Introduction
Ischemic cardiomyopathy (ICM) is one of the most
common cardiomyopathies with a high rate of sudden cardiac death;
>9 million people suffering from chronic stable angina in the
United States alone (1). It is
characterized by several clinical manifestations which manifest
clinically as dyspnea or chest pain, and has a pathophysiological
basis in myocardial ischemia and hypoxia due to coronary
atherosclerosis, as well as myocardial cell reduction, necrosis,
myocardial fibrosis and myocardial scar formation (2,3).
Advances in medical technology have made it possible to treat ICM
with a range of clinical treatments, including drugs, thrombus
recanalization, stent implantation and cardiac bypass surgery
(4). Restoring blood flow after
ischemia remains the best therapeutic option in limiting myocardial
infarct size and maintaining cardiac function (5). However, it can also trigger adverse
effects that damage myocardium known as ischemia/reperfusion (I/R)
injury (6). The pathogenesis of
ICM is complex and there is a lack of a definitive clinical
management strategy (7). Current
therapeutic strategies to prevent myocardial I/R injury are helping
to treat ICM (8). Therefore, it is
crucial to clarify the molecular biological mechanisms of ischemic
cardiomyopathy as well as new therapeutic strategies.
Ferroptosis is an iron-dependent form of regulated
cell death (9). Ferroptosis causes
oxidative damage to cell membranes, through the continuous
accumulation of lipid hydroperoxides (10). Studies have reported that most
diseases are associated with ferroptosis, such as cancer (11), traumatic brain injury (12), intracerebral hemorrhage (13) and I/R injury (14), and ferroptosis is an important form
of myocardial cell death in myocardial I/R injury (15). In clinical practice, patients with
myocardial infarction (MI) often experience left ventricular
remodeling accompanied by the accumulation of ferrous ions, and
large amounts of ferrous ions have been reported in the infarcted
tissue of patients with ST-segment elevation MI (16). Moreover, iron overload may cause
cardiomyocyte death, which is an important mechanism in the
development of cardiomyopathy (17). Investigating the molecular
mechanism of ferroptosis in I/R injury and understanding the
potential therapeutic targets related to ferroptosis may
effectively reduce myocardial I/R injury and avoid the advancement
of ICM during treatment.
In the present study, a systematic bioinformatics
analysis was performed to analyze the differential expression of
ferroptosis-related genes (FRGs) in ICM and their potential
crosstalk and functional pathways, which contributes to an in-depth
understanding of the mechanisms and new diagnostic and therapeutic
targets in ICM. A protein-protein interaction (PPI) network was
constructed to identify the hub genes. Subsequently, the expression
of hub genes and ferroptosis characteristics were verified in the
H9c2 anoxic reoxygenation (A/R) injury model, and then the
diagnostic ability of hub genes for ICM was evaluated. Finally,
potential ferroptosis-regulating drugs were predicted for ICM based
on key genes in the Drug Signatures Database (DSigDB), which will
help to improve the likelihood of translating the drugs into the
clinic.
Materials and methods
Data collection
The microarray datasets GSE26887(18) and GSE57338(19) were retrieved from the Gene
Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) (20). In the present study, data from 12
post-ischemic myocardium samples and 5 non-heart failure
heart-matched donor heart samples from the GSE26887 dataset were
subjected to RNA sequencing analysis and bioinformatic exploration.
For validation, data from 94 post-ischemic myocardium samples and
137 non-heart failure heart-matched donor heart samples from the
GSE57338 dataset were used.
Gene set enrichment analysis
(GSEA)
GSEA was performed using GSEA software (v4.2.3)
(http://software.broadinstitute.org/gsea/index.jsp)
(21) to observe the overall
correlation between ferroptosis and ICM. Genes in the GSE26887
dataset were scored using the ferroptosis custom dataset in GSEA.
The normalized enrichment score (NES) was calculated and P.adjust
<0.05 was considered to indicate a statistically significant
difference.
Weighted gene co-expression network
analysis (WGCNA) in GSE26887
The ‘WGCNA’ R package (v1.72-1) (22) was used and WGCNA was performed on
GSE26887, with abnormal samples removed to ensure reliable results
of the network construction. Firstly, an optimal soft threshold was
set to divide the data into different modules by the ‘flashClust’
(v1.01-2-3) (22) function in
WGCNA. The adjacency matrix consisted of continuous values between
0-1, thus the constructed network conformed to a power-law
distribution and represented the real biological state more
accurately. Secondly, the block module function was used to
construct scale-free networks and perform module partitioning
analysis. Each module was summarized by module identity genes, and
each module was illustrated in color. Ultimately, the module which
demonstrated the highest positive correlation with the ICM was
identified.
Identification of differentially
expressed genes (DEGs) and differentially expressed FRGs
(DEFRGs)
The ‘limma’ R package (v3.52.0) (23) was used to determine the DEGs
between ICM and normal hearts. The thresholds set for selecting the
DEGs were log2fold change (FC) ≥0.50 and false discovery
rate (FDR) <0.05. The volcanoes and heat maps of the DEGs were
plotted using the ‘ggplot2’ R package (v3.4.3) (24). A total of 484 FRGs were acquired
from the FerrDb database (http://www.zhounan.org/ferrdb/) (25), classified as Driver, Suppressor and
Marker genes (Table SI). The
online Venn diagram tool (v1.9.0) (https://jvenn.toulouse.inrae.fr/app/example.html)
(26) overlapped the most
ICM-correlated modular genes, DEGs in GSE26887 and FRGs. The
overlapping genes were defined as DEFRGs.
Gene ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) enrichment analyses
GO annotation and KEGG pathway enrichment analysis
of DEFRGs was performed using the ‘cluster profiler’ R package
(v4.8.3) (27) and visualized
using Sangerbox 3.0 (http://sangerbox.com/home.html) (28). P.adjust <0.05 was used to
indicate statistically significant enrichment by DEFRGs.
PPI network and identification of key
modules and hub genes
In the present study, the PPI network for DEFRGs was
constructed using the online tool provided by the Search Tool for
the Retrieval of Interacting Genes/Proteins (STRING) database
(v11.09) (https://string-db.org/) (29). To visualize the resulting PPI
network, Cytoscape (v3.7.2; Cytoscape Consortium) software
(30) was used. Furthermore, using
cytoHubba and the Maximal Clique Centrality (MCC) algorithm, and
the MCODE plugin (24) (Cytoscape
Consortium) (degree cutoff=2; node score cutoff=0.2; K-core=2), the
genes were screened and the top 10 hub genes and key functional
modules were identified.
Receiver operating characteristic
curve (ROC) analysis
To evaluate the diagnostic ability of the hub genes
identified, ROC curves were constructed using data from the
GSE26887 and GSE57338 databases. The area under the curve (AUC) was
calculated and hub genes with AUC >0.7 were considered to have
favorable diagnostic potential.
Cell culture and treatment
H9c2 cardiomyocytes were purchased from the Cell
Bank/Stem Cell Bank (Chinese Academy of Sciences). All cells were
cultured in DMEM (Hyclone; Cytiva) with 10% FBS (HyClone; Cytiva)
and placed in a 37˚C incubator under standard conditions (5%
CO2, 95% humidity and 21% O2 concentration)
for 24 h.
H9c2 cells were separated into three groups: i)
Control, ii) A/R and iii) A/R + ferrostatin-1 (Fer-1). The control
group cells were incubated with normal medium (37˚C; 5%
CO2; 95% humidity and 21% O2 concentration)
for 24 h. As with previous studies (31,32),
the A/R group was induced by 4 h anoxia and 4 h reoxygenation. The
H9c2 cells were exposed to anoxic conditions by adding fresh anoxia
medium (1 mM CaCl2, 10 mM KCI, 20 mM HEPES, 98.5 mM
NaCl, 1.2 mM MgSO4, 6 mM NaHCO3, 0.9 mM
NaH2PO4 and 40 mM sodium lactate; pH 6.8) and
then incubated in a 37˚C chamber with an atmosphere of 95%
N2 and 5% CO2 for 4 h. Following 4 h of
anoxia, the anoxia medium was removed from the cells and a
reoxygenation medium (1 mM CaCl2, 5.5 mM glucose, 5 mM
KCI, 20 mM HEPES, 129.5 mM NaCl, 1.2 mM MgSO4, 20 mM
NaHCO3 and 0.9 mM NaH2PO4; pH 7.4)
was added. The cells were then incubated in a 37˚C incubator with
an atmosphere of 95% O2 and 5% CO2 for 4 h.
Fer-1 (5 µM; MedChemExpress) pretreatment was added to the A/R +
Fer-1 group in a 37˚C chamber under standard conditions (5%
CO2; 95% humidity and 21% O2 concentration)
for 2 h to inhibit ferroptosis.
Evaluation of cell viability
Cell viability was assessed using the Cell Counting
Kit-8 (CCK-8) assay (cat. no. GK10001; GlpBio Technology, Inc.)
based on the suggested procedure. Briefly, H9c2 cells were plated
into a 96-well plate at a density of 5x103 cells/well
and CCK-8 solution was added to each well at a final concentration
of 10% and incubated for 1 h at room temperature. The optical
density (OD) value at 450 nm was measured using Spark®
multimode microplate reader (Thermo Fisher Scientific, Inc.).
Western blot analysis
Proteins were extracted from the lysates of the H9c2
cells using RIPA lysis buffer (Beijing Solarbio Science &
Technology Co., Ltd.), which contained 1% PMSF. The extracted
proteins were determined using a BCA kit (GlpBio Technology, Inc.).
Proteins were denatured through boiling for 5 min and then
separated using 12% SDS-PAGE with a protein mass of 40 µg per lane.
The separated proteins were then transferred onto PVDF membranes
(MilliporeSigma). The membranes were then blocked with 5% non-fat
dry milk for 2 h at room temperature, followed by incubation
overnight at 4˚C with the following primary antibodies:
Prostaglandin-endoperoxide synthase 2 (PTGS2; 1:500; cat. no.
