Introduction
Kidney cancer is a malignant cancer. According to a
recent report, there were predicted to be ~14,890 cases of kidney
cancer-associated mortalities in 2023(1). Renal cell carcinoma (RCC) originates
from renal tubular epithelial cells and accounts for >90% of
kidney cancer cases (2). Due to
genetic heterogeneity, different genetic factors dominate in
different cohorts of patients with RCC, resulting in difficulty
when administering effective therapies (3). For example, different expression of
CB2R could be used as a prognostic marker in RCC (4). Although patients with localized or
regional kidney cancer show a good five-year survival rate, the
5-year survival rate of those with metastasis remains at ~15%
(1). Additionally, ~20-30%
patients with RCC already present with metastasis at the time of
diagnosis (2). Therefore, it is of
importance to understand the intrinsic mechanisms underlying RCC
physiology for developing novel therapeutic strategies to treat
RCC.
Inflammation is associated with cancer. In an
inflammatory environment, normal cells can undergo malignant
transformation, where aberrant events may occur, ultimately leading
to the development of cancer (5).
Therefore, inflammation may be a promising target for cancer
therapy (6). Inhibition of
tumor-associated macrophages, such as M2-like macrophages, has been
reported to improve antitumor immune responses in solid cancer
(7). Inter-α-trypsin inhibitor
heavy chain 1 (ITIH1) is a gene located in chromosome 3p21.1
that encodes a preproprotein of the heavy chain of the
inter-α-trypsin inhibitor complex, which is involved in inhibition
of inflammatory diseases, such as sepsis (8). ITIH genes appear to serve a
tumor suppressor role that frequently exhibit decreased expression
in human cancers (9). Previous
studies have reported that the reduced expression of ITIH1 can
promote liver cancer progression, suggesting ITIH1 to be a
prognostic or diagnostic indicator of this malignancy (10,11).
In addition, Kopylov et al (12) reported that downregulation of ITIH1
expression was associated with the initial development of
colorectal cancer. However, the role of ITIH1 in the initiation or
progression of RCC remains unclear.
The hypo- or hyperactivation of certain growth or
death signaling pathways can occur in cancer (13). Of these pathways, hyperactivation
of the NF-κB signaling pathway is commonly observed in cancer
(14). In gastrointestinal cancer,
activation of NF-κB signaling has been reported to promote cancer
initiation and development (14).
In RCC, inhibition of NF-κB signaling has been reported to
sensitize cancer cells to tyrosine kinase inhibitors (15). Furthermore, NF-κB signaling was
considered a promising target for cancer therapy according to a
report by Yu et al (16).
However, it remains unclear if there is a relationship between
ITIH1 and NF-κB signaling in RCC.
To investigate the role and mechanism of ITIH1 in
RCC, the present study analyzed the effects of ITIH1 knockdown in
RCC cell lines and measured the protein expression levels of
downstream signaling molecules following ITIH1 knockdown. In
addition, experiments after ITIH1 overexpression were performed to
further investigate the signaling pathways regulated by ITIH1.
Materials and methods
Bioinformatics analysis
The mRNA expression profiles [transcripts per
million (TPM)] of the ITIH1 gene in clear cell renal cancer
tissues (n=110) and matched normal tissues (n=84) were obtained
from The Cancer Genome Atlas (TCGA) database (https://ualcan.path.uab.edu/analysis.html) (17). The survival rate of patients with
renal cancer was also extracted based on the clinical data in the
TCGA database from Ualcan. Briefly, we input the name of gene ITIH1
to the box ‘Enter gene symbol’ on the web and selected the type of
cancer ‘Kidney renal clear cell carcinoma’ in the box ‘TCGA
dataset’. The ‘Explore’ button was pressed and the website
(https://ualcan.path.uab.edu/cgi-bin/ualcan-res.pl)
appeared. Then the ‘Expression’ button was clicked and the
expression of ITIH1 gene was returned. ‘Survival’ was selected and
the association of ITIH1 expression with patients' survival
probability was returned. For survival analysis, the samples were
grouped as below: High expression (with TPM values above upper
quartile) and low/medium expression (with TPM values below upper
quartile) (17).
Cell culture
The A498, 786-O and ACHN RCC cell lines were
purchased from Shanghai Fuheng Biotechnology Co., Ltd. and cultured
in RPMI 1640 medium (Nanjing KeyGen Biotech Co., Ltd.) with 10% FBS
(Gibco; Thermo Fisher Scientific) at 37˚C under an atmosphere with
5% CO2. HK-2 cells were purchased from Cellverse
Bioscience Technology Co., Ltd. and cultured in DMEM (Nanjing
KeyGen Biotech Co., Ltd.) with 10% FBS.
Reverse transcription-quantitative
PCR
Total RNA was extracted from RCC cells (HK-2, A498,
786-O and ACHN) using the RNAeasy Kit (Beyotime Institute of
Biotechnology). Total RNA (0.1-1.0 µg) was used as a template to
transcribe the first strand of double-stranded complementary (c)
DNA with BeyoRT III cDNA kit (cat. no. D7178M; Beyotime Institute
of Biotechnology). The protocol for reverse transcription reaction
is as follows: 25˚C, 10 min; 40˚C, 50 min; and 80˚C, 10 min.
Subsequently, 1 µl the cDNA was used for the quantification of the
expression of genes of interest using the BeyoFast SYBR Green qPCR
Mix (Beyotime Institute of Biotechnology). The protocol is as
follows: 95˚C, 5 min; 95˚C, 15 sec; 60˚C, 20 sec for 40 cycles.
GAPDH was used as the internal reference control. The relative mRNA
expression levels of the ITIH1 gene were calculated using the
2-IICq method (18).
The primers used were as follows: ITIH1 forward (F),
5'-CTGCAGGGTTTCTACAGCCA-3' and reverse (R),
5'-CGCTCTCGGAGCAGTTTCTT-3'; and GAPDH F, 5'-GATGCTGGCGCTGAGTACG-3'
and R, 5'-GTTCACACCCATGACGA-3'.
Synthesis of small interfering (si)RNA
targeting ITIH1
SiRNAs targeting ITIH1 (siITIH1) or negative control
(NC) were synthesized by Beijing Tsingke Biotech Co., Ltd. Briefly,
siRNA (50 pmol) was mixed with Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) in 100 µl RPMI 1640 medium (Nanjing
KeyGen Biotech Co., Ltd.) with 10% FBS and incubated for 30 min at
room temperature. Subsequently, the mixture was delivered into the
RCC cells (A498 and ACHN) and incubated at 37˚C for 4 h. The
supernatant was then removed and substituted with fresh RPMI 1640
medium (Nanjing KeyGen Biotech Co., Ltd.) and incubated for 48 h at
37˚C before subsequent experimentation. The sequences of siITIH1
and NC used were as follows: si-#1 sense,
5'-UUAUGUCUCCGAUAAAUGCGU-3' and antisense,
5'-GCAUUUAUCGGAGACAUAAAG-3'; si-#2 sense,
5'-AACAUGAUGAGUAUUGAGGCA-3' and antisense,
5'-CCUCAAUACUCAUCAUGUUGA-3'; and NC sense,
5'-UUCUCCGTACGUGUCACGUTT-3' and antisense,
5'-ACGUGACACGUACGGAGAATT-3'.
