Introduction
Gynecological cancers accounted for ~94,000 new
cases annually in the United States between 2012 and 2016 (1,2).
Globally, gynecological cancers account for >3.6 million new
cases and 1.3 million mortalities per year, which represents
>40% of female cancer incidence and 30% of female cancer
mortality worldwide (3,4). Ovarian cancer (OC) is the most lethal
gynecological malignancy, with the highest mortality rate among
these types of cancer (3). Despite
advances in diagnostics and therapies, OC prognosis remains poor
because of the lack of sensitive and specific biomarkers for early
detection and prognosis. Novel OC biomarkers are urgently needed to
improve clinical outcomes.
Long noncoding RNAs (lncRNAs), RNA transcripts
>200 nucleotides long with limited protein-coding potential, are
critical regulators of tumor pathobiology (5). By modulating gene expression and key
oncogenic pathways, lncRNAs can function as tumor suppressors or
oncogenes and influence cancer cell chemoresistance, the
epithelial-mesenchymal transition, proliferation and other
malignant traits (6-8).
Studies have linked dysregulated lncRNA expression to the
pathogenesis and progression of various types of cancer, lncRNA
CASC15 drives the OC epithelial-mesenchymal transition by
regulating microRNA (miR)-23b and SMAD3(9), similarly, lncRNA-CDC6 and RMRP are
key regulators in breast and bladder cancer, respectively (10,11).
Additionally, lncRNA 01123 is an oncogenic lncRNA that promotes
hepatocellular carcinoma metastasis and proliferation through the
miR-34a-5p/TUFT1 pathway (12).
ZNF667-AS1 is a cancer-associated lncRNA located at
chromosomal region 19q13.43 in humans and it is silenced early in
malignant transformation due to promoter DNA hypermethylation and
loss of expression during cellular immortalization (13). ZNF667-AS1 is normally expressed in
healthy cells, but its expression is lost upon cellular
immortalization. This silencing is due to promoter DNA
hypermethylation following transformation. Studies have shown that
downregulation of ZNF667-AS1 occurs in different types of cancer,
which demonstrates a potential tumor-suppressive role (14,15).
Analysis of The Cancer Genome Atlas (TCGA) revealed
significant downregulation of ZNF667-AS1 in OC and its expression
was significantly associated with ovarian cancer prognosis.
However, the functional significance and mechanistic roles of
ZNF667-AS1 in OC pathogenesis are not fully defined. The present
study employed a multifaceted experimental approach, including
reverse transcription-quantitative (RT-q) PCR, RNA sequencing,
fluorescence in situ hybridization (FISH), cell function
assays and western blotting, to elucidate the tumor-suppressive
functions and molecular mechanisms of lncRNA ZNF667-AS1 in OC,
which pave the way for its development as a novel therapeutic
target.
Materials and methods
Pan-cancer analysis
Expression data for ZNF667-AS1 across 33 types of
cancer and corresponding normal tissues were extracted from TCGA
(http://sangerbox.com) and genotype-tissue
expression databases (http://sangerbox.com) and subsequent pan-cancer
analyses of ZNF667-AS1 concerning overall survival (OS),
disease-specific survival (DSS), immune infiltration (TIMER, EPIC,
IPS, MCPcounter, xCELL, QUANTISEQ and CIBERSORT), genomic
heterogeneity and stemness (DNA, EREG-METH, DMPs, ENHs, RNAss and
EREG-EXPs) were conducted via the SangerBox web tool (http://sangerbox.com) (16).
Cell cultures
The OC cell line SKOV3 used in the present study was
purchased from Procell Life Science & Technology Co., Ltd. The
IOSE80 and OVCA433 cells were a gift from the First Affiliated
Hospital of the Anhui Medical University (Anhui, China). SKOV3,
IOSE80 and OVCA433 cells were cultured in McCoy's 5A medium
(Procell Life Science & Technology Co., Ltd), RPMI-1640 medium,
or RPMI-1640 medium (Procell Life Science & Technology Co.,
Ltd), respectively, supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.). The cells were maintained
at 37˚C in a humidified 5% CO2 incubator and the medium
was renewed every 2 days.
