Introduction
Multiple myeloma (MM) is a malignant hematological
disease characterized by abnormal proliferation of clonal plasma
cells in the bone marrow, accompanied by the secretion of large
amounts of ineffective monoclonal immunoglobulins in the serum or
urine (1). This abnormal
proliferation ultimately leads to specific end-organ damage,
including hypercalcemia, renal insufficiency, anemia and osteolytic
lesions (2). There are ~140,000
new cases of MM are reported globally annually, with an increasing
trend per year (3). In most
countries, MM is the second most common malignant tumor of the
blood system and epidemiological studies have shown that it is
distributed regionally (4). In the
past decade, with the emergence of new chemotherapy drugs and
development of high-level antitumor regimens, the prognosis of
patients with MM has greatly improved (5). However, almost all patients will
eventually relapse, even those with a complete response to initial
treatment (6).
The intestinal microbiota is a complex ecosystem
composed of thousands of microorganisms that participate in
nutrient metabolism and absorption in the host gut, as well as
regulating host intestinal immunity through the mesenteric
lymphatic system (7). The immune
regulatory effect of the intestinal microbiota extends beyond the
gut, primarily through small molecules they produce (8). The anaerobic environment in the
gastrointestinal tract enables bacteria to ferment and produce
metabolites with immune activity that can enter the systemic
circulation, especially butyrate produced by Clostridium
butyricum (9). Disturbance of
the intestinal microbiota and an abnormal increase in its
metabolites can lead to continuous stimulation signals throughout
the entire gastrointestinal tract and trigger various diseases,
including tumors (10). Previous
studies have evaluated the relationship between intestinal
microbiota and other hematological tumors (11); however, MM has rarely been studied,
especially in high-altitude and cold regions.
Antibody-secreting plasma cells can survive in the
gut for a long time (12). The
composition of the intestinal microbiota can influence the degree
of antigen stimulation in these cells and may play a role in the
development of mutations and clonal evolution. Drugs used to treat
MM, such as proteasome inhibitors and alkylating agents, often
cause adverse gastrointestinal reactions (13). Several studies have shown changes
in the composition and abundance of intestinal microbiota in MM
patients undergoing hematopoietic stem cell transplantation (HSCT)
(14-16).
The present study hypothesized that certain intestinal
microorganisms and their metabolites play regulatory roles in the
progression of MM. The present study performed 16s rRNA
high-throughput sequencing to characterize the intestinal
microbiota of patients with MM and found changes in the abundance
of several bacteria. It also conducted correlation and Kyoto
Encyclopedia of Genes and Genomes (KEGG) function prediction
analyses. The present study provided a theoretical basis for
elucidating the pathogenesis of MM and a new approach for its
prevention and treatment.
Materials and methods
Patients
A total of 15 newly-diagnosed patients with MM, who
met the diagnostic criteria of the International Myeloma Working
Group (IMWG) (17), were enrolled
at the first hospital of Qiqihar between October 2021 and March
2023. All patients originated from the northwest of Heilongjiang
Province and the Inner Mongolia Autonomous Region, which are
high-altitude and cold regions of China. A total of 11 healthy
individuals from the spouses, children and parents of the patients
were recruited as controls, because they shared a common living
environment and dietary habits similar to those of the patients.
Patients with gastrointestinal diseases or diarrhea, infectious
diseases, or metabolic disorders; or those who had recently taken
antibiotics, gastrointestinal motility drugs, microecological
regulators, or immunosuppressants were excluded from the study. No
significant differences in age, sex and basal metabolic rate were
observed among the enrolled subjects (Table I).
 | Table IGeneral characteristics of the
subjects. |
Table I
General characteristics of the
subjects.
| Characteristic | MM (n=15) | Healthy control
(n=11) | P-value |
|---|
| Age, years | 66.13±1.89 | 62.09±2.36 | 0.189 |
| Sex
(Male/Female) | 11/4 | 7/4 | 0.683 |
| BMI, kg/m² | 26.87±0.93 | 25.74±1.56 | 0.446 |
| R-ISS Stage | I (3) | | |
| | II (6) | | |
| | III (6) | | |
| Disease
Subtype | IgG-κ (5) | | |
| | IgG-λ (3) | | |
| | IgM-κ (1) | | |
| | Light chain-λ
(4) | | |
| | Unknown (2) | | |
Sample collection
Fecal samples were collected from patients before
systemic treatment and from healthy controls in the early morning.
To avoid sample contamination, the patients were instructed to
defecate in a clean container. An appropriate sample was collected
using a disposable sample spoon and placed in a sterile closed
container. To avoid interference from environmental factors during
the experiment, the entire sampling process did not exceed 10 min.
Samples were stored at -80˚C until transport to the testing
laboratory with dry ice. Venous blood (5 ml) was collected from
patients and healthy volunteers on an empty stomach and centrifuged
at 2,000 x g and room temperature for 10 min. Serum was then
collected and was stored at -20˚C until further detection of
complement components, such as C1q, C3 and C4 (BN II; Siemens
AG).
Sample DNA extraction
Total genomic DNA in fecal samples from the two
groups was extracted using the PF Mag Bind Stool DNA Kit (Omega
Bio-Tek, Inc.) according to the manufacturer's instructions. The
integrity of the extracted genomic DNA was detected by 1% agarose
gel electrophoresis. The concentration of the DNA was determined
using NanoDrop2000 spectrophotometer (Thermo Fisher Scientific,
Inc.).