Construction of recombinant plasmid
overexpressing ITIH1
The coding sequence of the ITIH1 gene
(accession no. NM_001166434.3) was chemically synthesized and
subcloned into the pcDNA3.1 (General Biologicals Company) plasmid.
The recombinant plasmid pcDNA-ITIH1 (~2 µg) was transferred into
A498 and ACHN cells as aforementioned. The empty vector pcDNA3.1
was used as control.
Western blotting assay
The total proteins were extracted from A498 cells
treated with siITIH1 or NC for 48 h using the Nuclear and
Cytoplasmic Protein Extract kit (Beyotime Institute of
Biotechnology) and quantified on the UV Spectrophotometer (Hangzhou
Lifereal Biotechnology Co., Ltd.). Total protein (~10 µg) was
separated by 15% SDS-PAGE (Shanghai Yeasen Biotechnology Co.,
Ltd.), transferred onto PVDF membranes (Shanghai Yeasen
Biotechnology Co., Ltd.) and blocked with 5% non-fat milk for 1 h
at room temperature. Subsequently, the PVDF membranes were
incubated with primary antibodies overnight at 4˚C, washed with
TBST containing 0.5% Tween 20 (Tanon Science and Technology Co.,
Ltd.) three times and incubated with HRP-conjugated secondary
antibodies for 1 h at room temperature. After the PVDF membranes
were washed with PBST three times, they were visualized with an
enhanced ECL chemiluminescent kit (Shanghai Yeasen Biotechnology
Co., Ltd.) and analyzed by ImageJ software (version 1.51j8;
National Institutes of Health). The following antibodies were used
in the present study: ITIH1 (1:2,000; cat. no. ab233032; Abcam),
phosphorylated (p-) NF-κB (1:1,000; cat. no. ab194729; Abcam),
NF-κB (1:1,000; cat. no. AG3101; Beyotime Institute of
Biotechnology), IκB (1:2,000; cat. no. AG2737; Beyotime Institute
of Biotechnology), IKK (1:1,000; cat. no. AF0198; Beyotime
Institute of Biotechnology), cyclinD1 (1:1,000; cat. no. AF0126;
Beyotime Institute of Biotechnology), proliferating cell nuclear
antigen (PCNA; 1:1,000; cat. no. AG8029; Beyotime Institute of
Biotechnology), GAPDH (1:2,000; cat. no. ab9485; Abcam), α-SMA
(1:3,000; cat. no. AF1507; Beyotime Institute of Biotechnology) and
goat anti-rabbit HRP-conjugated IGG antibodies (1:20,000; cat. no.
ab6721; Abcam).
Cell Counting Kit-8 assay
RCC cells (A498 and ACHN) were seeded into 96-well
plates (3-5x103 cells/well) and transfected with siITIH1
or NC. After the cells were cultured for 24, 48, 72 and 96 h at
37˚C,10% CCK-8 reagent (Beyotime Institute of Biotechnology) was
added to each well and incubated for 1 h at 37˚C. The absorbance
value at 450 nm of each well was measured using a microplate-reader
(Hangzhou Allsheng Instruments Co., Ltd.). For rescue experiments,
the NF-κB signaling pathway inhibitor JSH-23 (cat. no. HY-13982;
MedChemExpress) was used for 48 h at 37˚C after cells were treated
with siITIH1 at 10 µM for 24 h.
Cell migration and invasion
assays
RCC cells (A498 and ACHN) were seeded into Transwell
inserts (8-µm pore size; Wuxi NEST Biotechnology Co., Ltd.) at
2-3x104 cells/well, placed into 24-well plates and
transfected with siITIH1 or NC. In the upper chamber, culture
medium without serum was added and culture medium with 10% fetal
bovine serum was added to the bottom of the inserts. After
incubating for 48 h at 37˚C, the inserts were removed, the cells on
the upper surface were scraped away whereas cells on the lower
surface were fixed using 4% formaldehyde for 10 min at room
temperature and stained with 0.1% crystal violet dye (Beyotime
Institute of Biotechnology) for 10 min at room temperature. After
washing with PBS three times, the inserts were imaged using a light
microscope and the number of positively stained cells at three
fields of view were manually calculated as a percentage of total
cells.
For the cell invasion assay, the Transwell inserts
were pre-treated with 20 µl Matrigel (1 mg/ml per chamber; Corning,
Inc.) overnight at 4˚C before being treated as aforementioned.
Apoptosis assay using flow
cytometry
RCC cells (A498 and ACHN) were seeded into 6-well
plates (1x104 cells/well) and transfected with siITIH1
or NC. After incubation for 48 h at 37˚C, the cells were collected
at 250 x g for 5 min at room temperature and a total of
6x104 cells were stained with 100 µl dye buffer
containing 5 µl Annexin V/FITC-A reagent (Shanghai Yeasen
Biotechnology Co., Ltd.) for 15 min at room temperature in the
dark. Stained cells were washed with cold PBS before the positively
stained cells were detected using a flow cytometer equipment
(FACSCelesta; BD Biosciences) and analyzed by Flowjo V10.7.1 (BD
Biosciences).
Statistical analysis
Data were analyzed using SPSS software (version
16.0; SPSS, Inc.) and were presented as the mean ± standard
deviation (n=3). Differences between two groups were analyzed using
the unpaired Student's t-test, whilst differences among >2
groups were analyzed by one-way ANOVA followed by Tukey's post hoc
test. Kaplan-Meier survival curve analysis was used to analyze
survival data followed by log-rank test. P<0.05 was considered
to indicate a statistically significant difference.