Cell transfection
In vitro transfection of OC cells was
performed using Lipofectamine® 2000 reagent (Thermo
Fisher Scientific, Inc.) with a PCDH vector expressing lncRNA
ZNF667-AS1 (2.5 µg of lncRNA ZNF667-AS1 and 5 µl of
Lipofectamine® 2000 reagent), ZNF667-AS1 small
interfering (si)RNA (10 µl of siRNA and 5 µl of
Lipofectamine® 2000 reagent), or corresponding negative
controls (Wuhan Miaoling Biotech Science Co., Ltd.) and the
transfection mass of the control group was the same as that of the
experimental group. The sequences of plasmid were shown in Table SI. Transfections were conducted at
60% confluence in 6-well plates following the manufacturer's
protocol. Following transfection, the experiment was continued for
24-48 h in a 37˚C cell incubator.
RT-qPCR
Total RNA was extracted from 2x106 cells
using TRIzol® reagent (Thermo Fisher Scientific, Inc.)
and detection of RNA concentration and purity (260/280) was
performed using nucleic acid protein analyzer. RNA was reverse
transcribed into cDNA with a Promega reverse transcription system
(Promega Corporation). RT-qPCR was performed with SYBR Green Master
Mix (Vazyme Biotech Co., Ltd.) on a real-time PCR detection system:
95˚C predenaturation 30 sec, 40 cycles of 95˚C 10 sec and 60˚C 30
sec. Relative lncRNA ZNF667-AS1 levels were quantified by the
2-ΔΔCq method and normalized to the level of GAPDH
(17). The RT-qPCR primers used
are listed in Table SI. In
studies that do not involve metabolic regulation, GAPDH, as a
widely expressed steward gene, is an internal reference gene with
stable expression and is often used in lncRNA studies (18). RNA extraction, cDNA synthesis, and
qPCR performed according to the manufacturer's protocols and these
experiments were replicated three times.
5-Ethynyl-2'-deoxyuridine (EdU)
assay
Cell proliferation was assessed using the EdU
incorporation assay. Briefly, cells grown on coverslips in 24-well
plates were incubated with EdU medium for 2 h at 37˚C. Following
fixation in 4% paraformaldehyde at room temperature for 15 min and
permeabilization in 0.3% Triton X-100 (Biosharp Life Sciences), EdU
labelling was performed via a click reaction cocktail for 30 min,
following the manufacturer's instructions (Beyotime Institute of
Biotechnology). The cells were counterstained with Hoechst 33342
(Beyotime Institute of Biotechnology) at room temperature for 10
min and visualized under a fluorescence microscope (magnification,
x100).
Colony formation assay
The cells were seeded in 6-well plates and cultured
for 10-14 days at 37˚C with 5% CO2. Colonies were fixed
in 4% paraformaldehyde at room temperature for 15 min, stained with
0.1% crystal violet solution (Beyotime Institute of Biotechnology)
at room temperature for 15 min and counted manually. A total of
three wells in each group were counted and used as separate
experiments.
Scratch wound healing assay
Confluent cell monolayers in 6-well plates were
scratched with a 200 µl pipette tip to create wound gaps, washed
three times with PBS and incubated in serum-free medium. Wound
closure was monitored by taking phase contrast images at 0 and 24 h
post wounding using an inverted microscope (magnification,
x100).
Transwell assay
Cell migratory and invasive capacities were
evaluated via Transwell assays. For invasion, the upper chamber was
precoated with Matrigel matrix at 37˚C for 2 h (BD Biosciences).