16s rRNA sequencing and paired-end (PE) library
construction. Using the extracted DNA as a template, the V3-V4
hypervariable region of the 16s rRNA gene was amplified with primer
pairs 338F (5'-ACTCCTACGGGGGGGGCAG-3') and 806R
(5'-GACTACHVGGGTWTCTAAT-3') using the ABI GeneAmp 7500 PCR
thermocycler (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Amplicons were recovered using 2% agarose gel, purified using the
AxyPrep DNA Gel Extraction Kit (Axygen Biosciences; Corning, Inc.)
and quantified using a Quantus Fluorometer (Promega Corporation). A
PE library was constructed using the NEXTFLEX Rapid DNA-Seq Kit
(BioScientific, Inc.) and PE sequencing was performed on an
Illumina MiSeq PE300 platform (Illumina, Inc.). The raw sequencing
data were uploaded to the Majorbio Cloud platform (https://cloud.majorbio.com; Majorbio Bio-Pharm
Technology Co. Ltd) for bioinformatics analysis.
Bioinformatics analysis
Raw sequencing data were subjected to quality
control and splicing using the Fastp software (https://github.com/OpenGene/fastp, version
0.19.6) and Flash software (https://ccb.jhu.edu/software/FLASH/index.shtml,
version 1.2.11), respectively. The optimized sequences were
clustered into operational taxonomic units (OTUs) based on 97%
similarity using the UPARSE software (http://www.drive5.com/uparse/, version 11). Sequences
annotated as chloroplasts and mitochondria were removed from all
samples and the number of 16s rRNA gene sequences in all samples
was rarefied to 20,000. OTU taxonomic annotation was performed
based on the Silva 16s rRNA gene database (https://www.arb-silva.de/, version 138) using the RDP
classifier software (https://sourceforge.net/projects/rdp-classifier/,
version 11.5). Functional prediction analysis was performed using
PICRUSt2 (http://picrust.github.io/picrust/, version 2.2.0). All
bioinformatic analyses were conducted using the Majorbio Cloud
platform (https://cloud.majorbio.com; Majorbio
Bio-Pharm Technology Co. Ltd).
Statistics analysis
The comparison of age and BMI among the general
characteristics of the subjects was conducted by t-test and the sex
by chi-square test. The ACE, Chao, Sobs and Shannon indices were