Results
Clinical significance of ITIH1 in
renal cancer
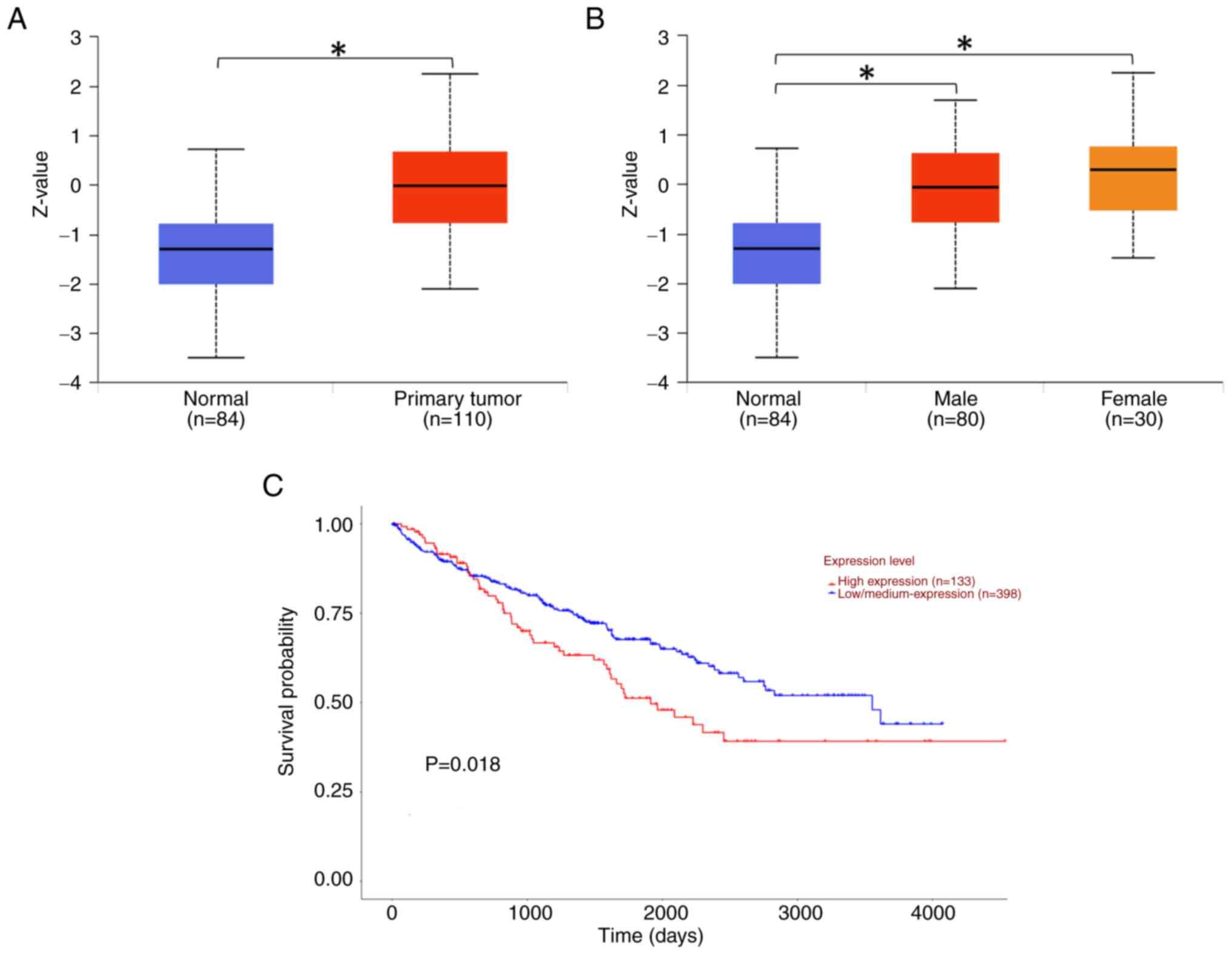
According to the data obtained from the TCGA
database, the protein expression level of ITIH1 in primary RCC
tissues (n=110) was significantly higher compared with that in the
normal tissues (n=84) (Fig. 1A).
The expression levels of ITIH1 in female (n=30) and male (n=80)
patients with RCC was significantly higher compared with that in
normal tissues (n=84) (Fig. 1B).
Moreover, patients with high expression levels of ITIH1 (n=133)
exhibited a significantly decreased 5-year survival rate compared
with those with low/medium expression levels of ITIH1 (n=398;
Fig. 1C). However, the mRNA and
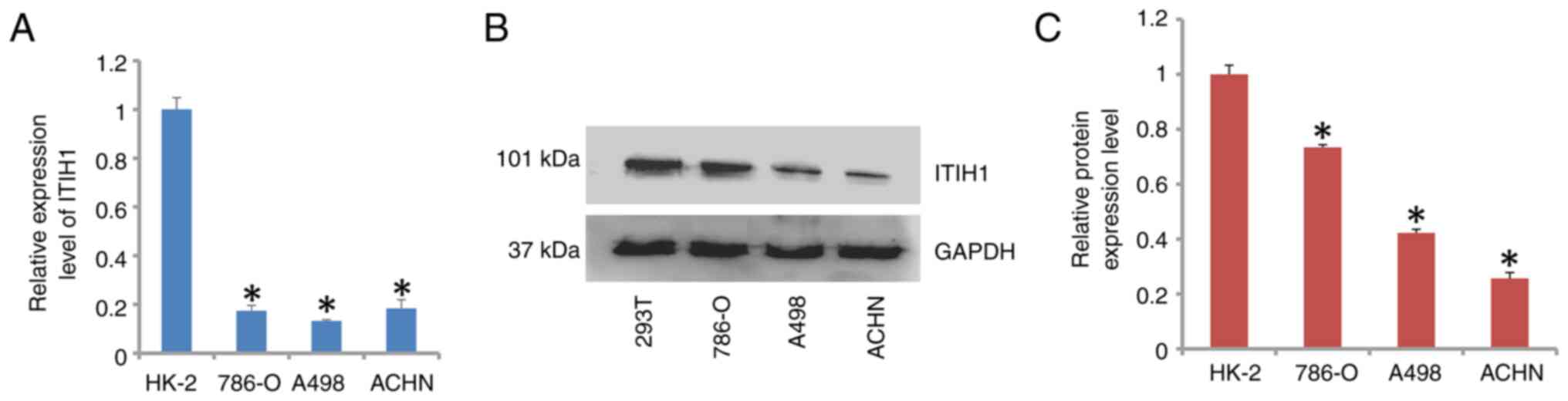
protein expression levels of ITIH1 in the ACHN, A498 and 786-O RCC
cell lines were significantly lower compared with that in the
control HK-2 cells (Fig. 2A-C).
Therefore, the role of ITIH1 in renal cancer requires further
investigation.