Cells in serum-free medium were added to the upper chamber, while
complete medium supplemented with 10% FBS as a chemoattractant was
added to the lower well. Following 24-48 h of incubation, the cells
on the lower surface were fixed in 4% paraformaldehyde at room
temperature for 15 min and stained with crystal violet solution
(Beyotime Institute of Biotechnology). The cells were imaged and
quantified under a light microscope (magnification, x100). For the
migration assays, the upper chamber was without the Matrigel
matrix.
RNA sequencing (RNA-seq)
RNA extracted from three pairs of stably transfected
ZNF667-AS1 overexpressing and control OC cells was subjected to
RNA-seq analysis by Genergy Bio-Technology (Shanghai) Co.
Differentially expressed genes were identified and analyzed for
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes
(KEGG) enrichment. The gene set for the KEGG pathway analysis was
obtained from the Molecular Signatures Database (curated gene sets,
canonical pathways, KEGG analysis for human gene symbols; v2022.1;
https://www.gsea-msigdb.org/gsea/index.jsp).
Fluorescence in situ hybridization
(FISH)
FISH was performed using a Shanghai GenePharma Co.,
Ltd. kit, according to the manufacturer's protocol. OC cells grown
on coverslips were hybridized with a Cy3-labelled ZNF667-AS1 probe,
counterstained with DAPI and visualized via confocal microscopy
(magnification, x400). The ZNF667-AS1 probe was designed and
synthesized by Shanghai GenePharma Co., Ltd.
Western blotting
Total protein was extracted from the cells with RIPA
lysate (Beyotime Institute of Biotechnology) and determined by BCA.
The proteins (20-100 µg) were subjected to SDS-PAGE (10%) and then
transferred to a polyvinylidene difluoride (PVDF) membrane. The
membrane was blocked with 5% skimmed milk for 1 h at room
temperature and then incubated with primary and secondary
antibodies. Bound antibodies were detected via enhanced
chemiluminescence (ECL). The specific antibodies employed in this
experiment were GAPDH (cat. no. 200306-7E4; OriGene Technologies,
Inc.), Tubulin (cat. no. 250009; OriGene Technologies, Inc.), PCNA
(cat. no. 10205-2-AP; Proteintech Group, Inc.), MMP2 (cat. no.
10373-2-AP; Proteintech Group, Inc.), MMP9 (cat. no. 10375-2-AP;
Proteintech Group, Inc.), rabbit second antibody (cat. no.
SA00001-2; Proteintech Group, Inc.) and mouse second antibody (cat.
no. SA00001-1; Proteintech Group, Inc.). All primary antibodies
were diluted 1:1,000 and incubated at 4˚C for 12 h, while goat
anti-rabbit and anti-mouse IgG H&L (HRP) second antibodies were
diluted 1:5,000 and incubated at room temperature for 1 h. Finally,
ImageJ (National Institutes of Health) was used to calculate the
gray value of the protein strip.
Statistical analysis
The data are presented as the means ± standard
deviation. Statistical comparisons between groups were conducted
via unpaired Student's t-test in GraphPad Prism 8.0 (Dotmatics) and
R version 4.1.2 (http://www.R-project.org/). P<0.05 was considered
to indicate a statistically significant difference.