used to evaluate alpha diversity and the Wilcoxon rank-sum test was
used to analyze intergroup differences. Principal coordinate
analysis (PCoA) based on the Bray-Curtis distance algorithm was
used for beta diversity analysis and analysis of similarities
(ANOSIM) was used to determine statistical significance. Linear
discriminant analysis (LDA) effect size (LEfSe) was used to
identify bacterial groups with significant differences (LDA>2;
P<0.05) in abundance from phylum to genus levels. Disease
diagnosis model constructed by Random Forest (RF) analysis, and ROC
curve analysis was used to verify the accuracy of the constructed
model. The Mantel test and distance-based redundancy analysis
(db-RDA) were used for environmental factor correlation analysis to
investigate the effects of clinical indicators on the composition
of the intestinal microbiota. Network analysis was performed using
Spearman correlation (ρ>0.6; P<0.05). PICRUSt2 was used to
predict KEGG functional genes. P<0.05 was considered to indicate
a statistically significant difference.
Results
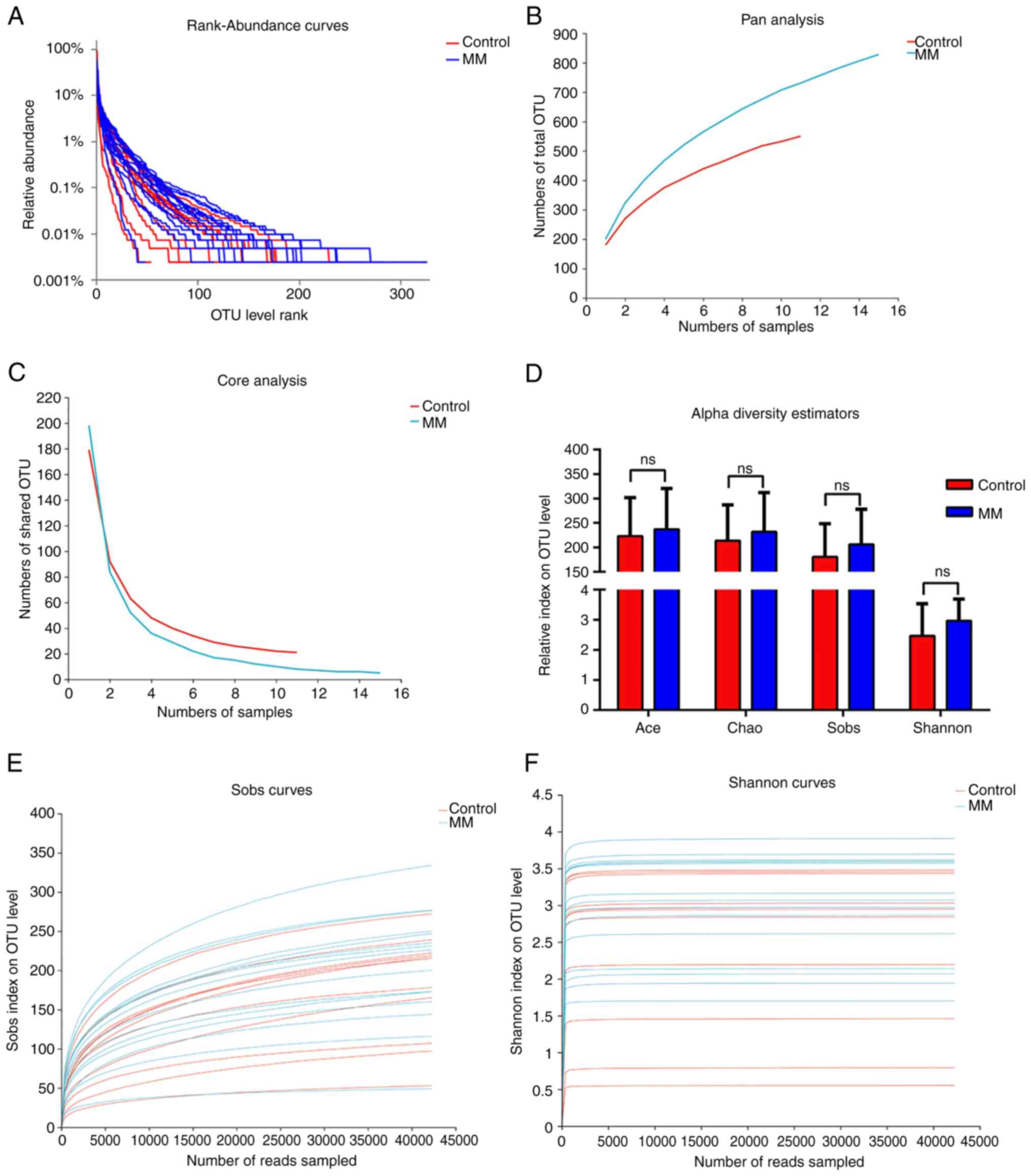
Microbial diversity analysis
The present study obtained taxonomic information on
the microbiota through OTU analysis and evaluated species abundance
and distribution uniformity using rank-abundance curves. Analysis
of the rank-abundance curve showed that species richness and
evenness in the MM group were higher compared with the healthy
group; however, the proportion of dominant bacteria in the MM group
was lower (Fig. 1A). Pan analysis
showed that, as the number of samples increased, the total number
of fecal species increased, especially in the MM group, indicating
that the sample size in this study was relatively small (Fig. 1B). However, as the sample size
increased, the decrease in shared OTU tended to plateau through
core analysis, indicating that the sample size of this experiment
was acceptable (Fig. 1C). The Ace,
Chao, Sobs and Shannon indices were calculated to evaluate the
alpha diversity in the two groups. The Wilcoxon rank-sum test
showed that alpha diversity in the MM group was higher compared
with that in the control group, but the difference was not
significant (Fig. 1D). Dilution