R23969; Chengdu Zen-Bioscience Co., Ltd), glutathione peroxidase 4
(GPX4; 1:1,000; cat. no. 381958; Chengdu Zen-Bioscience Co., Ltd.)
and β-actin (1:1,000; cat. no. GB113225-100; Wuhan Servicebio
Technology Co., Ltd.). Subsequently, the membranes were incubated
with horseradish peroxidase-conjugated secondary antibodies
(1:2,000; cat. no. A0239; Beyotime Institute of Biotechnology) for
2 h at room temperature. Positive blots were visualized using the
Ultra High Sensitivity ECL kit (cat. no. GK10008; GlpBio
Technology, Inc.) and imaged using the FluorChem FC3 System
(ProteinSimple). Finally, the densitometric scanning of the blots
were calculated using ImageJ software (v1.8.0.345; National
Institutes of Health). β-actin was selected as the internal
reference.
Measurement of intracellular reactive
oxygen species (ROS)
To determine the intracellular levels of ROS, the
ROS Detection Kit (cat. no. S0033M; Beyotime Institute of
Biotechnology) was used. H9c2 cells were seeded in a 6-well plate
at a density of 1x105 cells per well. After incubation
and treatment as aforementioned, H9c2 cells were incubated with
2',7'-Dichlorodihydrofluorescein diacetate for 30 min at 37˚C in
the dark according to the manufacturer's instructions.
Subsequently, a fluorescence microscope (Olympus Corporation) was
used to observe the associated alterations.
Iron content detection
Total iron content in H9c2 cells was obtained using
a Total Iron Content Colorimetric Assay Kit (cat. no. E1042;
Applygen Technologies, Inc.). H9c2 cells were lysed at 4˚C for 20
min, the cell lysate was centrifuged at 12,000 x g for 5 min at 4˚C
and the total iron ion content was measured according to the
manufacturer's instructions. The OD value was determined using the
Spark multimode microplate reader (Thermo Fisher Scientific,
Inc.).
Detection of antioxidant enzymes
activities and lipid peroxidation
Treated H9c2 cells were lysed at 4˚C for 20 min and
centrifuged at 12,000 x g for 10 or 5 min at 4˚C to remove cell
debris. The malondialdehyde (MDA), superoxide dismutase (SOD),
glutathione (GSH) and glutathione disulfide (GSSG) contents, and
the GSH/GSSG ratio in H9c2 cells were measured according to the
manufacturer's instructions of the Lipid Peroxidation MDA Assay Kit
(cat. no. S01031S; Beyotime Institute of Biotechnology), Total
Superoxide Dismutase Assay Kit with WST-8 (cat. no. S0101S;
Beyotime Institute of Biotechnology), GSH and GSSG Assay Kit (cat.
no. S0053; Beyotime Institute of Biotechnology), respectively. The
OD value was determined using the Spark multimode microplate reader
(Thermo Fisher Scientific, Inc.).
Reverse transcription
(RT)-quantitative (q)PCR
H9c2 cells were subjected to RNA extraction using
the TRIzol™ reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) to obtain total RNA. The quality and yield of the
isolated RNA were assessed to confirm its suitability for
evaluating mRNA expression. cDNA was produced by RT of total RNA
using the HiScript® II 1st Strand cDNA Synthesis Kit
(cat. no. R211-01; Vazyme Biotech Co., Ltd.). Briefly, 20 µl RT
system (1 µg RNA, 10 µl RT mix, 2 µl HiScript II Enzyme Mix, 1 µl
oligo, 1 µl random hexamers and RNase-free double-distilled
H2O) was converted to cDNA using the Bio-Rad PCR T100
Thermal Cycler (Bio-Rad Laboratories, Inc.) at 25˚C for 5 min, 50˚C
for 15 min and 85˚C for 2 min. Subsequently, using the SYBR Green
qPCR Master Mix (cat. no. B21203; Bimake.com), the
level of gene expression was determined by qPCR and normalized to
the level of β-actin. Briefly, 20 µl qPCR system (2 µl cDNA, 10 µl
SYBR Green qPCR Master Mix, 1 µl forward primers, 1 µl reverse
primers and 6 µl RNase-free double-distilled H2O) was
amplified by BIO-RAD CFX Connect Real-Time PCR Detection Systems
(Bio-Rad Laboratories, Inc.). The procedure was as follows:
Denaturation at 95˚C for 10 min, followed by thermal cycling at
95˚C for 15 sec and denaturation at 60˚C for 1 min, repeated for 40
cycles. Relative mRNA expression levels were calculated using the
2-ΔΔCq method (33). The primers used in the present
study were designed and synthesized by Sangon Biotech Co., Ltd.
(Shanghai, China). Table SII
provides details on the primers used.
Potential therapeutic drug
prediction
Data on protein-drug interactions from the DSigDB
(34) were used to predict the
potential impact of drugs on ferroptosis in the context of ICM. The
Drugbank database (35) was used
to identify the chemical structures of the predicted drugs.
Statistical analysis
The results are presented as either the mean ±
standard deviation or the mean ± standard error of the mean.
Statistical analyses were performed in GraphPad Prism 8 (v8.0.2;
Dotmatics). To compare differences between two groups, the unpaired
two-tailed Student's t-test was used. For comparisons between ≥3
groups, one-way analysis of variance and Dunnett's post hoc
multiple comparison test was used. P<0.05 was used to indicate a
statistically significant difference.
Results
Overall study protocol
The present study followed the flowchart presented
in Fig. 1. Transcriptomic data
obtained from the GSE26887 dataset available in the GEO public
database, with data from 12 samples of post-ischemic myocardium and
5 samples of non-heart failure heart-matched donor heart.
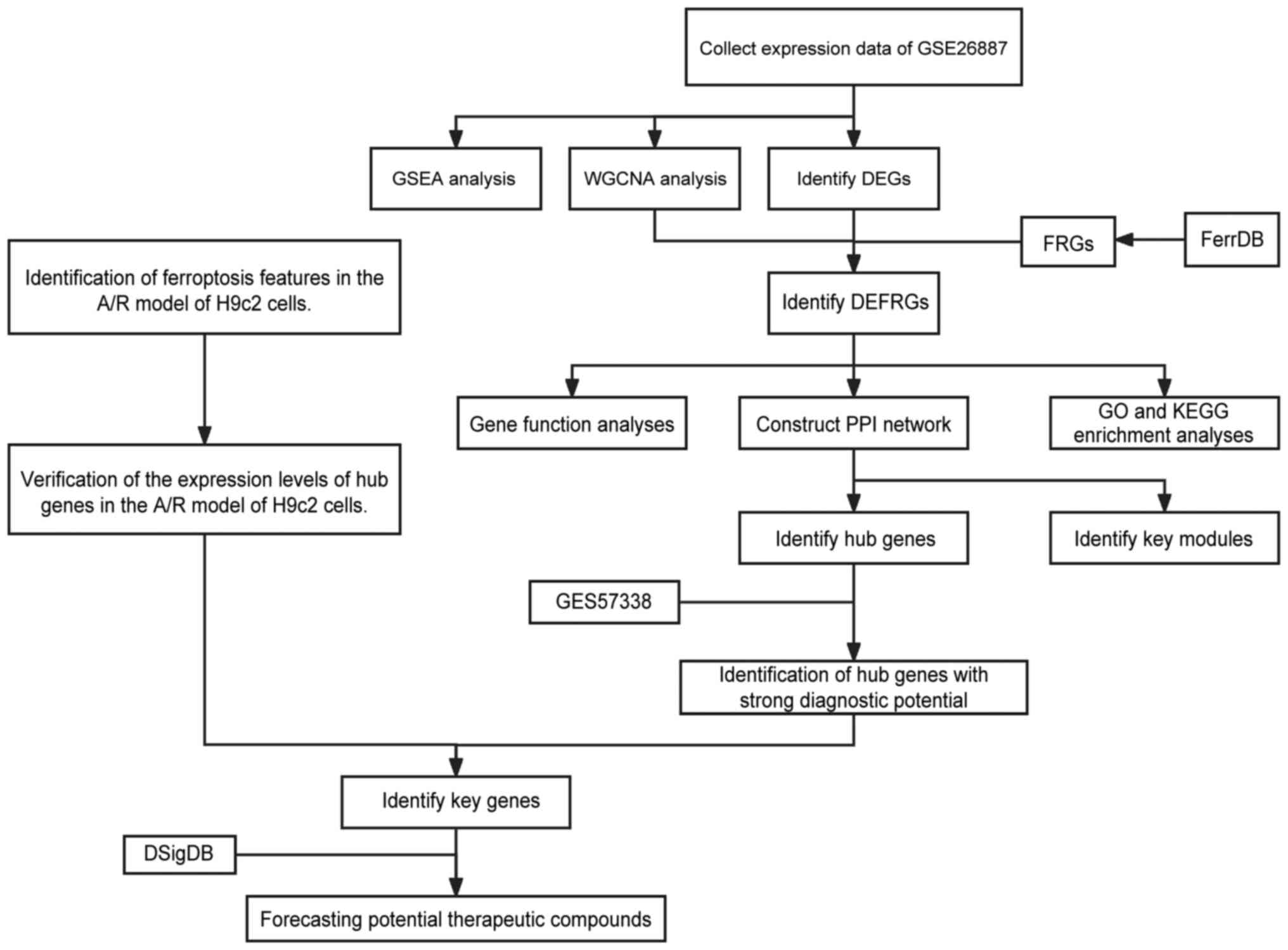 | Figure 1Overall protocol of the present
study. GSEA, gene set enrichment analysis; WGCNA, weighted gene
co-expression network analysis; DEG, differentially expressed
genes; A/R, anoxic reoxygenation; FRG, ferroptosis-related gene;
DEFRG, differentially expressed FRG; PPI, protein-protein
interaction; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes
and Genomes; DSigDB, Drug Signatures Database. |
Altered transcriptome in ICM is
associated with the dysregulation of genes related to
ferroptosis
GSEA is a widely-utilized method for evaluating the
pertinence of defined gene sets with respect to the disease
phenotype in disease transcriptomes (36). GSEA was used to assess the
variation of FRGs in the transcriptome of individuals with ICM vs.