ITIH1 contributes to the inhibition of
renal cancer cell proliferation
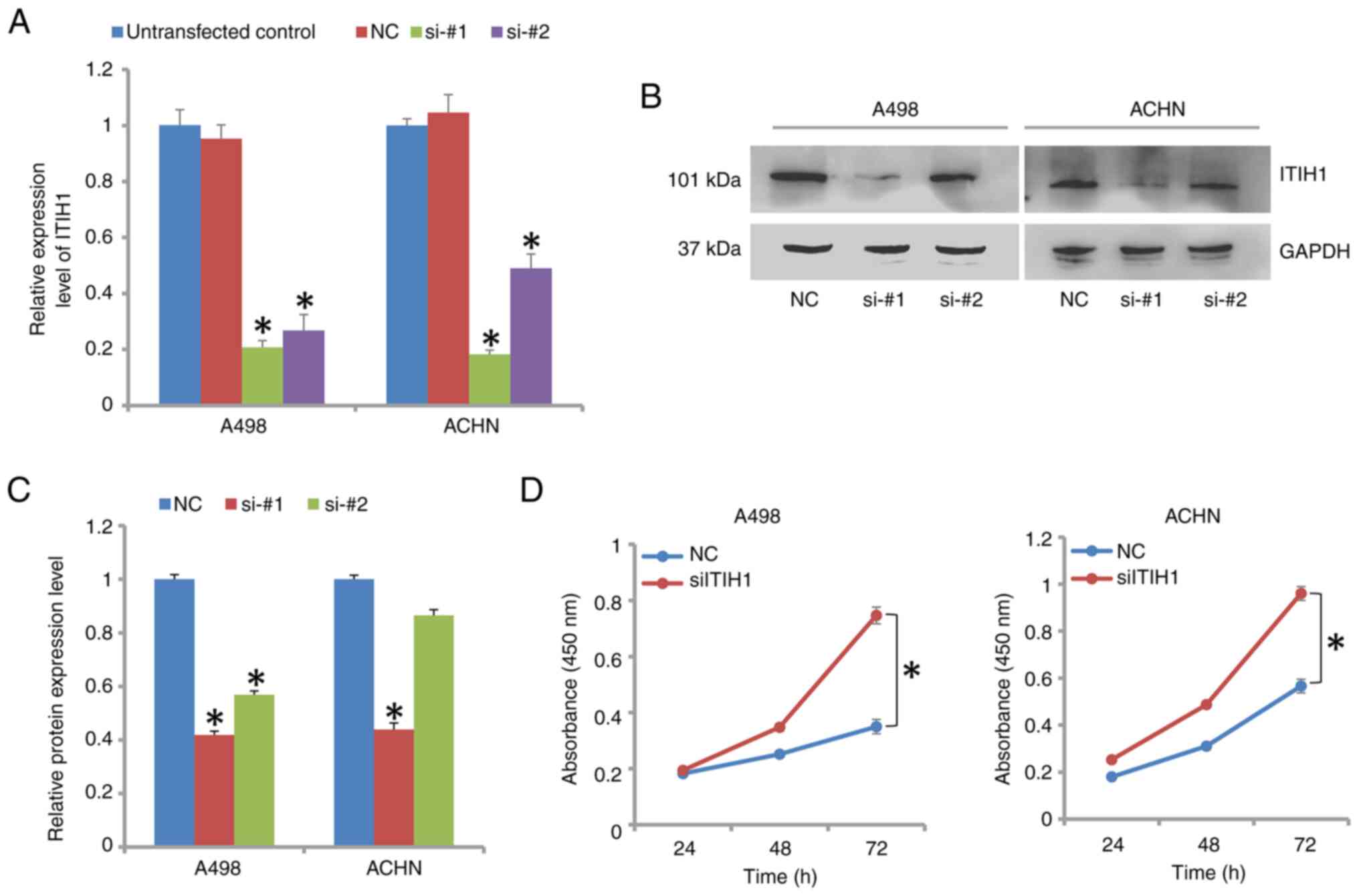
The mRNA and protein expression levels of ITIH1 in
A498 and ACHN cells were found to be significantly decreased
following transfection with siRNAs targeting ITIH1 (Fig. 3A-C). The knockdown efficiency of
si-#1 in both cell lines was >70% at mRNA level. On protein
level, si-#1 was also more effective at decreasing the protein
expression levels of ITIH1 compared with si-#2. Therefore,
subsequent transfection experiments used si-#1 (siITIH1). Cells
transfected with siITIH1 exhibited significantly increased
proliferative capabilities compared with those by NC cells
(Fig. 3D). In 786-O cells,
knockdown of ITIH1 expression also significantly increased
proliferation (Fig. S1A and
B). By contrast, overexpression
of ITIH1 significantly decreased the proliferation of A498, ACHN
and 786-O cells (Fig. S2A and
B). Therefore, it could be
suggested that ITIH1 negatively regulated proliferation of RCC
cells.
ITIH1 negatively regulates cell
migration and invasion in renal cancer cells
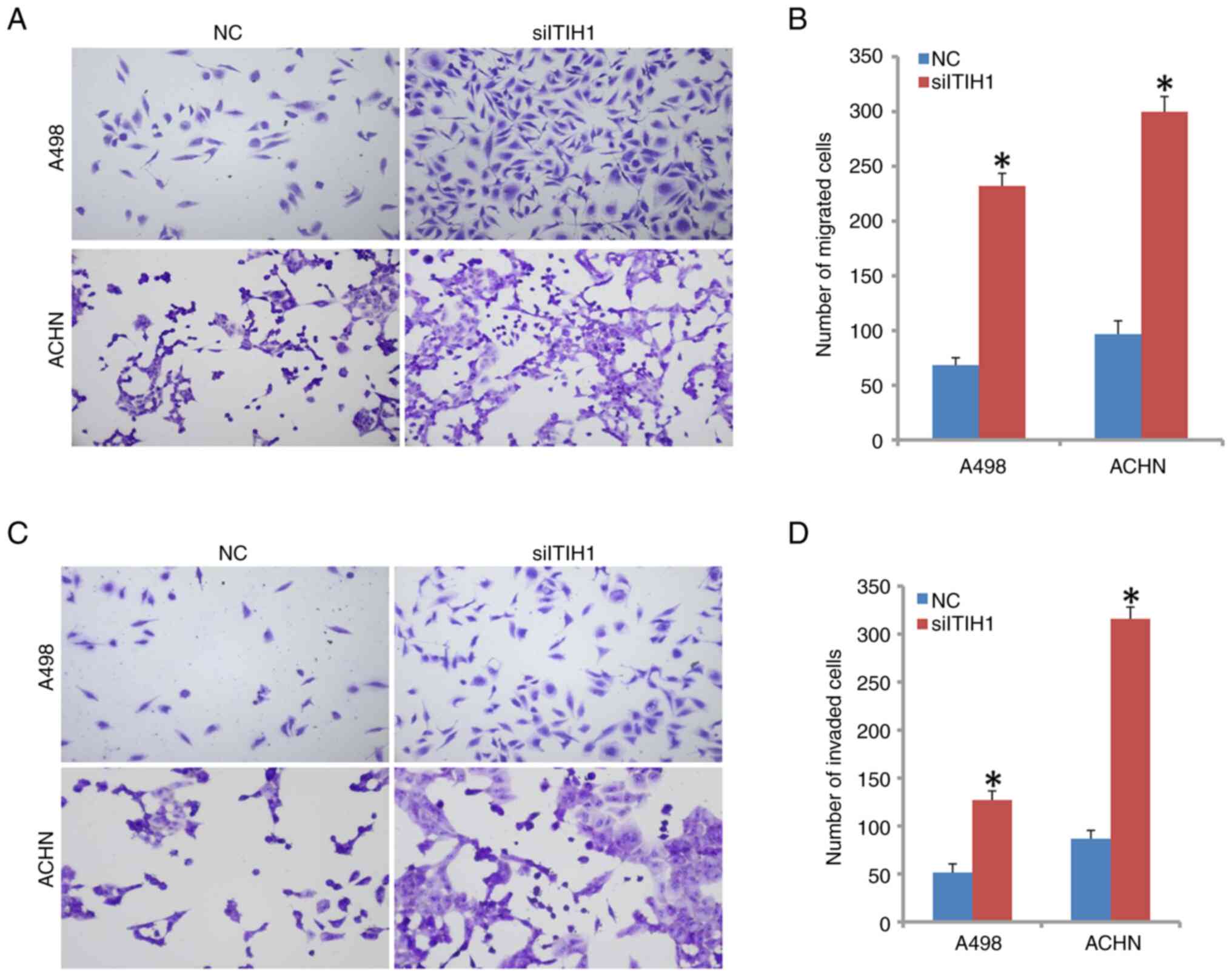
Significantly increased cell migratory ability was
observed in siITIH1-transfected A498 and ACHN cells compared with
that in NC cells (Fig. 4A and
B). The number of successfully
invading cells in the siITIH1 group was also found to be
significantly higher compared with that in the NC group in A498,
ACHN and 786-O cells (Figs. 4C,
D, S1C and D). However, the number of invading cells
was significantly reduced in A498, ACHN and 786-O cells following
the overexpression of ITIH1 compared with the vector group
(Fig. S2C and D). These findings suggest that ITIH1 can
suppress migration and invasion by renal cancer cells.