curve analysis, including Sobs and Shannon, showed that the curve
reached a plateau, indicating that this sequencing covered almost
all the bacteria in the sample (Fig.
1E and F), thus ensuring the
rationality of subsequent analysis.
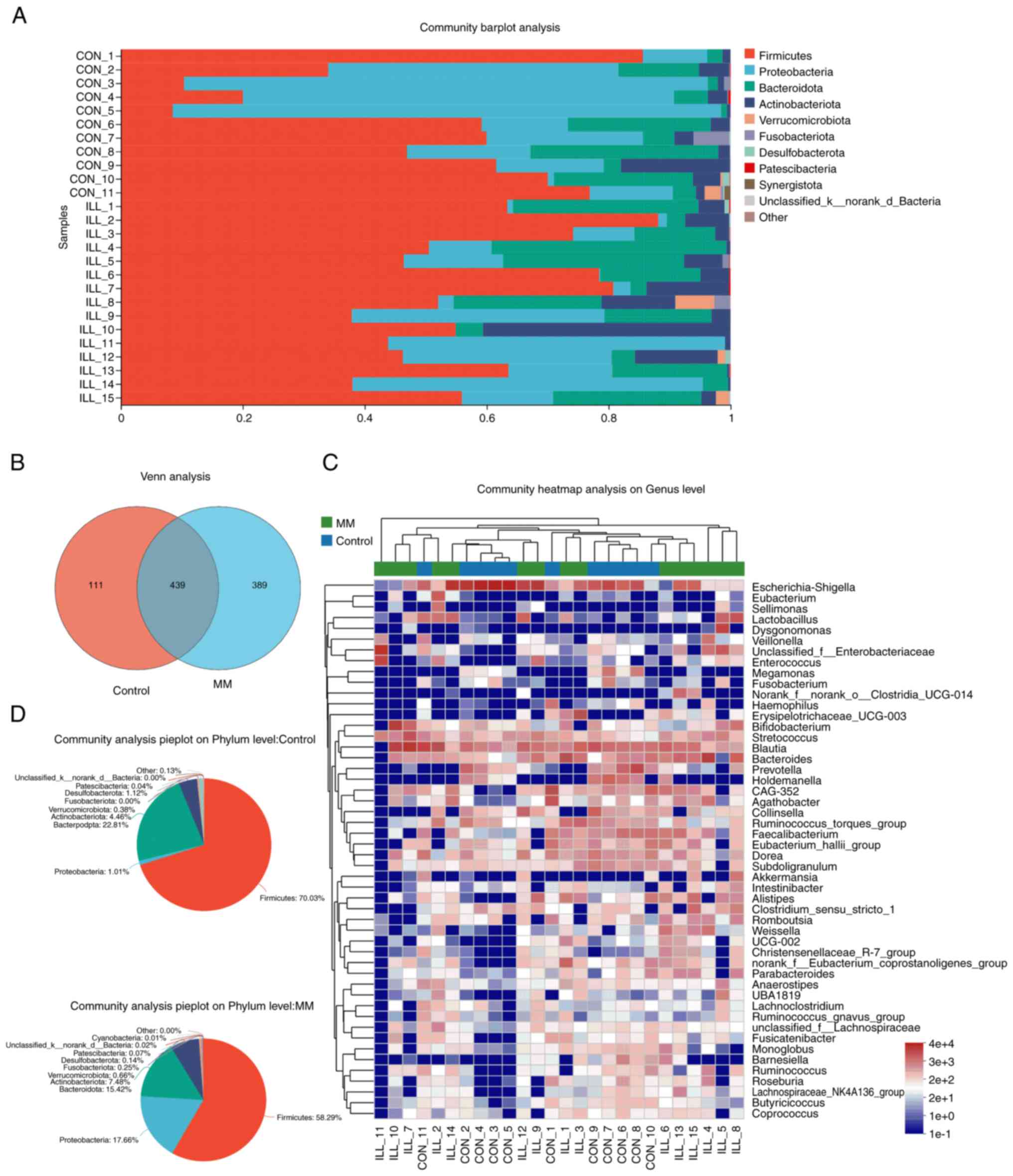
Microbial composition analysis
The representative sequences of the OTUs were
compared with the microbial reference database to obtain species
information. To facilitate the search for microbial disease
markers, a Venn analysis was performed to count the number of
species (such as OTUs) that were common and unique to the MM and
control groups. The results showed that 439 OTUs were shared
between groups and 389 OTUs were unique to MM (Fig. 2B). To understand the composition of
the microbial communities, those significantly enriched were
displayed through community bar plot analysis (Fig. 2A). At the phylum level, Firmicutes,
Proteobacteria, Bacteroides, Actinobacteria, Verrucomicrobiota,
Fusobacterota, Desulfobacterota, Patescibacteria and Synergistota
were abundant in both groups. Furthermore, pie plots were used to
show the proportion of the dominant species in the microbial
community (Fig. 2D). Community
heatmap analysis was used to display significantly enriched taxa at
the genus level, with Escherichia-Shigella, Blautia, Bacteroides,
Bifidobacterium and Streptococcus the most abundant (Fig. 2C). Next, significant group-specific
species were searched for.
Differential species analysis
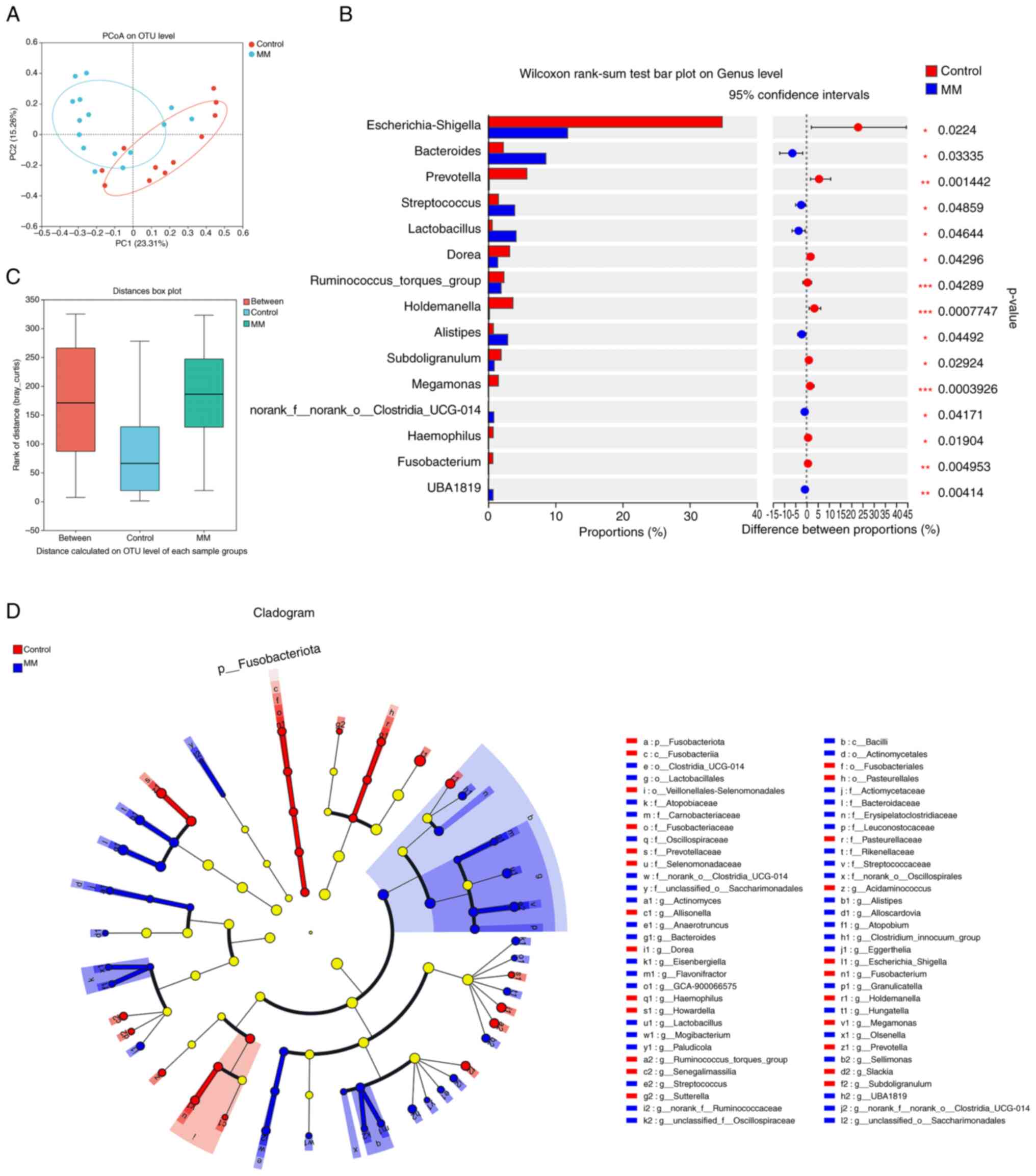
The present study performed beta diversity analysis
using PCoA to evaluate the differences between the MM and control
groups (Fig. 3A). PCoA is a
non-binding dimensionality reduction analysis method that can be
used to study similarities or differences in the composition of
sample communities. Each sample is represented as one point and the
closer the two points are, the more similar the species
composition. Moreover, the present study conducted an ANOSIM on the
grouped samples to test the significance of the differences between
the two groups. ANOSIM analysis showed a significant difference in
the microbial composition between the MM and control groups
(R=0.1446; P<0.05; Fig. 3C).