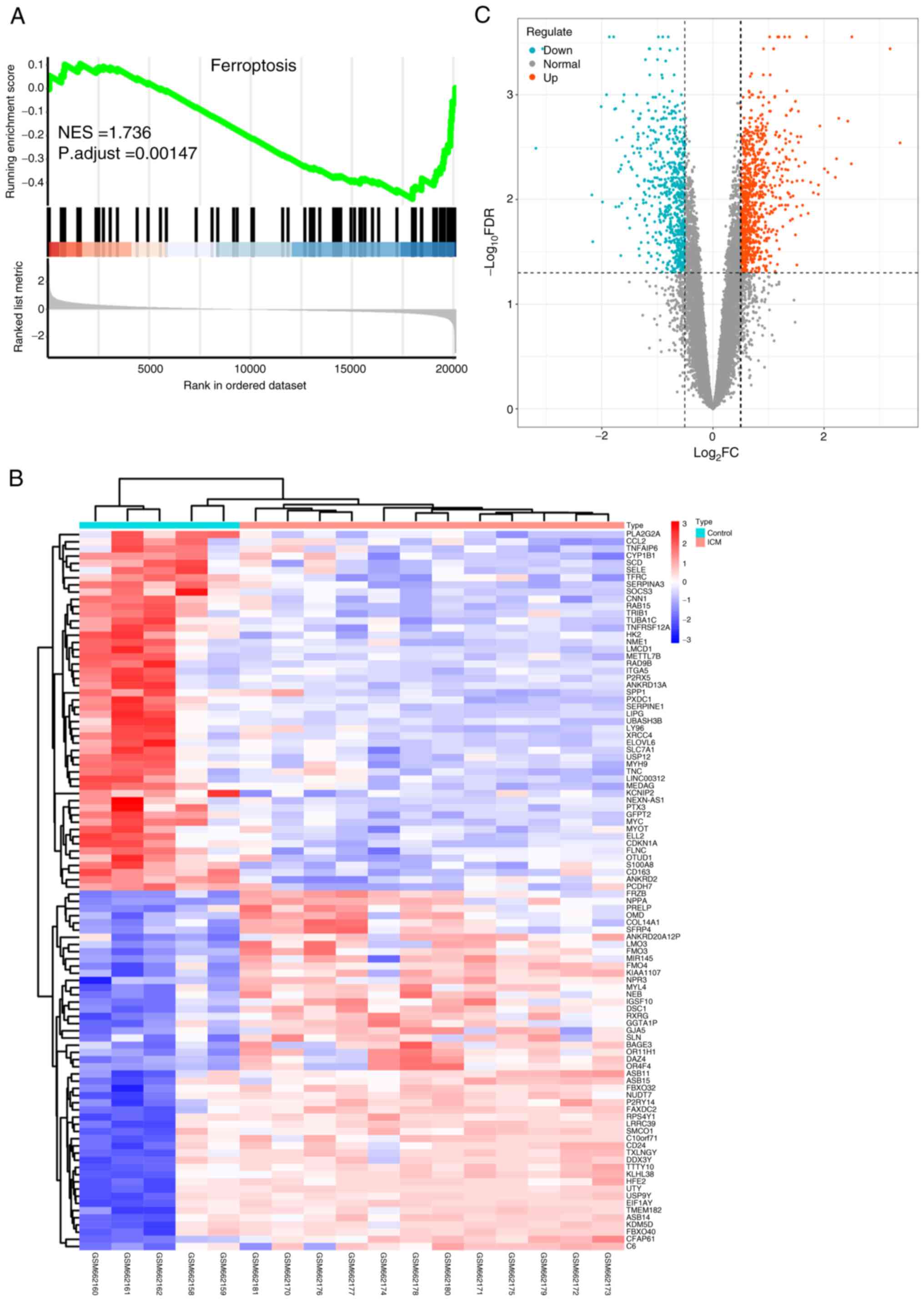
the control group, as presented in Fig. 2. GSEA revealed significant
dysregulation of FRGs between the two groups (NES=1.736; P.adjust
<0.005; Fig. 2A). This suggests
that FRGs are a crucial feature of ICM and their dysregulation
provides evidence for the association between ICM and
ferroptosis.
Identification of differentially
expressed genes
Following the division of data from GSE26887 into
normal and ICM groups, a differential analysis was performed using
the ‘limma’ R package and 1,524 DEGs were identified with
significance set at FDR <0.05 and log2FC >0.5
(Table SIII). Among these, 669
exhibited low expression and 855 showed high expression. These
results are presented using a heat map (Fig. 2B) and a volcano plot (Fig. 2C), revealing distinct differential
expression profiles of the DEGs between the ICM and control
samples.
WGCNA
WGCNA is an analytical method used to analyze gene
expression patterns of multiple samples, which can cluster genes
with similar expression patterns. It has been widely used in
several studies such as in gene association analysis (37).
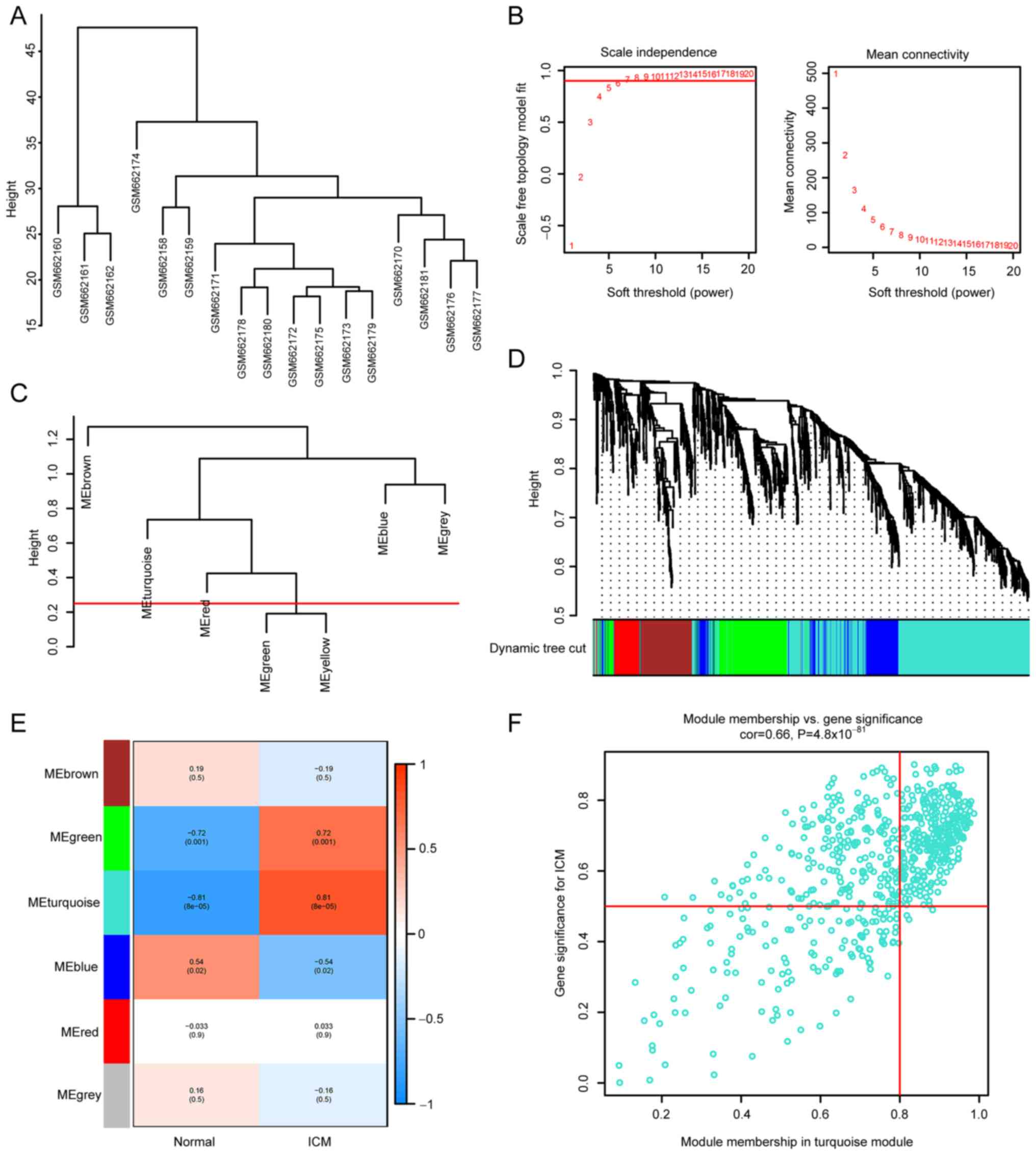
Firstly, cluster analysis was performed using the
‘flashClust’ (v1.01-2-3) (22)
function in WGCNA package with a threshold of 70, and cluster 1 was
found to contain 17 samples. The sample clustering tree is shown in
Fig. 3A, and the normal control
and ICM groups were clustered separately. Secondly, a soft
threshold of β=7 (scale-free R2=9; Fig. 3B) was used to establish a
scale-free network. Thirdly, the threshold was set at 0.25 and the
minimum number of modules was set at 50 to amalgamate comparable
modules in a cluster. A total of six modules were created, each
containing genes with comparable patterns of expression (Fig. 3C). The analysis of module-trait
associations revealed that several modules were linked to ICM. The
turquoise module, consisting of 683 genes, was found to have the
strongest correlation with ICM (r=0.81; P=8x10-5;
Fig. 3D and E). The correlation between the genes
within the turquoise module and the ICM genes was significant
(correlation=0.66; P=4.8x10-81; Fig. 3F).
