chen B, Yang J, Xie
X, Wang X, Sun J and Wang J: The prognostic value and mechanisms of
TMEM16A in human cancer. Front Mol Biosci. 8(542156)2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Guo S, Chen Y, Shi S, Wang X, Zhang H,
Zhan Y and An H: Arctigenin, a novel TMEM16A inhibitor for lung
adenocarcinoma therapy. Pharmacol Res. 155(104721)2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Chaudhary NI, Schnapp A and Park JE:
Pharmacologic differentiation of inflammation and fibrosis in the
rat bleomycin model. Am J Respir Crit Care Med. 173:769–776.
2006.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Vats A and Chaturvedi P: The regenerative
power of stem cells: Treating bleomycin-induced lung fibrosis. Stem
Cells Cloning. 16:43–59. 2023.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Qian W, Cai X, Qian Q, Zhang W and Wang D:
Astragaloside IV modulates TGF-β1-dependent epithelial-mesenchymal
transition in bleomycin-induced pulmonary fibrosis. J Cell Mol Med.
22:4354–4365. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Cutroneo KR, White SL, Phan SH and Ehrlich
HP: Therapies for bleomycin induced lung fibrosis through
regulation of TGF-beta1 induced collagen gene expression. J Cell
Physiol. 211:585–589. 2007.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Huang C, Zhao L, Xiao Y, Tang Z, Jing L,
Guo K, Tian L and Zong C: M2 macrophage-derived exosomes carry
miR-142-3p to restore the differentiation balance of irradiated
BMMSCs by targeting TGF-β1. Mol Cell Biochem: Jun 3, 2023 (Epub
ahead of print).
|
|
45
|
Ni N, Ma W, Tao Y, Liu J, Hua H, Cheng J,
Wang J, Zhou B and Luo D: Exosomal MiR-769-5p exacerbates
ultraviolet-induced bystander effect by targeting TGFBR1. Front
Physiol. 11(603081)2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Cao Q, Liu F, Ji K, Liu N, He Y, Zhang W
and Wang L: MicroRNA-381 inhibits the metastasis of gastric cancer
by targeting TMEM16A expression. J Exp Clin Cancer Res.
36(29)2017.PubMed/NCBI View Article : Google Scholar
|