Intergroup differences at the genus level were then analyzed. LEfSe
analysis revealed significant differential bacteria from the phylum
to genus levels between the two groups (Fig. 3D). Furthermore, bacterial genera
with significant differences between the MM and control groups were
specifically identified at the genus level using the Wilcoxon
rank-sum test. A total of 15 bacterial genera were significantly
different between the two groups, among which Bacteroides,
Streptococcus, Lactobacillus and Alistipes were significantly
enriched in the MM group (Fig.
3B).
Correlation and functional prediction
analyses
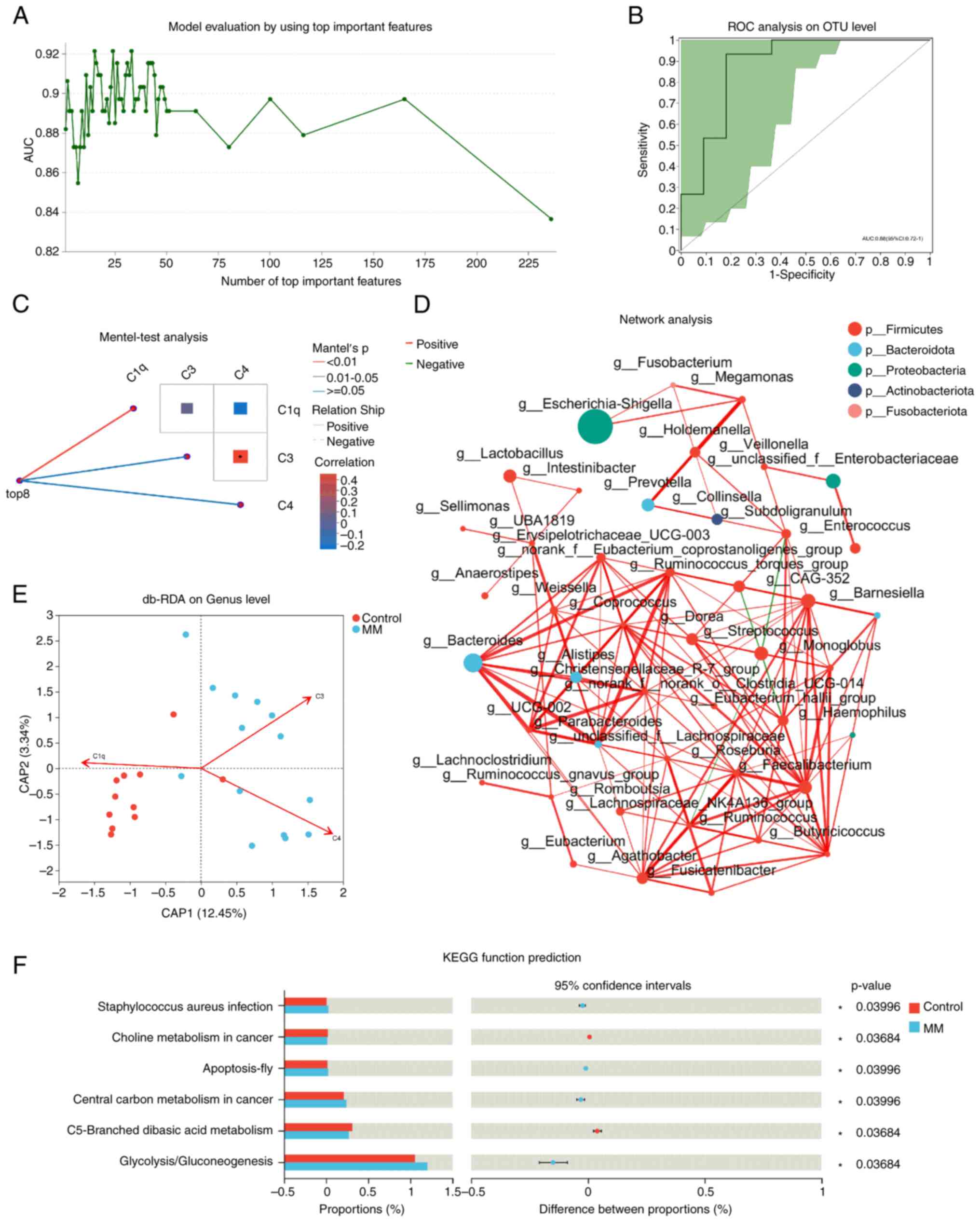
To investigate the role of different species in
disease diagnosis, Random Forest analysis was used to construct a
disease diagnostic model (Fig. 4A)
and then receiver operating characteristic (ROC) analysis was used
to verify the accuracy of the constructed model (Fig. 4B). The results showed an area under
curve (AUC) of 0.88 [95% confidence interval (CI): 0.72-1.00],
indicating that this diagnostic model was accurate. To understand
the effect of clinical indicators on the intestinal microbiota, The
Mantel test was used to verify the correlation between the
community distance matrix and the complement system. Correlation
analysis showed that the top eight differential species were
related to the content of serum complement C1q, while a significant
correlation was observed between C3 and C4, two clinical indicators
(Fig. 4C). Additionally, db-RDA
analysis showed a positive correlation between the levels of
complement C3 and C4 and intestinal microbiota species composition
in the MM group and a negative correlation with C1q, while the
control group showed the opposite result (Fig. 4E).