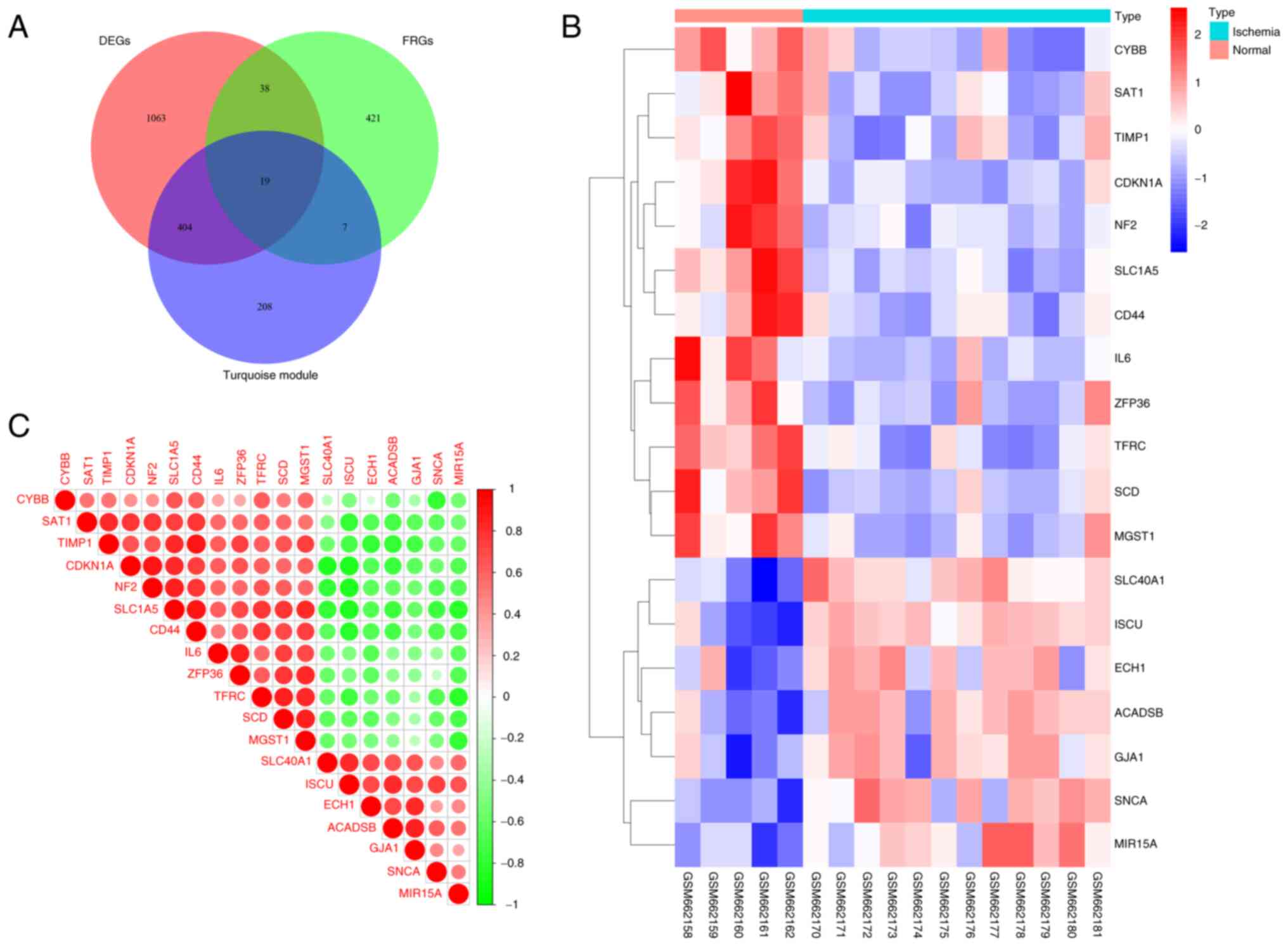
Identification of DEFRGs
After the aforementioned screening process, 1,524
DEGs, 638 turquoise module genes identified by WGCNA, and 484 FRGs
were overlapped, resulting in 19 genes (Table SIV) for further analysis (Fig. 4A). A clustered heatmap (Fig. 4B) and a correlation heatmap
(Fig. 4C) were generated to
display the differences in expression and correlation of the 19
DEFRGs between ischemic cardiac tissue and normal control cardiac
tissue.
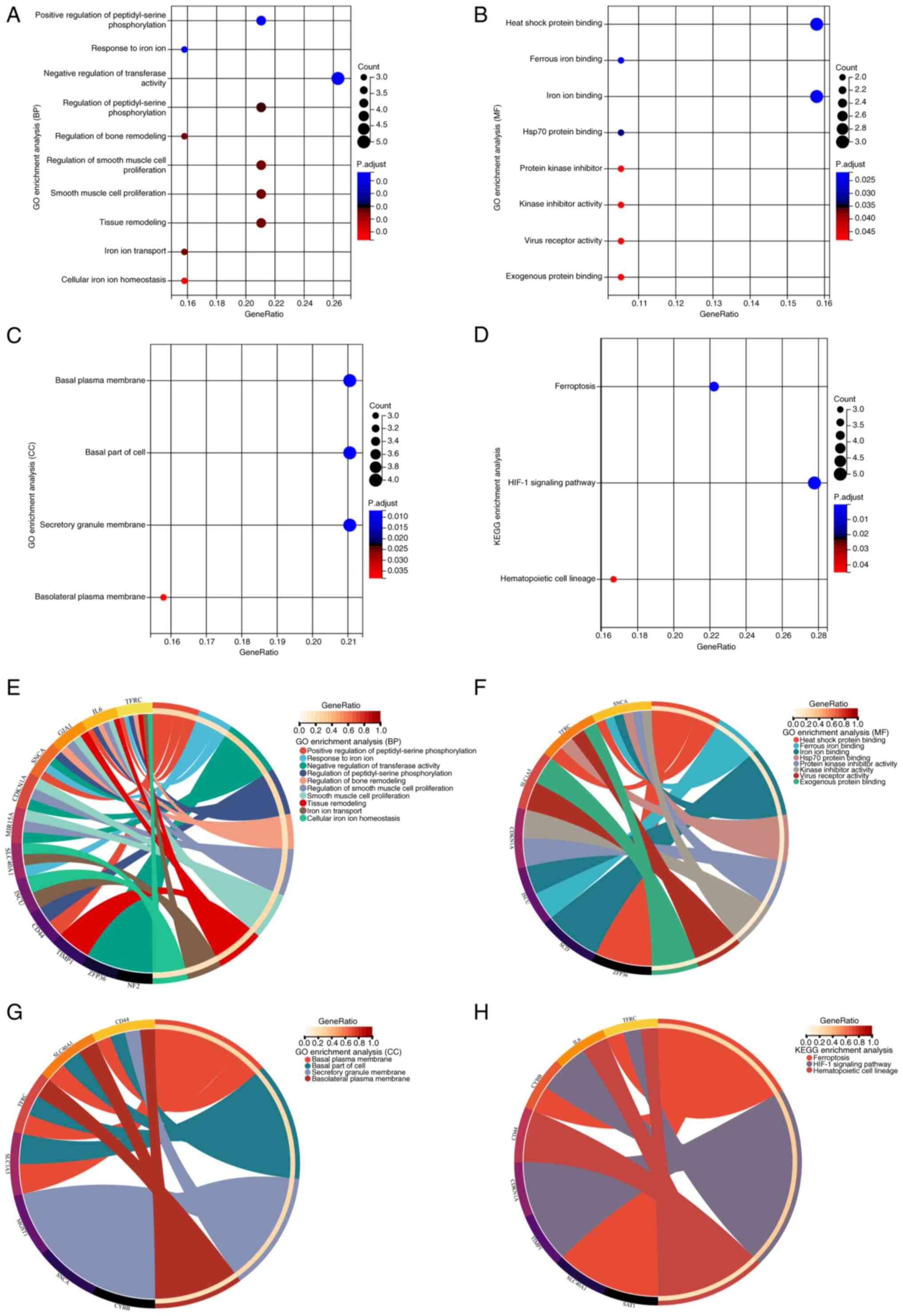
GO and KEGG analyses
To understand the functions and pathways associated
with DEFRGs, enrichment analyses were performed for GO and KEGG
pathways. The DEFRGs were primarily associated with the biological
processes of cellular iron ion homeostasis and negative regulation
of transferase activity, positive regulation of peptidyl serine
phosphorylation, smooth muscle cell proliferation, and tissue
remodeling (Fig. 5A). Among the
molecular functions, the DEFRGs were also involved in other
processes such as heat shock protein and iron ion binding, protein
kinase inhibitor activity, virus receptor activity and exogenous
protein binding (Fig. 5B). The
DEFRGs were found to be localized in several cellular components,
such as the basal plasma membrane, cellular basal fraction and
secretory granule membrane (Fig.
5C). KEGG enrichment analysis revealed that the DEFRGs were
mainly associated with ferroptosis, hypoxia-inducible factor
(HIF)-1 signaling and hematopoietic cell lineage pathways (Fig. 5D). Associations between genes and
different functions or pathways were identified in gene and pathway
cross-talk mapping and these findings indicate that multiple genes
and pathways may be dysregulated in ICM (Fig. 5E-H).