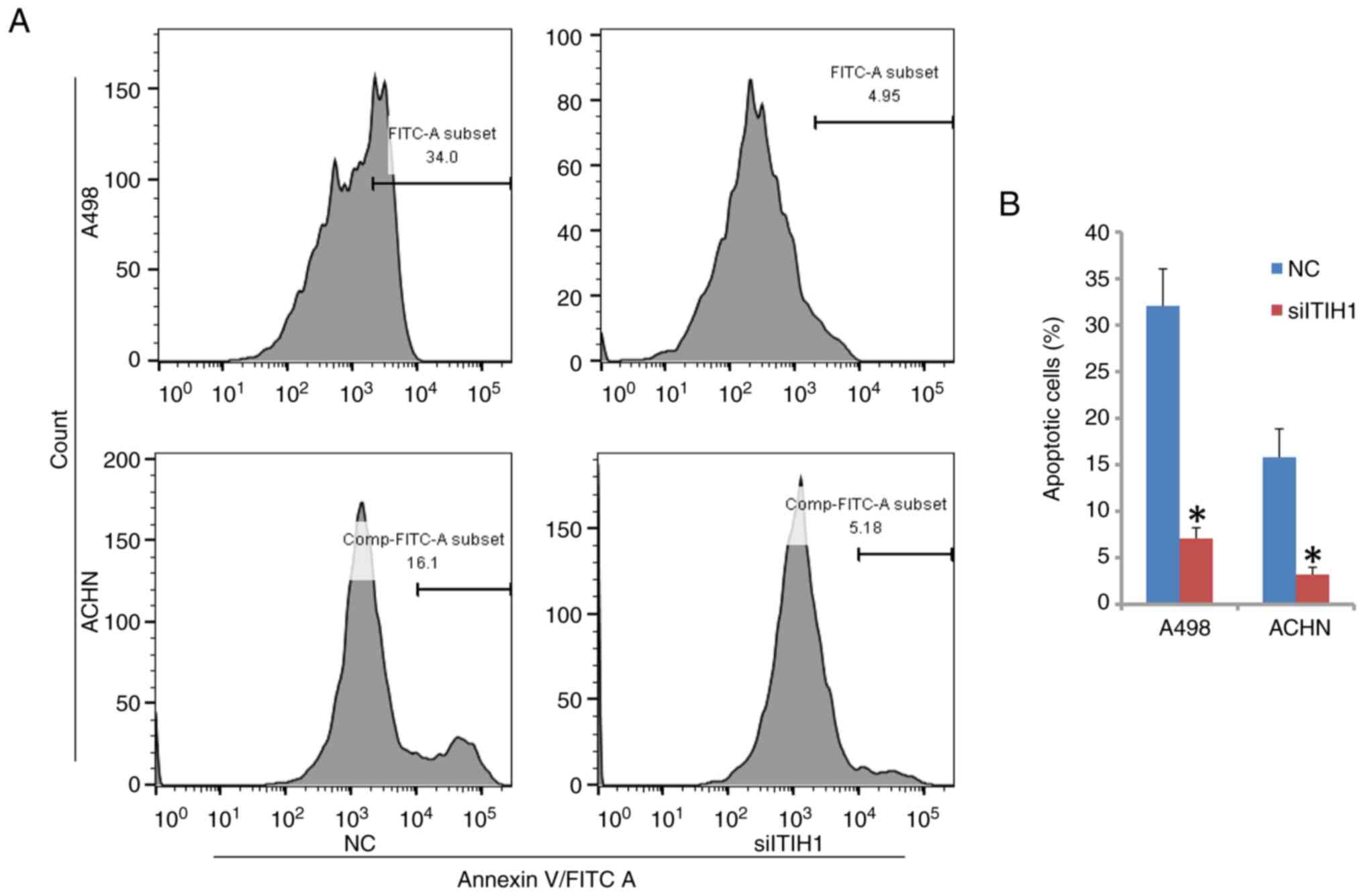
ITIH1 enhances the apoptosis of renal
cancer cells
The percentage of apoptotic cells of A498 and ACHN
was found to be significantly reduced when ITIH1 expression was
knocked down compared with that in NC cells (Fig. 5A and B). The percentage of apoptotic cells in
A498 cells in the NC group was 32.07%, whereas it was 7.08% in the
siITIH1 group. In ACHN cells, the percentage of apoptotic cells in
the NC group was 15.81%, whilst it was 3.20% in the siITIH1 group.
Therefore, these results suggest that ITIH1 can promote apoptosis
in renal cancer cells.
ITIH1 regulates the NF-κB signaling
pathway
NF-κB signaling was activated when ITIH1 expression
was knocked down in A498 cells (Fig.
6A and B). Specifically, the
phosphorylation level of NF-κB was observed to be significantly
increased in the siITIH1 group compared with that in the NC group
(Fig. 6C). Furthermore, the
protein expression level of IκB was downregulated. By contrast, the
protein expression level of IKK, the negative regulator of IκB
(19), was upregulated.
Additionally, the protein expression levels of proliferation
markers cyclin D1 and PCNA, in addition to those of the migration
marker α-smooth muscle actin (α-SMA), were significantly increased
in the siITIH1 group compared with those in the NC group (Fig. 6A and B).