Single-factor correlation network analysis was
performed to determine the interactions between the species in the
sample. The 50 most abundant species were selected at the genus
level and the Spearman correlation coefficient (ρ) was calculated
to examine correlations among the species (r>0.6; P<0.05).
The size of the nodes represents species abundance, the red line
represents a positive correlation, the green line represents a
negative correlation and the thickness of the line represents the
size of the correlation coefficient (Fig. 4D). the KEGG database was used to
predict and compare the functional genes related to metabolic
pathways of microbial communities in different sample groups.
Compared with the control group, gene expression involved in
Staphylococcus aureus infection, apoptosis, central carbon
metabolism in cancer and glycolysis/gluconeogenesis were
significantly increased in the MM group, while those in choline
metabolism in cancer and C5 branched basic acid metabolism were
significantly reduced. The KEGG function prediction was performed
using PICRUSt2 software package that is based on 16S amplicon
sequencing results. Unlike metagenomic sequencing, KEGG function
prediction using PICRUSt2 does not have gene count thresholds
(Fig. 4F).
Discussion
Plasma cells are major participants in adaptive
immunity owing to their ability to produce immunoglobulins and
resist microbial infections (18).
When plasma cells mutate into clones, abnormal globulins
(monoclonal proteins) are produced and these immunogenic proteins
can drive the immune system to resist the host (19). Researchers from the University of
Oslo in Norway found that plasma cells, especially those lacking
CD19 and CD45 expression, can survive in the gut for decades
(12). The intestinal
microenvironment is rich in immune stimuli from normal flora and
external pathogens and long-term exposure to these antigens
increases the chance of genetic mutations that can cause clonal
plasma cell generation and plasma cell tumors such as multiple
myeloma (20). The present study
indicated that the occurrence of MM may be related to changes in
the intestinal microbiota and multiple correlation and prediction
analyses were conducted.
Monoclonal gammopathy of undetermined significance
(MGUS) is a precursor disease of MM (21). Pepeljugoski et al (22) performed the first intestinal
microbiota detection in patients with MGUS and MM. Compared with
healthy controls, Odoribacter and Lactobacillus were
the most enriched genera in patients with MM, whereas
Blautia and Faecalibacterium were the most reduced
genera. Compared with the MGUS group, Kluyvera and
Bacteroides had the highest abundance, whereas
Blautia and Parabacteroides had the lowest abundance
in the MM group. Moreover, the microbial diversity in patients with
MM and MGUS was higher than that in healthy individuals. Their
study confirmed the distinct differences between MM and its
precursor disease in the intestinal microbiota at the genus level,
suggesting that an imbalance in the intestinal microbiome may be
related to MM progression. However, a study by Zhang et al
(23) showed that the Shannon
index in patients with MM was lower compared with that in healthy
patients, indicating that the diversity of the intestinal
microflora in patients decreased. Compared with healthy controls,
the proportions of Bacteroides, Faecalibacterium and
Roseburia in MM increased at the genus level. Similarly, the
present study showed that Bacteroides was the most abundant
species in MM. Compared with patients with MM in plain areas,
Streptococcus, Lactobacillus and Streptomyces were
also significantly more abundant, but an increase in the proportion
of Faecalibacterium and Roseburia was not observed.
In addition, the alpha diversity of the intestinal microbiota in MM
patients living in high-altitude areas was higher compared with
that in the healthy control group, but the difference was not
significant. Random Forest analysis was also used to select
important microorganisms (biomarkers) to construct a disease
diagnostic model and validated the accuracy of the constructed
model using ROC analysis. The AUC was 0.88 (95% CI: 0.72-1.00),
indicating that this diagnostic model has a certain accuracy.
Regarding prognosis, the enrolled patients are being tracked. The
effect of differences in intestinal microbiota between MM patients
in high-altitude and plain areas on disease prognosis will be in
future research.
To explore the mechanism underlying the association
between intestinal microbiota and MM, Calcinotto et al
(24) used Vk*MYC mice to simulate
human MM. They found that Prevotella heparinolytica, a gut
commensal bacterium, can promote the differentiation of Th17 cells
that produce IL-17 and that Th17 cells can migrate to the bone
marrow to promote disease progression. This study suggests that
commensal bacteria in the gut unleash a paracrine signaling network
between innate and adaptive immunity that promotes the progression
of MM. The complement system is the major participator of adaptive
immunity and previous studies have shown differences in complement
levels between patients with MM and healthy populations (25-27).
Similarly, the clinical indicator correlation analysis of the
present study suggested that complement components are related to
the composition of the intestinal microbiota in patients with MM.