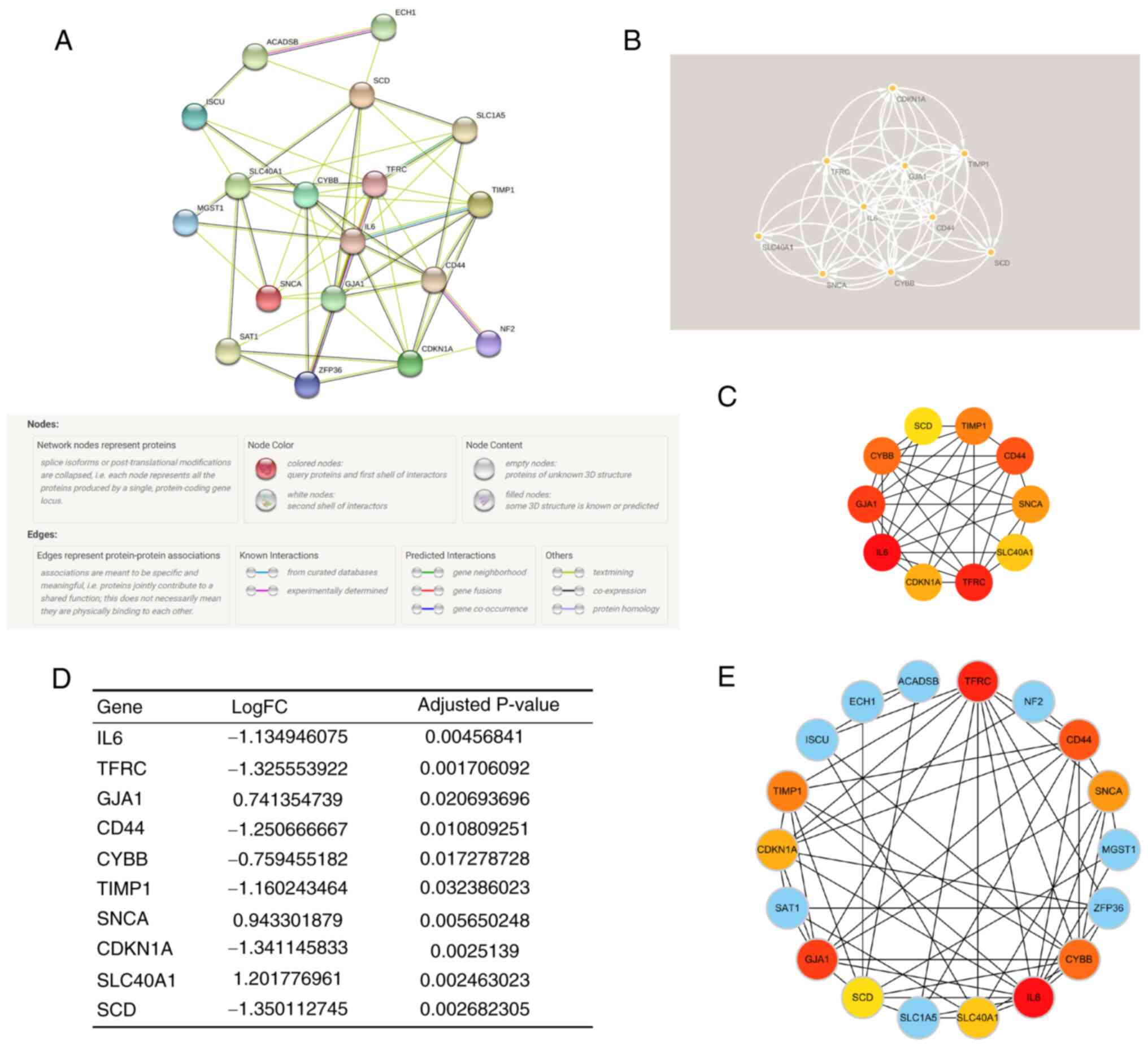
Construction of PPI network and
identification of key module and hub genes
As the regulatory role of FRGs in ICM involves the
cross-talk of multiple gene functions and pathways, a PPI network
of DEFRGs was constructed (Fig.
6A) using the STRING database and functional modules and key
genes in the PPI network were searched for using Cytoscape
software. Using cytoHubba and the MCC algorithm, and the MCODE
plugin for gene screening, the key functional and top 10 hub genes
were identified: Interleukin-6 (IL6), Transferrin receptor protein
1 (TFRC), Gap junction α-1 protein (GJA1), CD44 antigen (CD44),
Cytochrome b-245 heavy chain (CYBB), Metalloproteinase inhibitor 1
(TIMP1), α-synuclein (SNCA), Cyclin-dependent kinase inhibitor 1
(CDKN1A), Solute carrier family 40 member 1 (SLC40A1), Stearoyl-CoA
desaturase (SCD) (Fig. 6C), and
key modules (Fig. 6B). Notably,
the key modules completely overlapped with the hub genes. Fig. 6D shows the differential expression
of hub genes in ICM. Moreover, it was found that the hub genes were
associated with other DEFRGs in ICM (Fig. 6E), which indicated that the hub
genes may regulate ferroptosis through multiple genes in ICM.
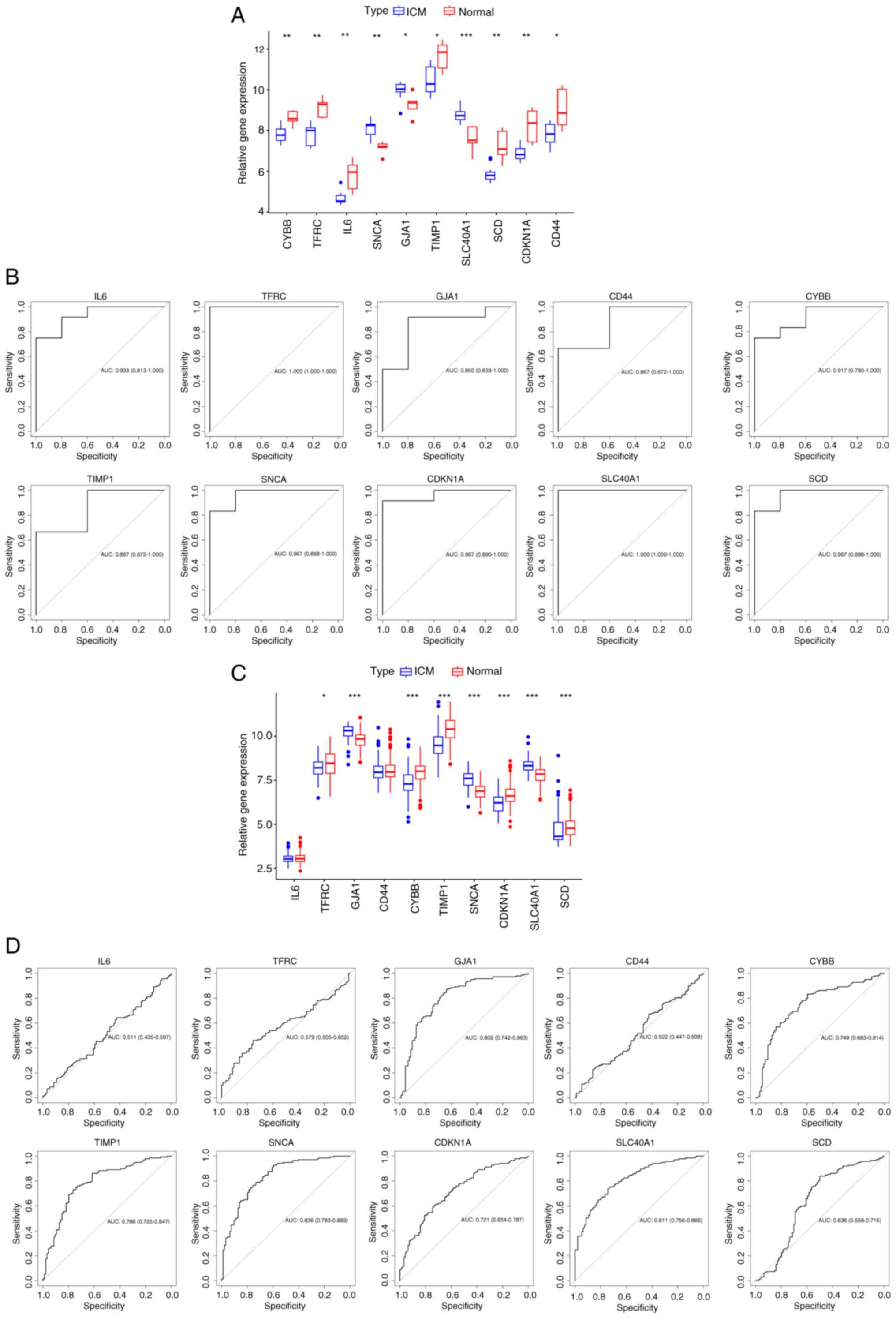
Exploration of the diagnostic
capability of hub genes
The diagnostic capability of hub genes in the
GSE26887 and GSE57338 datasets were assessed. In the GSE26887
dataset, the expression of hub genes was compared, and the screened
hub genes were demonstrated to be significantly different in both
ICM and normal heart tissues (Fig.
7A). ROC analysis results confirmed the association between the
hub genes and ICM. Additionally, the diagnostic potential of these
genes was favorable (AUC >0.7; Fig.
7B) (38). Subsequently,
validation of the hub genes was performed based on the GSE57338
dataset. Fig. 7C demonstrates that
IL-6 and CD44 were not elevated in the samples with
ICM and did not significantly differ from normal samples; however,
GJA1, SNCA, and SLC40A1 had significantly
higher gene expression in ICM samples, and TFRC,
TIMP1, CDKN1A, SCD and CYBB had
significantly lower gene expression in ICM samples compared with
the normal samples. The diagnostic ability of the hub genes was
then further validated, in which TFRC, IL-6 and
CD44 were unable to effectively diagnose ICM and the
diagnostic potential of SCD was weak; however, the other hub
genes demonstrated good diagnostic ability (Fig. 7D). The aforementioned genes were
validated by multiple steps, and most of the FRGs demonstrated good
diagnostic ability, suggesting that there may be a connection
between ferroptosis and ICM. Nevertheless, the significance of hub
genes needs to be continued to be explored in subsequent
studies.
Validation of hub gene expression in
the H9c2 A/R injury model
To confirm whether the ferroptosis inhibitor Fer-1
pretreatment can attenuate ferroptosis and protect cardiomyocytes
from the impact of A/R injury, a A/R cellular injury model was
established using H9c2 cells. The H9c2 cells were subjected to A/R
treatment to simulate myocardial I/R injury, and the extent of
ferroptosis in the model, as well as the changes in FRGs, were
detected. Firstly, the changes in GPX4 and PTGS2 were assessed at
the protein level, which are widely used as indicators for
detecting the occurrence of ferroptosis (10). The protein level of GPX4 in A/R
treated H9c2 cells significantly decreased in comparison with
control, whilst the protein level of PTGS2 in A/R treated H9c2
cells significantly increased in comparison with control
correspondingly; however, these changes were reversed by
pretreatment with Fer-1 (Fig. 8A
and B). Consistently, H9c2 cells
subjected to A/R treatment exhibited notably higher levels of
intracellular ROS in comparison with control; however, intervention
with Fer-1 resulted in a marked decrease in ROS levels in
comparison with A/R (Fig. 8C). In
subsequent experiments, the cell viability assay revealed that A/R
treated H9c2 cells had lower cell viability compared with the
control; however, Fer-1 intervention increased the cell viability
of H9c2 cells compared with A/R (Fig.