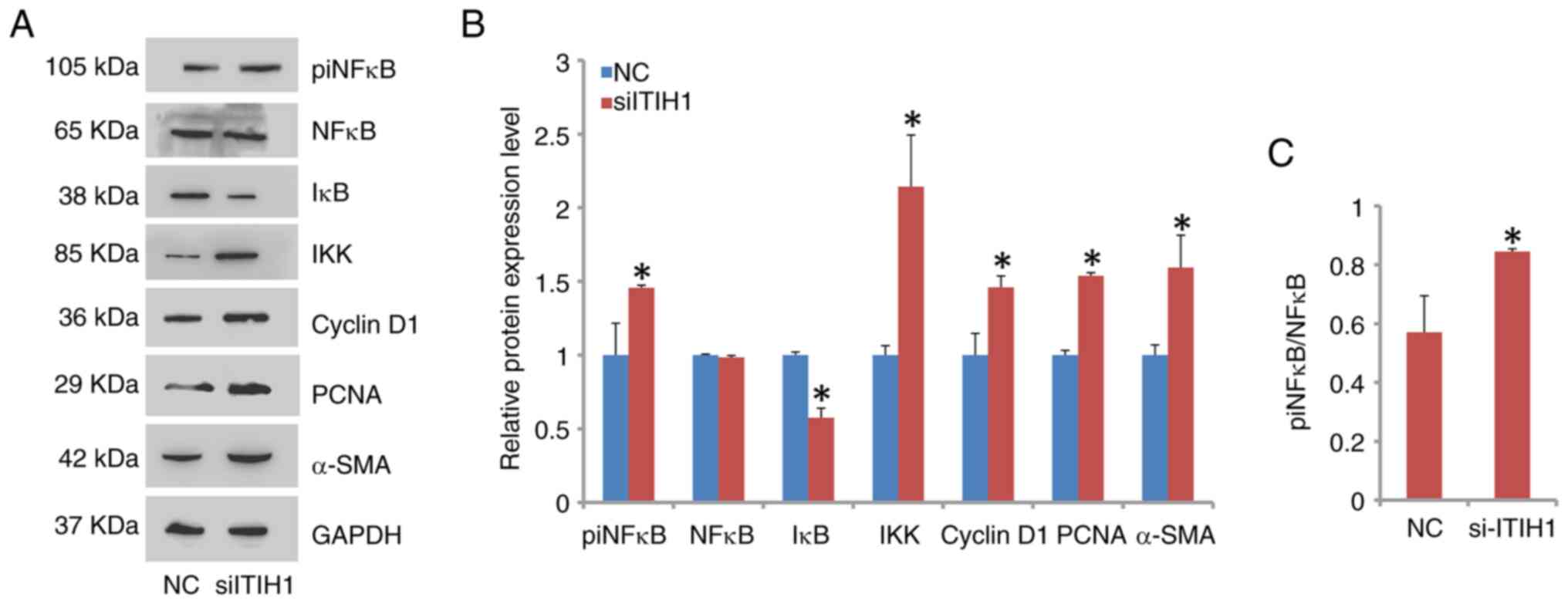 | Figure 6NF-κB pathway is activated by ITIH1
knockdown. (A) Western blotting analysis of the knockdown of
siITHI1 on the expression of components in the NF-κB pathway,
showing that the protein levels of (B) piNF-κB, IKK, Cyclin D1,
PCNA and α-SMA were significantly increased whereas those of IκB
were significantly decreased compared with NC cells. (C) A
significant increase in the phosphorylation of NF-κB was
demonstrated in siITIH1 cells compared with that in NC cells.
*P<0.05 vs. NC. ITIH1, inter-α-trypsin inhibitor
heavy chain 1; si, small interfering RNA; NC, negative control; pi,
phosphorylated; PCNA, proliferating cell nuclear antigen; α-SMA,
α-smooth muscle actin. |
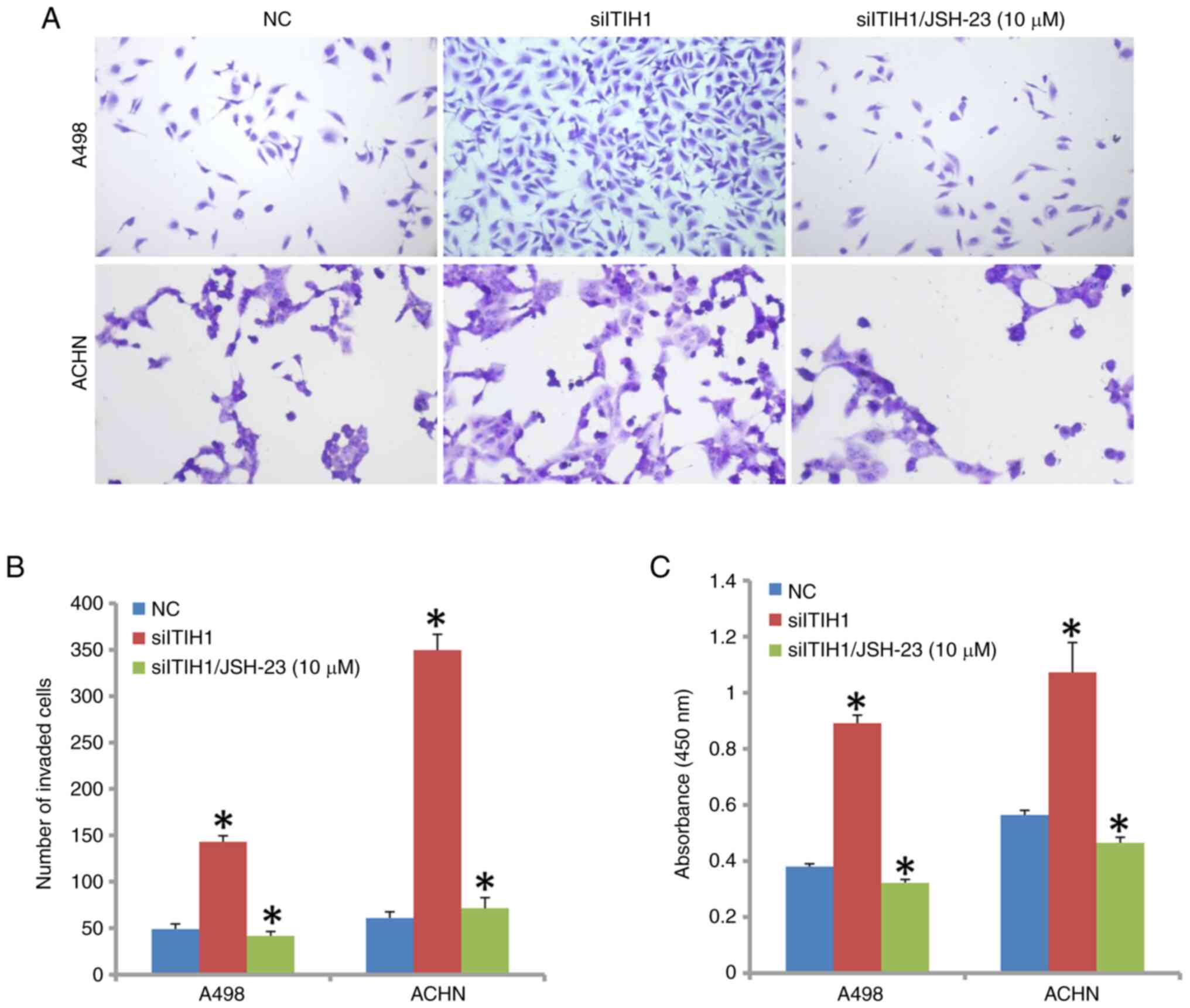
Treatment of cells with JSH-23, an inhibitor of
NF-κB signaling, in addition to siITIH1 transfection, significantly
decreased A498 and ACHN cell invasion ability compared with that in
the siITIH1 transfection-only group (Fig. 7A and B). In addition, A498 and ACHN cell
proliferation was also significantly inhibited when JSH-23 was used
in combination with siITIH1 compared with that in the siITIH1
transfection-only group (Fig.
7C).
Discussion
Renal cell carcinoma is a major subtype of kidney
cancer (1). Treating RCC
effectively remains a challenge at present, particularly for
patients with distant metastasis (2). The survival and quality of life of
patients with RCC has been improved in recent years due to medical
improvements. However, there are currently few successful treatment
options for patients with late-stage disease (20). An obstacle for the complete
remission of such patients is that RCC is a highly heterogeneous
cancer and the mechanisms underlying its occurrence remain unknown.
Therefore, it is of importance to determine the mechanism driving
the initiation and progression of RCC.