Jian et al (28) found that
the accumulation of urea nitrogen due to renal damage during MM
progression may result in the enrichment of nitrogen-cycling
bacteria such as Klebsiella and Streptococcus,
suggesting strong metabolic interactions between intestinal
bacteria and the host. Patients with MM achieve negative minimal
residual disease (MRD) after early treatment and often have
improved outcomes than those who remain as MRD+. Pianko
et al (29) observed a
higher relative abundance of Eubacterium hallii and
Faecalibacterium prausnitzii in 16 MRD- patients
compared with 18 MRD+ patients, which suggests that
there may be a relationship between intestinal microbiota
composition and treatment outcome in MM. Functionally, these two
bacteria produce short-chain fatty acids (mainly butyric acid) that
play anti-inflammatory roles. El Jurdi et al (30) found that the composition and
abundance of the gastrointestinal microbiome changed following HSCT
in patients with MM. Enrichment of Blautia and
Ruminococcus was associated with a higher incidence of
diarrhea, nausea and vomiting following transplantation. It is
noteworthy that these two bacteria are anaerobic and do not produce
butyric acid. Hu et al (31) showed significant temporal
differences in the diversity and abundance of
Bifidobacterium, Prevotella, Sutterella and
Collinsella between MM patients with complete or partial
remission after chimeric antigen receptor T cell therapy.
Metabolomic analysis showed that intermediates involved in multiple
amino acid metabolic pathways, such as choline, L-cysteine,
rosmarinic acid, L-phenylalanine and 2-phenylacetamide were
significantly enriched in patients in complete remission. The
present study predicted multiple metabolic pathways related to MM
through KEGG functional analysis, including not only central carbon
and choline metabolism in cancer, but also pathways such as dibasic
acid metabolism, apoptosis, infection and
glycolysis/gluconeogenesis. The present study further enriched the
signaling and metabolic pathways of the intestinal microbiota that
affect MM progression.
In recent years, the introduction of new drugs such
as proteasome inhibitors has extended the survival time of most
patients with MM, which remains incurable in most cases. However,
research on the pathogenesis of MM is limited. No substantive
breakthroughs in treatment methods have been achieved to date. The
present study conducted a diversity analysis and differential
bacterial screening of the intestinal microbiota of patients
through 16s rRNA high-throughput sequencing, which may aid in the
development of clinical microbial markers. Moreover, the present
study predicted the signaling and metabolic pathways of the
intestinal microbiota related to MM using functional analyses.
However, there are some shortcomings in the present
study. Due to the small number of enrolled patients, it did not
classify and compare MM patients of different subtypes, nor did it
analyze the relationship between different stages and prognosis in
MM. The present study only described the differences between MM
patients and healthy individuals and these deficiencies will
remedied in future work. Although the present study represented a
step forward in understanding the pathogenesis of MM, conclusive
evidence is lacking. Metagenomics and metabolomics research will be
conducted in the future to elucidate the molecular mechanisms by
which the intestinal microbiota affects the occurrence and
development of MM and to identify therapeutic targets for MM. With
an in-depth study of intestinal microbiota, the diagnosis and
treatment of MM should lead to substantive breakthroughs.
Acknowledgements
The authors would like to thank Majorbio Bio-Pharm
Technology Co. Ltd. (Shanghai, China) for their assistance in
bioinformatics analysis.
Funding
Funding: The present study was supported by the science and
technology plan of Heilongjiang Provincial Health Commission (grant
no. 20211111000006).
Availability of data and materials
The data generated in the present study may be found
in the NCBI Sequence Read Archive (SRA) database under accession
number (PRJNA1090483) or at the following URL: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1090483.
Authors' contributions
All authors contributed to the conception and design
of this study. Material preparations were performed by XuG and HJ.
Sample collection was performed by LS and YW. Related experiments
were performed by LD and XiG. Data collection and analysis were
performed by YK. The manuscript was written by XL. Funding and
supervision were conducted by HG. XL and HG confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First Hospital of Qiqihar (approval no.
2021-KY-007-04). All patients signed written informed consent in
accordance with the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
van de Donk NWCJ, Pawlyn C and Yong KL:
Multiple myeloma. Lancet. 397:410–427. 2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Pawlyn C and Morgan GJ: Evolutionary
biology of high-risk multiple myeloma. Nat Rev Cancer. 17:543–556.
2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Cowan AJ, Allen C, Barac A, Basaleem H,
Bensenor I, Curado MP, Foreman K, Gupta R, Harvey J, Hosgood HD, et
al: Global Burden of multiple myeloma. JAMA Oncol. 4:1221–1227.
2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Zhou L, Yu Q, Wei G, Wang L, Huang Y, Hu
K, Hu Y and Huang H: Measuring the global, regional, and national
burden of multiple myeloma from 1990 to 2019. BMC Cancer.
21(606)2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Moreau P, Kumar SK, San Miguel J, Davies
F, Zamagni E, Bahlis N, Ludwig H, Mikhael J, Terpos E, Schjesvold
F, et al: Treatment of relapsed and refractory multiple myeloma:
Recommendations from the international myeloma working group.
Lancet Oncol. 22:e105–e118. 2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Laubach J, Garderet L, Mahindra A, Gahrton
G, Caers J, Sezer O, Voorhees P, Leleu X, Johnsen HE, Streetly M,
et al: Management of relapsed multiple myeloma: Recommendations of
the international myeloma working group. Leukemia. 30:1005–1017.
2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Rooks MG and Garrett WS: Gut microbiota,
metabolites and host immunity. Nat Rev Immunol. 16:341–352.
2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Adak A and Khan MR: An insight into gut
microbiota and its functionalities. Cell Mol Life Sci. 76:473–493.