8D). In addition, significantly increased levels of total iron
(Fig. 8E), MDA (Fig. 8F) and GSSG (Fig. 8G), significantly decreased levels
of GSH (Fig. 8H), GSH/GSSG ratio
(Fig. 8I) and SOD (Fig. 8J) were observed in A/R treated H9c2
cells compared with the control. These findings indicate the
presence of ferroptosis during the process of A/R.
 | Figure 8Altered ferroptosis level and
assessment of hub gene expression in the H9c2 A/R injury model. (A)
Representative western blotting bands of PTGS2 and GPX4. (B)
Relative protein expression levels of PTGS2 and GPX4, estimated
using ImageJ software. (C) Representative images of ROS content in
H9c2 myofibroblasts. (D) Cell viability was assessed using the Cell
Counting Kit-8 assay. H9c2 cell levels of (E) total iron, (F) MDA,
(G) GSSG, (H) GSH, (I) GSH/GSSG ratios and (J) SOD in each group.
Reverse transcription-quantitative PCR results of (K) SCD, (L)
GJA1, (M) CYBB, (N) SLC40A1, (O) SNCA, (P) CDKN1A and (Q) TIMP1.
n≥3. *P<0.05; **P<0.01;
***P<0.001; ****P<0.0001. A/R, anoxic
reoxygenation; PTGS2, prostaglandin-endoperoxide synthase 2; GPX4,
glutathione peroxidase 4; ROS, reactive oxygen species; SOD,
superoxide dismutase; GSH, glutathione; GSSG, glutathione
disulfide; MDA, malondialdehyde; ns, not significant; CON, control;
Fer-1, ferrostatin-1; SCD, Stearoyl-CoA desaturase; GJA1, Gap
junction alpha-1 protein; CYBB, Cytochrome b-245 heavy chain;
SLC40A1, Solute carrier family 40 member 1; SNCA, Alpha-synuclein;
CDKN1A, Cyclin-dependent kinase inhibitor 1; TIMP1,
Metalloproteinase inhibitor 1. |
Disruption of FRGs was also observed in the
transcriptomics of human ischemic myocardium samples. The
transcription levels of hub genes in the A/R model were assessed
and the qPCR results showed that 3/7 hub genes (GJA1,
SCL40A1 and SNCA; Fig.
8K-Q) were significantly differentially elevated in the model
group compared with the control and were significantly alleviated
by the addition of Fer-1 intervention, indicating that they were
involved in the regulation of ICM ferroptosis. Thus, it was
demonstrated that GJA1, SCL40A1 and SNCA are
key genes regulating ferroptosis in ICM.
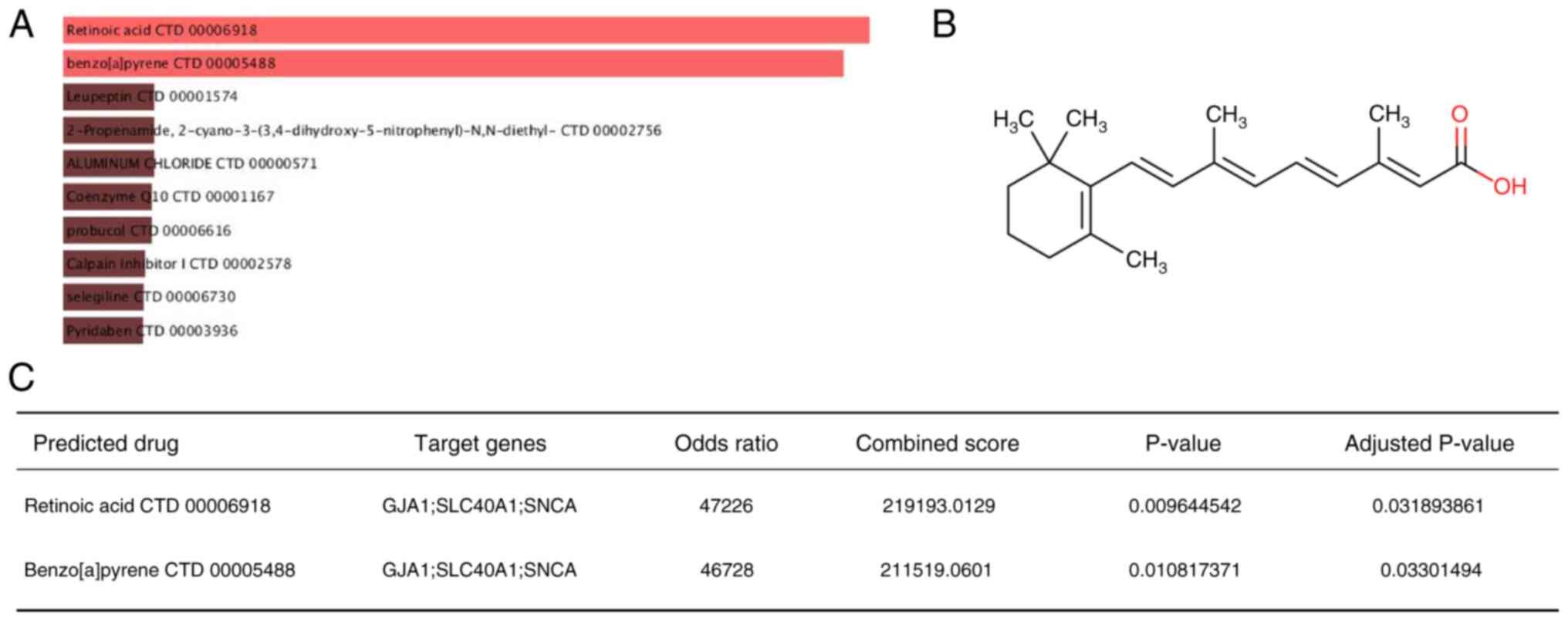
Potential ferroptosis-regulating drugs
prediction
The DSigDB was used to predict potential
ferroptosis-regulating drugs associated with key genes that may
treat ICM by modulating ferroptosis. A final total of 233 drugs was
obtained and their combination scores and corresponding target
genes are listed in Table SV. The
drugs with the highest combined scores were retinoic acid and
benzo[a]pyrene (Fig. 9A). Retinoic
acid (combined scores=219193; Fig.
9B) had a strong drug-target association (P.adjust <0.05;
Fig. 9C). By contrast,
benzo[a]pyrene is strongly toxicogenic and may accelerate the
process of ferroptosis, requiring protection in daily life species
(39).
Discussion
Globally, ICM is a principal contributor to
mortality, sickness and incapacity arising from cardiovascular
disease (40,41). Therefore, it is urgent to identify
the pathophysiological mechanisms of ICM and provide meaningful
diagnostic and therapeutic targets for clinical work (7).
Ferroptosis is an iron-dependent and distinct form
of regulated cell death, differing from apoptosis, necrosis and
autophagy (9). It is characterized
by the accumulation of lipid hydroperoxides that reach lethal
levels, causing oxidative damage to the cellular membrane (9,10).
In the past decade, an increasing number of studies have supported
the important pathophysiological role of ferroptosis in the
development of cardiovascular diseases such as doxorubicin-induced
cardiomyopathy, myocardial I/R injury, myocardial infarction and
heart failure (42). High levels
of ferroptosis, mediated by certain signaling and metabolic
pathways, may result in more severe injury to ICM, and the
regulatory molecular mechanisms and signaling pathway cross-talk
remain unclear (43). The rapid
advancement of transcriptomics has offered new insights into the
pathological process of ferroptosis in ICM (44). The present study comprehensively
analyzed the dysregulated ferroptosis-related genes in the
transcriptome of ICM, explored related functions and pathways
through GO and KEGG enrichment analysis, predicted hub genes
through the PPI network, analyzed their diagnostic ability through
ROC curves, and assessed hub genes in in vitro models.
Finally, potential ferroptosis-targeted drugs were obtained from
the DSigDB, providing several potential choices for the clinical
treatment of ICM.
In the present study, GSEA demonstrated a
correlation between ferroptosis and ICM, which is consistent with
previous research results, indicating the presence of ferroptosis
in the pathological process of ICM (44-46).
WGCNA and differential analysis identified clustered modules and
dysregulated FRGs in ICM, and subsequent GO enrichment analysis
revealed that the dysregulated FRGs in the ICM are involved in the
regulation of ferroptosis through several pathways.
Iron is a crucial micronutrient within the human
body as it serves a vital role in biological processes, including
the transportation, storage and utilization of oxygen (47). Iron homeostasis serves a crucial
role in the function of the heart. Most iron ions are transported
through transport proteins on the basal membrane, and intracellular
iron ions can also bind to ferritin and are secreted through the
multivesicular body-exosome pathway (48). FRGs negatively regulate cellular
iron homeostasis and transferase activity, causing iron ions to
accumulate in the mitochondria, increasing oxidative stress and
ultimately leading to mitochondrial dysfunction (49). Due to the limited regenerative
capacity of cardiomyocytes, a stable scar is formed immediately
after myocardial ischemia. However, under the stimulation of
chronic ischemia, the excessive deposition of collagen-rich
extracellular matrix determines the pathological remodeling of the
myocardium and causes an increase in mechanical strain in the
border zone, which may lead to expansion of the fibrotic area,
decreased tissue compliance and increased cardiac afterload
(50-54).