TCGA database has been a useful public resource for
cancer research over the past decade (21). It provides data from patients with
various types of cancer. In particular, the genetic information
available in TGCA in each cancer, such as gene expression changes
and alterations, have already been explored, making this database a
valuable asset for drug development and gene therapy research.
Based on the data from TCGA, the present study demonstrated that
the expression level of ITIH1 was significantly higher in RCC tumor
tissues compared with that in normal tissues. However, in RCC cell
lines, ITIH1 exhibited lower expression levels compared with that
in the HK-2 control cell line. This expression pattern was the
opposite of that demonstrated by data obtained from the TCGA. This
may be due to the selected cases of clinical specimens or the small
sample size used. The ITIH protein family has been previously
reported to be responsible for extracellular matrix stability, the
expression of which was frequently lost in certain types of solid
tumors, such as breast cancer (9).
Additionally, Hamm et al (9) previously showed that ITIH1 expression
was lost in kidney cancer. In a number of types of liver and
colorectal cancers, ITIH1 was reported to serve a suppressor role
in tumor progression and was associated with good clinical
prognosis (11,12). In RCC cell lines, it was
demonstrated that ITIH1 was significantly downregulated compared
with HK-2 cells, which was consistent with the findings reported by
Hamm et al (9).
Furthermore, based on the TCGA database, patients with RCC with
high ITIH1 expression exhibited a poorer survival rate. This is
opposite to the role of ITIH1 in HCC (11). Therefore, the role of ITIH1 in the
development of RCC requires further exploration.
ITIH1 knockdown in RCC cells was found to
significantly increase the proliferation of cells compared with
that in the NC group. Additionally, both the migratory and invasion
capabilities of RCC cells were significantly increased by ITIH1
knockdown compared with those by NC cells. Tumors can be
characterized by aggressive proliferation and expansion (22). Tumor cells will typically lose the
contact inhibition feature exhibited by normal cells and
proliferate without inhibitions as long as the energy supply is
sufficient, which results in the formation of solid tumor spheres
(23). In addition, cancer cells
can secrete MMPs, which digest the extracellular matrix, causing
the leakage of tumor cells (24).
This allows cancer cells to invade the surrounding tissues to form
new tumor foci (25). In clinical
studies, drugs targeting cell proliferation or metastasis have been
reported to be efficient in controlling the cancer progression. AKT
inhibitors have been documented to potently inhibit cell
proliferation and survival in cancer, such as breast cancer and
ovarian cancer (26). Furthermore,
Traditional Chinese Medicine has been found to effectively suppress
cell invasion and migration in certain types of cancers, such as
lung cancer and gastric cancer (27). In the present study, ITIH1
knockdown increased cell proliferation and invasion in
vitro. Therefore, ITIH1 may serve an important role in the
progression of RCC. Inducing apoptosis is a main aim of therapy
targeting cancer cells (28).
Cancer cells prevent apoptosis by changing the expression levels of
critical molecules, such as caspase-3 in the caspase cascade
response signaling pathway (22).
The present study demonstrated that ITIH1 expression induced
apoptosis in RCC cells, suggesting that ITIH1 is a critical
molecule in mediating apoptosis of RCC cells.
NF-κB signaling is important for normal embryonic
development and physiological activities, such as the inflammatory
response (29). However, aberrant
activation of NF-κB signaling has been reported in a number of
diseases, including certain types of cancers. NF-κB signaling was
found to be increased in cancers, such as lung cancer, where
targeting the NF-κB pathway may serve as a promising method for
therapy (16). Yan et al
(30) previously reported that
activation of NF-κB by S1P promoted RCC progression. In the present
study, it was demonstrated that ITIH1 was a negative regulator of
the NF-κB pathway, since this pathway was activated when ITIH1
expression was knocked down in RCC cells. The protein expression
levels of proliferative markers Cyclin D1 and PCNA were also found
to be upregulated after ITIH1 knockdown in the present study. In
addition, the protein expression level of α-SMA was also increased.
α-SMA is a gene that positively contributes tO cell migration
processes (31). α-SMA was
previously reported to accelerate the progression of liver and
colorectal cancer by facilitating metastasis (32,33).
Therefore, NF-κB signaling may induce the expression of Cyclin D1,
PCNA and α-SMA, in turn promoting cell proliferation and
metastasis. Treatment of RCC cells with JSH-23, an inhibitor of
NF-κB signaling, in addition to siITIH1 transfection, decreased the
proliferative and invasion ability of cells compared with the
siITIH1 group alone. This further suggested that NF-κB signaling is
an important pathway downstream of ITIH1. Akt signaling pathway is
another important pathway during development and aberration could
lead to cancer (34). The present
study also detected the expression of critical members in Akt
signaling such as Akt and PI3K. However, no significant alterations
of these members occurred. It may be possible that the clinical
specimens of the TCGA data are not of a large enough sample size. A
larger cohort of samples encompassing more subtypes of RCC may
support the findings of the present study. Therefore, the lack of
clinical specimens, which could've been used to detect the
expression pattern of ITIH1 and analyze the association of ITIH1
expression with clinicopathological factors, is a limitation of the
present study. These data could be used to validate the conclusions
reported in the present study. Further research is required to
fully elucidate the role of ITIH1 expression in clinical specimens
and the underlying mechanism of action of ITIH1 in RCC.
In conclusion, data from the present study suggest
that ITIH1 can negatively regulate cell proliferation, migration
and invasion by RCC cells. NF-κB signaling may be an important
signaling pathway regulated by ITIH1 in RCC. The results of the
present study may provide a potential avenue of treatment for
patients with RCC.
Supplementary Material
Knockdown of ITIH1 promotes cell
proliferation and invasion in 786-O cells. (a) Knockdown efficiency
of si-#1 was higher compared with that of si-#2 in 786-O cells. (b)
Proliferative ability of 786-O cells increased significantly in the
siITIH1-transfected group compared with that in the NC group. (c)
Transwell assay images demonstrated that (d) the number of invading
cells was significantly higher in cells treated with siITIH1
compared with that by NC cells. Magnification, x100.
*P<0.05 vs. NC. ITIH1, inter-α-trypsin inhibitor
heavy chain 1; si, small interfering RNA; NC, negative
control.
Overexpression of ITIH1 inhibits cell
proliferation and invasion in RCC. (a) Transfection of RCC cells
with oeITIH1 significantly increased the expression level of ITIH1
compared with that in the vector group. (b) Proliferative ability
of cells decreased significantly in the oeITIH1-treated group
compared with that in the vector group. (c) Transwell assay images
demonstrated that (d) the number of invaded cells was significantly
lower in the oeITIH1 group compared with that in the vector group.
Magnification, x100. *P<0.05 vs. vector. ITIH1,
inter-α-trypsin inhibitor heavy chain 1; oe, overexpression.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
SY and JG designed the study, supervised the
experiments and reviewed the paper. GYu, JG and YY drafted the
paper and performed the experiments. WH, WW and DH performed the
data analysis and contributed to the draft of this paper. GYa and
JW reviewed the draft and contributed to the data analysis. All
authors read and approved the final version of the manuscript. JG
and SQY confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Authors' information
Dr Shiquan Yang ORCID iD: 0009-0005-7416-5603.