2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Jasiński M, Biliński J and Basak GW: The
role of the gut microbiome in pathogenesis, biology, and treatment
of plasma cell dyscrasias. Front Oncol. 11(741376)2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Cani PD: Human gut microbiome: Hopes,
threats and promises. Gut. 67:1716–1725. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
D'Angelo CR, Sudakaran S and Callander NS:
Clinical effects and applications of the gut microbiome in
hematologic malignancies. Cancer. 127:679–687. 2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Landsverk OJ, Snir O, Casado RB, Richter
L, Mold JE, Réu P, Horneland R, Paulsen V, Yaqub S, Aandahl EM, et
al: Antibody-secreting plasma cells persist for decades in human
intestine. J Exp Med. 214:309–317. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Stansborough RL and Gibson RJ: Proteasome
inhibitor-induced gastrointestinal toxicity. Curr Opin Support
Palliat Care. 11:133–137. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Peled JU, Devlin SM, Staffas A, Lumish M,
Khanin R, Littmann ER, Ling L, Kosuri S, Maloy M, Slingerland JB,
et al: Intestinal microbiota and relapse after hematopoietic-cell
transplantation. J Clin Oncol. 35:1650–1659. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kusakabe S, Fukushima K, Maeda T, Motooka
D, Nakamura S, Fujita J, Yokota T, Shibayama H, Oritani K and
Kanakura Y: Pre- and post-serial metagenomic analysis of gut
microbiota as a prognostic factor in patients undergoing
haematopoietic stem cell transplantation. Br J Haematol.
188:438–449. 2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
D'Angelo C, Sudakaran S, Asimakopoulos F,
Hematti P, El-Gamal D, Safdar N and Callander N: Perturbation of
the gut microbiome and association with outcomes following
autologous stem cell transplantation in patients with multiple
myeloma. Leuk Lymphoma. 64:87–97. 2023.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Rajkumar SV, Dimopoulos MA, Palumbo A,
Blade J, Merlini G, Mateos MV, Kumar S, Hillengass J, Kastritis E,
Richardson P, et al: International myeloma working group updated
criteria for the diagnosis of multiple myeloma. Lancet Oncol.
15:e538–e548. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Cancro MP and Tomayko MM: Memory B cells
and plasma cells: The differentiative continuum of humoral
immunity. Immunol Rev. 303:72–82. 2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Perini T, Materozzi M and Milan E: The
immunity-malignancy equilibrium in multiple myeloma: Lessons from
oncogenic events in plasma cells. FEBS J. 289:4383–4397.
2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Alkharabsheh O, Sidiqi MH, Aljama MA,
Gertz MA and Frankel AE: The human microbiota in multiple myeloma
and proteasome inhibitors. Acta Haematol. 143:118–123.
2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ghobrial IM, Detappe A, Anderson KC and
Steensma DP: The bone-marrow niche in MDS and MGUS: Implications
for AML and MM. Nat Rev Clin Oncol. 15:219–233. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Pepeljugoski AC, Morgan G and Braunstein
M: Analysis of intestinal microbiome in multiple myeloma reveals
progressive dysbiosis compared to MGUS and healthy individuals.
Blood. 134:3076. 2019.
|
|
23
|
Zhang B, Gu J, Liu J, Huang B and Li J:
Fecal microbiota taxonomic shifts in chinese multiple myeloma
patients analyzed by quantitative polimerase chain reaction (QPCR)
and 16S rRNA high-throughput sequencing. Med Sci Monit.
25:8269–8280. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Calcinotto A, Brevi A, Chesi M, Ferrarese
R, Garcia Perez L, Grioni M, Kumar S, Garbitt VM, Sharik ME,
Henderson KJ, et al: Microbiota-driven interleukin-17-producing
cells and eosinophils synergize to accelerate multiple myeloma
progression. Nat Commun. 9(4832)2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Yang R, Huang J, Ma H, Li S, Gao X, Liu Y,
Shen J and Liao A: Is complement C1q a potential marker for tumor
burden and immunodeficiency in multiple myeloma? Leukemia Lymphoma.
60:1812–1818. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang L, Ling X, Li F, Yang T, Shi K, Zhao
S, Yu L, Li Z and He H: Complement 4 aids in the prediction of
newly diagnosed multiple myeloma outcome in patients. Clin Med
Insights Oncol. 16(11795549221079171)2022.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Liu X, Zhou Z and Sun D: Values of
immunoglobulin and complements for evaluating treatment outcomes of
patients with multiple myeloma. Clin Lab Nov. 1(69)2023.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Jian X, Zhu Y, Ouyang J, Wang Y, Lei Q,
Xia J, Guan Y, Zhang J, Guo J, He Y, et al: Alterations of gut
microbiome accelerate multiple myeloma progression by increasing
the relative abundances of nitrogen-recycling bacteria. Microbiome.
8(74)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Pianko MJ, Devlin SM, Littmann ER,
Chansakul A, Mastey D, Salcedo M, Fontana E, Ling L, Tavitian E,
Slingerland JB, et al: Minimal residual disease negativity in
multiple myeloma is associated with intestinal microbiota
composition. Blood Adv. 3:2040–2044. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
El Jurdi N, Filali-Mouhim A, Salem I,
Retuerto M, Dambrosio NM, Baer L, Lazarus HM, Caimi P, Cooper B,
Tomlinson B, et al: Gastrointestinal microbiome and mycobiome
changes during autologous transplantation for multiple myeloma:
Results of a prospective pilot study. Biol Blood Marrow Transplant.
25:1511–1519. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Hu Y, Li J, Ni F, Yang Z, Gui X, Bao Z,
Zhao H, Wei G, Wang Y, Zhang M, et al: CAR-T cell therapy-related
cytokine release syndrome and therapeutic response is modulated by
the gut microbiome in hematologic malignancies. Nat Commun.
13(5313)2022.PubMed/NCBI View Article : Google Scholar
|