Existing research indicates that regulating FRGs can effectively
reduce cardiac injury, fibrosis and pathological remodeling during
ischemia (55). In the present
study, the DEFRGs were found to be enriched in iron metabolism and
the HIF-1 signaling pathway. Additionally, they were linked to
certain pathways of hematopoietic cell lines, which could be
associated with the transportation of iron ions. These findings
indicate that the regulation of ferroptosis in ICM involves unknown
key molecules and pathways that require further exploration. To
gain a more thorough understanding of the mechanism behind iron
metabolism in ICM, additional research is necessary.
By constructing the PPI interaction network to
identify the top 10 hub genes based on the MCC algorithm. The
diagnostic ability of hub genes was determined by ROC, and genes
with weak diagnostic ability were excluded. In order to more
realistically verify the expression of the remaining hub genes, the
H9c2 A/R injury model was further constructed to verify whether the
expression trend of the remaining hub genes was consistent with the
transcriptomics data. In the present study, GJA1,
SCL40A1 and SNCA showed promise as therapeutic
targets for ICM. The upregulation of SNCA leads to cells
being subjected to abnormal ROS generation and glutathione
utilization, resulting in oxidative stress of lipid peroxidation
and eventual cell death via ferroptosis (56). Notably, upon activation of
ferroptosis, GJA1 is markedly upregulated as a negative
regulatory gene. However, the mechanism of GJA1 in the
ferroptosis pathway remains incompletely understood, potentially as
a result of its bidirectional regulatory effect on ferroptosis
which has yet to be acknowledged (57,58).
Further research is necessary to supplement this. Furthermore, as a
positive regulatory gene, SCL40A1 notably increased in an
A/R model, resulting in cell iron overload and ferroptosis
(59). The aforementioned hub
genes are expected to become new targets for the treatment of ICM,
and further investigation into their specific regulatory mechanisms
is required.
The present study aimed to regulate dysregulated
genes in ICM for treatment and clinical transformation. According
to the selected hub genes in the present study, potential
therapeutic drugs were predicted for ICM. Among them, retinoic acid
and benzo[a]pyrene demonstrated a high drug target association.
Retinoic acid is essential for maintaining tissue homeostasis and
has been shown to ameliorate I/R injury and several instances of
drug-induced cardiotoxicity (60).
These effects are achieved through the inhibition of oxidative
stress, prevention of cardiomyocyte apoptosis and attenuation of
p38 MAPK, JNK and NF-κB activation (61-63).
Benzo[a]pyrene is a polycyclic aromatic hydrocarbon present in
tobacco smoke and indoor air pollution. It possesses attributes
such as lipophilicity, refractoriness, bioaccumulation,
cytotoxicity, mutagenicity and carcinogenicity (64,65).
It may have a negative regulatory effect on I/R injury and lacks
protective properties (39). As a
result, it should be avoided in everyday circumstances. Similar
protective medicines, such as retinoic acid, may regulate
ferroptosis to reduce damage to the ICM. However, their
pharmacological mechanisms have not yet been explored (66).
The analyses in the present study confirmed the
presence of numerous dysregulated FGRs in ICM, providing evidence
for subsequent exploration; however, it is worth noting that the
present study has certain limitations. First, the present study
observed that ferroptosis features in the H9c2 cell injury model,
including intracellular ROS aggregation, ferroptosis marker protein
changes and lipid peroxidation (44-46).
Nevertheless, it did not investigate how dysregulation of DEFRGs
regulates ferroptosis in ICM, which is a limitation of the present
study. In addition, the present study did not directly observe the
characteristics of ferroptosis in human ICM. To establish more
dependable diagnostic and therapeutic targets, and to translate the
findings of the present study into clinical outcomes, in-depth
mechanism and functional studies are imperative for comprehending
the regulation of ferroptosis in ICM by the FGRs.
Overall, extensive dysregulation of
ferroptosis-related genes was identified in ICM. These are situated
in the basement membrane and secretory granules. They trigger
intracellular iron overload through the transport function of iron
ions, which may lead to positive regulatory peptide serine
phosphorylation, smooth muscle cell proliferation and tissue
remodeling. Furthermore, the expression of hub FRGs (GJA1,
SCL40A1, SNCA) were assessed in an A/R model and a
statistically significant association between GJA1,
SCL40A1, SNCA and the occurrence of ICM was
identified. In addition, the present research predicted that
retinoic acid may provide a protective mechanism by regulating
ferroptosis in ICM. The current study offers new insights and
evidence regarding the involvement of ferroptosis in ICM. This is
important for discovering the pathological mechanisms of ICM and
identifying its diagnostic and therapeutic targets.
Supplementary Material
Ferroptosis-related genes were
obtained from the FerrDb database.
Base sequences of the forward and
backward primers for the hub gene.
Differentially expressed genes of
GSE26887.
Overlap of genes between
differentially expressed genes, turquoise module genes and
ferroptosis-related genes (differentially expressed
ferroptosis-related genes).
Detailed information of the predicted
potential ferroptosis-regulating drugs by DSigDB.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Nature
Science Foundation of China (grant nos. 81860082 and 82160073) and
Jiangxi Provincial Natural Science Foundation (grant nos.
20212ACB206011, 20224ACB206002 and 20232BAB206009).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
STZ, ZCQ, RYZ performed the cellular experiments
and participated in writing and data analysis. HXZ provided the
experimental design. ZCQ and RYZ confirm the authenticity of all
the raw data. RBQ and HZP analyzed the experimental data. LFZ and
ZQX provided software support. SQL and LW designed the experiments
and provided financial assistance. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Moroni F, Gertz Z and Azzalini L: Relief
of ischemia in ischemic cardiomyopathy. Curr Cardiol Rep.
23(80)2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Cabac-Pogorevici I, Muk B, Rustamova Y,
Kalogeropoulos A, Tzeis S and Vardas P: Ischaemic cardiomyopathy.
Pathophysiological insights, diagnostic management and the roles of
revascularisation and device treatment. Gaps and dilemmas in the
era of advanced technology. Eur J Heart Fail. 22:789–799.
2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Li Y, Du Y, Cao J, Gao Q, Li H, Chen Y and
Lu N: MiR-130a inhibition protects rat cardiac myocytes from
hypoxia-triggered apoptosis by targeting Smad4. Kardiol Pol.
76:993–1001. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Xu Q, Liu S, Gong Q, Zhu R, Liu J, Wu Q
and Zhou X: Notch1 protects against ischemic-reperfusion injury by
suppressing PTEN-Pink1-Mediated mitochondrial dysfunction and
mitophagy. Cells. 12(137)2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Wang X, Xie W, Zhang Y, Lin P, Han L, Han
P, Wang Y, Chen Z, Ji G, Zheng M, et al: Cardioprotection of
ischemia/reperfusion injury by cholesterol-dependent MG53-mediated
membrane repair. Circ Res. 107:76–83. 2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Gao C, Wang R, Li B, Guo Y, Yin T, Xia Y,
Zhang F, Lian K, Liu Y, Wang H, et al: TXNIP/Redd1 signalling and
excessive autophagy: A novel mechanism of myocardial
ischaemia/reperfusion injury in mice. Cardiovasc Res. 116:645–657.
2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hausenloy DJ and Yellon DM: Myocardial
ischemia-reperfusion injury: A neglected therapeutic target. J Clin
Invest. 123:92–100. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hao L, Wang J and Liu N: Long noncoding
RNA TALNEC2 regulates myocardial ischemic injury in H9c2 cells by
regulating miR-21/PDCD4-medited activation of Wnt/β-catenin
pathway. J Cell Biochem. 120:12912–12923. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta
R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS,
et al: Ferroptosis: An iron-dependent form of nonapoptotic cell
death. Cell. 149:1060–1072. 2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Stockwell BR, Friedmann Angeli JP, Bayir
H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK,
Kagan VE, et al: Ferroptosis: A regulated cell death nexus linking
metabolism, redox biology, and disease. Cell. 171:273–285.
2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wang M, Mao C, Ouyang L, Liu Y, Lai W, Liu
N, Shi Y, Chen L, Xiao D, Yu F, et al: Long noncoding RNA LINC00336
inhibits ferroptosis in lung cancer by functioning as a competing
endogenous RNA. Cell Death Differ. 26:2329–2343. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kenny EM, Fidan E, Yang Q, Anthonymuthu
TS, New LA, Meyer EA, Wang H, Kochanek PM, Dixon CE, Kagan VE and
Bayir H: Ferroptosis contributes to neuronal death and functional
outcome after traumatic brain injury. Crit Care Med. 47:410–418.
2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Chen B, Chen Z, Liu M, Gao X, Cheng Y, Wei
Y, Wu Z, Cui D and Shang H: Inhibition of neuronal ferroptosis in
the acute phase of intracerebral hemorrhage shows long-term
cerebroprotective effects. Brain Res Bull. 153:122–132.
2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Guan X, Li X, Yang X, Yan J, Shi P, Ba L,
Cao Y and Wang P: The neuroprotective effects of carvacrol on
ischemia/reperfusion-induced hippocampal neuronal impairment by
ferroptosis mitigation. Life Sci. 235(116795)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kobayashi M, Suhara T, Baba Y, Kawasaki
NK, Higa JK and Matsui T: Pathological roles of iron in
cardiovascular disease. Curr Drug Targets. 19:1068–1076.
2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bulluck H, Rosmini S, Abdel-Gadir A, White
SK, Bhuva AN, Treibel TA, Fontana M, Ramlall M, Hamarneh A, Sirker
A, et al: Residual myocardial iron following intramyocardial
hemorrhage during the convalescent phase of reperfused
ST-Segment-Elevation myocardial infarction and adverse left
ventricular remodeling. Circ Cardiovasc Imaging.
9(e004940)2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li D, Pi W, Sun Z, Liu X and Jiang J:
Ferroptosis and its role in cardiomyopathy. Biomed Pharmacother.
153(113279)2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Greco S, Fasanaro P, Castelvecchio S,
D'Alessandra Y, Arcelli D, Di Donato M, Malavazos A, Capogrossi MC,
Menicanti L and Martelli F: MicroRNA dysregulation in diabetic
ischemic heart failure patients. Diabetes. 61:1633–1641.