References
|
1
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48.
2023.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bahadoram S, Davoodi M, Hassanzadeh S,
Bahadoram M, Barahman M and Mafakher L: Renal cell carcinoma: An
overview of the epidemiology, diagnosis, and treatment. G Ital
Nefrol. 39:2022–vol3. 2022.PubMed/NCBI
|
|
3
|
Acosta PH, Panwar V, Jarmale V, Christie
A, Jasti J, Margulis V, Rakheja D, Cheville J, Leibovich BC, Parker
A, et al: Intratumoral resolution of driver gene mutation
heterogeneity in renal cancer using deep learning. Cancer Res.
82:2792–2806. 2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Deligiannis D, Anastasiou I, Mitropoulos
D, Mitsos P and Theocharis S: Clinical importance of cannabinoid
type 1 receptor (CB1R) and cannabinoid type 2 receptor CB2R)
expression in renal cell carcinoma. Cureus.
16(e55121)2024.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Singh N, Baby D, Rajguru JP, Patil PB,
Thakkannavar SS and Pujari VB: Inflammation and cancer. Ann Afr
Med. 18:121–126. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Marelli G, Sica A, Vannucci L and Allavena
P: Inflammation as target in cancer therapy. Curr Opin Pharmacol.
35:57–65. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Anfray C, Ummarino A, Andón FT and
Allavena P: Current strategies to target
tumor-associated-macrophages to improve anti-tumor immune
responses. Cells. 9(46)2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Stober VP, Lim YP, Opal S, Zhuo L, Kimata
K and Garantziotis S: Inter-α-inhibitor ameliorates endothelial
inflammation in sepsis. Lung. 197:361–369. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hamm A, Veeck J, Bektas N, Wild PJ,
Hartmann A, Heindrichs U, Kristiansen G, Werbowetski-Ogilvie T,
Maestro RD, Knuechel R, et al: Frequent expression loss of
Inter-alpha-trypsin inhibitor heavy chain (ITIH) genes in multiple
human solid tumors: A systematic expression analysis. BMC Cancer.
8(25)2008.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Qian X, Bao ZM, Yao D and Shi Y: Lysine
demethylase 5C epigenetically reduces transcription of ITIH1 that
results in augmented progression of liver hepatocellular carcinoma.
Kaohsiung J Med Sci. 38:437–446. 2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chang QH, Mao T, Tao Y, Dong T, Tang XX,
Ge GH and Xu ZJ: Pan-cancer analysis identifies ITIH1 as a
novel prognostic indicator for hepatocellular carcinoma. Aging
(Albany NY). 13:11096–11119. 2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kopylov AT, Stepanov AA, Malsagova KA,
Soni D, Kushlinsky NE, Enikeev DV, Potoldykova NV, Listitsa AV and
Kaysheva AL: Revelation of proteomic indicators for colorectal
cancer in initial stages of development. Molecules.
25(619)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Vaghari-Tabari M, Ferns GA, Qujeq D,
Andevari AN, Sabahi Z and Moein S: Signaling, metabolism, and
cancer: An important relationship for therapeutic intervention. J
Cell Physiol. 236:5512–5532. 2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Peng C, Ouyang Y, Lu N and Li N: The NF-κB
signaling pathway, the microbiota, and gastrointestinal
tumorigenesis: Recent advances. Front Immunol.
11(1387)2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Liu W, Yan B, Yu H, Ren J, Peng M, Zhu L,
Wang Y, Jin X and Yi L: OTUD1 stabilizes PTEN to inhibit the
PI3K/AKT and TNF-alpha/NF-kappaB signaling pathways and sensitize
ccRCC to TKIs. Int J Biol Sci. 18:1401–1414. 2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yu H, Lin L, Zhang Z, Zhang H and Hu H:
Targeting NF-κB pathway for the therapy of diseases: Mechanism and
clinical study. Signal Transduct Target Ther. 5(209)2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658.
2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chen J and Chen ZJ: Regulation of NF-κB by
ubiquitination. Curr Opin Immunol. 25:4–12. 2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chowdhury N and Drake CG: Kidney cancer:
An overview of current therapeutic approaches. Urol Clin North Am.
47:419–431. 2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The cancer genome atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ribatti D: A revisited concept: Contact
inhibition of growth. From cell biology to malignancy. Exp Cell
Res. 359:17–19. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Niland S, Riscanevo AX and Eble JA: Matrix
metalloproteinases shape the tumor microenvironment in cancer
progression. Int J Mol Sci. 23(146)2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Li X, Li Y, Lu W, Chen M, Ye W and Zhang
D: The tumor vessel targeting strategy: A double-edged sword in
tumor metastasis. Cells. 8(1602)2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Shariati M and Meric-Bernstam F: Targeting
AKT for cancer therapy. Expert Opin Invesig Drugs. 28:977–988.
2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wang K, Chen Q, Shao Y, Yin S, Liu C, Liu
Y, Wang R, Wang T, Qiu Y and Yu H: Anticancer activities of TCM and
their active components against tumor metastasis. Biomed
Pharmacother. 133(111044)2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Carneiro BA and EI-Deiry WS: Targeting
apoptosis in cancer therapy. Nat Rev Clin Oncol. 17:395–417.
2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
O'Dea E and Hoffmann A: NF-κB signaling.
Wiley Interdiscip Rev Syst Biol Med. 1:107–115. 2009.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Yan YL, Bao G, Pei J, Cao Y, Zhang C, Zhao
P, Zhang Y and Damirin A: NF-κB and EGFR participate in
S1PR3-mediated human renal cell carcinomas progression. Biochim
Biophys Acta Mol Basis Dis. 1868(166401)2022.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Akkoc Y, Dalci K, Karakas HE, Erbil-Bilir
S, Yalav O, Sakman G, Celik F, Arikan S, Zeybek U, Ergin M, et al:
Tumor-derived CTF1 (cardiotrophin 1) is a critical mediator of
stroma-assisted and autophagy-dependent breast cancer cell
migration, invasion and metastasis. Autophagy. 19:306–323.
2023.PubMed/NCBI View Article : Google Scholar
|
|
32
|
De Marco M, Del Papa N, Reppucci F, Iorio
V, Basile A, Falco A, Iaccarino R, Brongo S, De Caro F, Capunzo M,
et al: BAG3 induces α-SMA expression in human fibroblasts and its
over-expression correlates with poorer survival in fibrotic cancer
patients. J Cell Biochem. 123:91–101. 2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Valcz G, Sipos F, Krenács T, Molnár J,
Patai AV, Leiszter K, Tóth K, Wichmann B, Molnár B and Tulassay Z:
Increase of α-SMA(+) and CK (+) cells as an early sign of
epithelial-mesenchymal transition during colorectal carcinogenesis.
Pathol Oncol Res. 18:371–376. 2012.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Nitulescu GM, Van De Venter M, Nitulescu
G, Ungurianu A, Juzenas P, Peng Q, Olaru OT, Grădinaru D, Tsatsakis
A, Tsoukalas D, et al: The Akt pathway in oncology therapy and
beyond (Review). Int J Oncol. 53:2319–2331. 2018.PubMed/NCBI View Article : Google Scholar
|