2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liu Y, Morley M, Brandimarto J,
Hannenhalli S, Hu Y, Ashley EA, Tang WH, Moravec CS, Margulies KB,
Cappola TP, et al: RNA-Seq identifies novel myocardial gene
expression signatures of heart failure. Genomics. 105:83–89.
2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9(559)2008.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43(e47)2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4)(S11)2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhou N and Bao J: FerrDb: A manually
curated resource for regulators and markers of ferroptosis and
ferroptosis-disease associations. Database (Oxford).
2020(baaa021)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Bardou P, Mariette J, Escudié F, Djemiel C
and Klopp C: jvenn: An interactive Venn diagram viewer. BMC
Bioinformatics. 15(293)2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z,
Feng T, Zhou L, Tang W, Zhan L, et al: clusterProfiler 4.0: A
universal enrichment tool for interpreting omics data. Innovation
(Camb). 2(100141)2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Shen W, Song Z, Zhong X, Huang M, Shen D,
Gao P, Qian X, Wang M, Li S, Song X, et al: Sangerbox: A
comprehensive, interaction-friendly clinical bioinformatics
analysis platform. iMeta. 1(e36)2022.
|
|
29
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613.
2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Wen L, Cheng X, Fan Q, Chen Z, Luo Z, Xu
T, He M and He H: TanshinoneⅡA inhibits excessive autophagy and
protects myocardium against ischemia/reperfusion injury via
14-3-3η/Akt/Beclin1 pathway. Eur J Pharmacol.
954(175865)2023.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hu T, Zou HX, Le SY, Wang YR, Qiao YM,
Yuan Y, Liu JC, Lai SQ and Huang H: Tanshinone IIA confers
protection against myocardial ischemia/reperfusion injury by
inhibiting ferroptosis and apoptosis via VDAC1. Int J Mol Med.
52(109)2023.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Yoo M, Shin J, Kim J, Ryall KA, Lee K, Lee
S, Jeon M, Kang J and Tan AC: DSigDB: Drug signatures database for
gene set analysis. Bioinformatics. 31:3069–3071. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Wishart DS, Feunang YD, Guo AC, Lo EJ,
Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, et al:
DrugBank 5.0: A major update to the DrugBank database for 2018.
Nucleic Acids Res. 46:D1074–D1082. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zou HX, Hu T, Zhao JY, Qiu BQ, Zou CC, Xu
QR, Liu JC, Lai SQ and Huang H: Exploring Dysregulated
ferroptosis-related genes in septic myocardial injury based on
human heart transcriptomes: Evidence and new insights. J Inflamm
Res. 16:995–1015. 2023.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Liu K, Chen S and Lu R: Identification of
important genes related to ferroptosis and hypoxia in acute
myocardial infarction based on WGCNA. Bioengineered. 12:7950–7963.
2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wang Q, Liu B, Wang Y, Bai B, Yu T and Chu
XM: The biomarkers of key miRNAs and target genes associated with
acute myocardial infarction. PeerJ. 8(e9129)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Tarasco M, Gavaia PJ, Bensimon-Brito A,
Cardeira-da-Silva J, Ramkumar S, Cordelières FP, Günther S,
Bebianno MJ, Stainier DYR, Cancela ML and Laizé V: New insights
into benzo[α]pyrene osteotoxicity in zebrafish. Ecotoxicol Environ
Saf. 226(112838)2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Alissa EM and Ferns GA: Heavy metal
poisoning and cardiovascular disease. J Toxicol.
2011(870125)2011.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Wu X, Li Y, Zhang S and Zhou X:
Ferroptosis as a novel therapeutic target for cardiovascular
disease. Theranostics. 11:3052–3059. 2021.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Del Re DP, Amgalan D, Linkermann A, Liu Q
and Kitsis RN: Fundamental mechanisms of regulated cell death and
implications for heart disease. Physiol Rev. 99:1765–1817.
2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Fang X, Ardehali H, Min J and Wang F: The
molecular and metabolic landscape of iron and ferroptosis in
cardiovascular disease. Nat Rev Cardiol. 20:7–23. 2023.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Feng Y, Madungwe NB, Imam Aliagan AD,
Tombo N and Bopassa JC: Liproxstatin-1 protects the mouse
myocardium against ischemia/reperfusion injury by decreasing VDAC1
levels and restoring GPX4 levels. Biochem Biophys Res Commun.
520:606–611. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Li W, Li W, Leng Y, Xiong Y and Xia Z:
Ferroptosis is involved in diabetes myocardial ischemia/reperfusion
injury through endoplasmic reticulum stress. DNA Cell Biol.
39:210–225. 2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Baba Y, Higa JK, Shimada BK, Horiuchi KM,
Suhara T, Kobayashi M, Woo JD, Aoyagi H, Marh KS, Kitaoka H and
Matsui T: Protective effects of the mechanistic target of rapamycin
against excess iron and ferroptosis in cardiomyocytes. Am J Physiol
Heart Circ Physiol. 314:H659–H668. 2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Andrews NC: Forging a field: The golden
age of iron biology. Blood. 112:219–230. 2008.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Gao G, Li J, Zhang Y and Chang YZ:
Cellular Iron Metabolism and Regulation. Adv Exp Med Biol.
1173:21–32. 2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Paterek A, Mackiewicz U and Mączewski M:
Iron and the heart: A paradigm shift from systemic to cardiomyocyte
abnormalities. J Cell Physiol. 234:21613–21629. 2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Zhang QJ, He Y, Li Y, Shen H, Lin L, Zhu
M, Wang Z, Luo X, Hill JA, Cao D, et al: Matricellular protein
cilp1 promotes myocardial fibrosis in response to myocardial
infarction. Circ Res. 129:1021–1035. 2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Schroer AK, Bersi MR, Clark CR, Zhang Q,
Sanders LH, Hatzopoulos AK, Force TL, Majka SM, Lal H and Merryman
WD: Cadherin-11 blockade reduces inflammation-driven fibrotic
remodeling and improves outcomes after myocardial infarction. JCI
Insight. 4(e131545)2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Snider JC, Riley LA, Mallory NT, Litsky
AS, Stoodley P, Swinehart SD, Duerr RA, Kaeding CC and Flanigan DC:
Targeting 5-HT2B receptor signaling prevents border zone expansion
and improves microstructural remodeling after myocardial
infarction. Circulation. 143:1317–1330. 2021.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Humeres C and Frangogiannis NG:
Fibroblasts in the Infarcted, remodeling, and failing heart. JACC
Basic Transl Sci. 4:449–467. 2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Talman V and Ruskoaho H: Cardiac fibrosis
in myocardial infarction-from repair and remodeling to
regeneration. Cell Tissue Res. 365:563–581. 2016.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Chen Y, Li X, Wang S, Miao R and Zhong J:
Targeting iron metabolism and ferroptosis as novel therapeutic
approaches in cardiovascular diseases. Nutrients.
15(59)2023.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Angelova PR, Choi ML, Berezhnov AV,
Horrocks MH, Hughes CD, De S, Rodrigues M, Yapom R, Little D, Dolt
KS, et al: Alpha synuclein aggregation drives ferroptosis: An
interplay of iron, calcium and lipid peroxidation. Cell Death
Differ. 27:2781–2796. 2020.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Yu M, Lin Z, Tian X, Chen S, Liang X, Qin
M, Zhu Q, Wu Y and Zhong S: Downregulation of Cx43 reduces
cisplatin-induced acute renal injury by inhibiting ferroptosis.
Food Chem Toxicol. 158(112672)2021.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Huang Q, Sha W, Gu Q, Wang J, Zhu Y, Xu T,
Xu Z, Yan F, Lin X and Tian S: Inhibition of Connexin43 improves
the recovery of spinal cord injury against ferroptosis via the
SLC7A11/GPX4 pathway. Neuroscience. 526:121–134. 2023.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Beutler E, Barton JC, Felitti VJ, Gelbart
T, West C, Lee PL, Waalen J and Vulpe C: Ferroportin 1 (SCL40A1)
variant associated with iron overload in African-Americans. Blood
Cells Mol Dis. 31:305–309. 2003.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Wiggert B, Bergsma DR, Helmsen R and
Chader GJ: Vitamin A receptors. Retinoic acid binding in ocular
tissues. Biochem J. 169:87–94. 1978.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Choudhary R, Baker KM and Pan J: All-trans
retinoic acid prevents angiotensin II- and mechanical
stretch-induced reactive oxygen species generation and
cardiomyocyte apoptosis. J Cell Physiol. 215:172–181.
2008.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Lou S, Zhong L, Yang X, Xue T, Gai R, Zhu
D, Zhao Y, Yang B, Ying M and He Q: Efficacy of all-trans retinoid
acid in preventing nickel induced cardiotoxicity in myocardial
cells of rats. Food Chem Toxicol. 51:251–258. 2013.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Nizamutdinova IT, Guleria RS, Singh AB,
Kendall JA, Baker KM and Pan J: Retinoic acid protects
cardiomyocytes from high glucose-induced apoptosis through
inhibition of NF-κB signaling pathway. J Cell Physiol. 228:380–392.
2013.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Yamaguchi A, Uchida M, Ishibashi H, Hirano
M, Ichikawa N, Arizono K, Koyama J and Tominaga N: Potential
mechanisms underlying embryonic developmental toxicity caused by
benzo[a]pyrene in Japanese medaka (Oryzias latipes). Chemosphere.
242(125243)2020.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Feng B, Li L, Xu H, Wang T, Wu R, Chen J,
Zhang Y, Liu S, Ho SSH, Cao J and Huang W: PM2.5-bound polycyclic
aromatic hydrocarbons (PAHs) in Beijing: Seasonal variations,
sources, and risk assessment. J Environ Sci (China). 77:11–19.
2019.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Wu Y, Huang T, Li X, Shen C, Ren H, Wang
H, Wu T, Fu X, Deng S, Feng Z, et al: Retinol dehydrogenase 10
reduction mediated retinol metabolism disorder promotes diabetic
cardiomyopathy in male mice. Nat Commun. 14(1181)2023.PubMed/NCBI View Article : Google Scholar
|