Results
lncRNA ZNF667-AS1 in pan-cancer
analysis
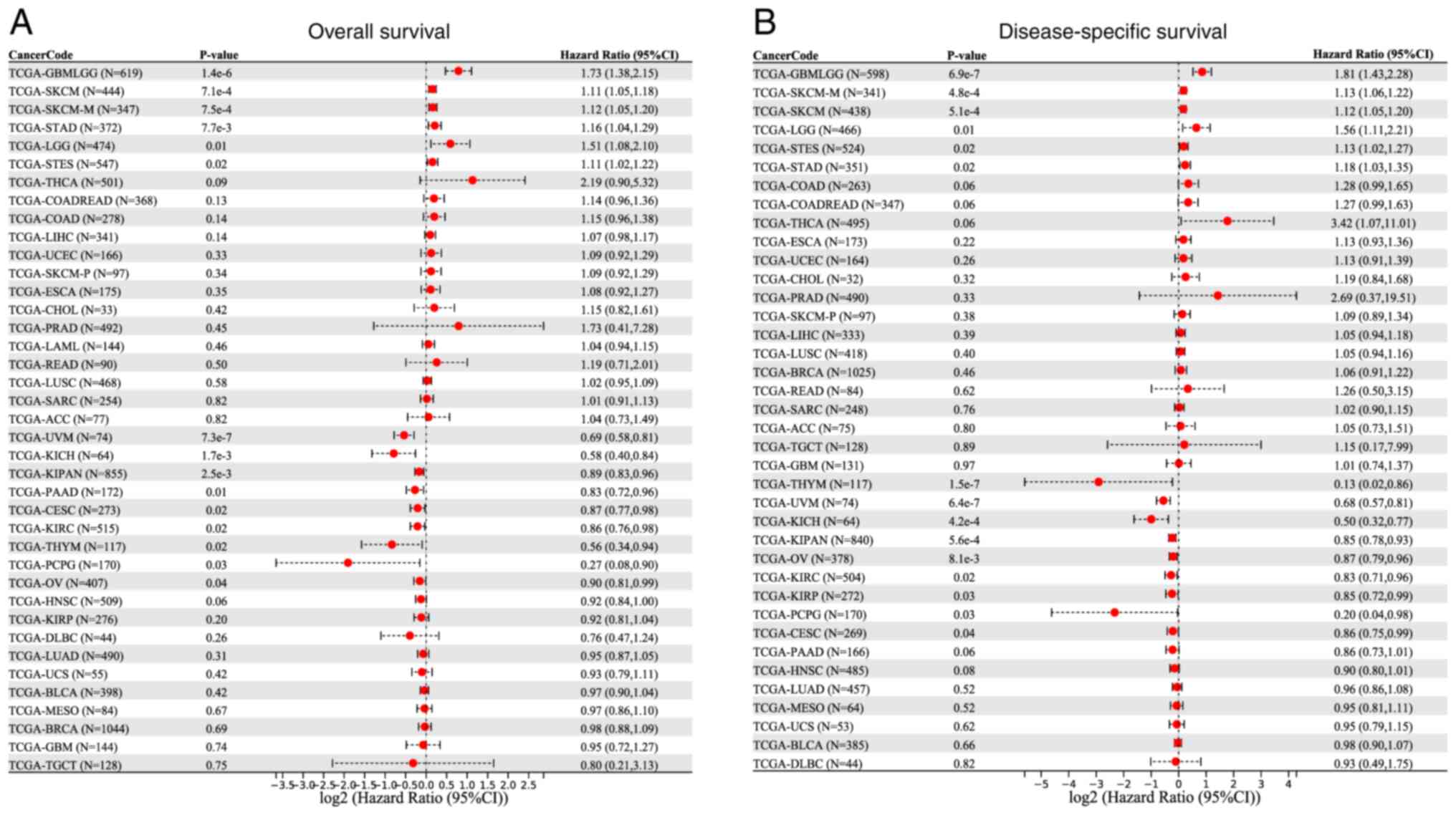
The present study explored the prognostic
significance of lncRNA ZNF667-AS1 in different types of cancer by
using data from the SangerBox website. The analysis revealed that
high expression of lncRNA ZNF667-AS1 was associated with poor
prognosis in several types of cancer, particularly glioblastoma
(GBM) and skin cutaneous melanoma. For example, in GBM patients,
elevated lncRNA ZNF667-AS1 is associated with reduced OS, as shown
in Fig. 1A, where a forest plot
demonstrates a significant difference (P<0.05) between the high-
and low-expression groups. By contrast, in uveal melanoma and OC,
high expression of lncRNA ZNF667-AS1 is linked to an improved
prognosis and the forest plot in Fig.
1B demonstrated improved outcomes in patients with elevated
expression.
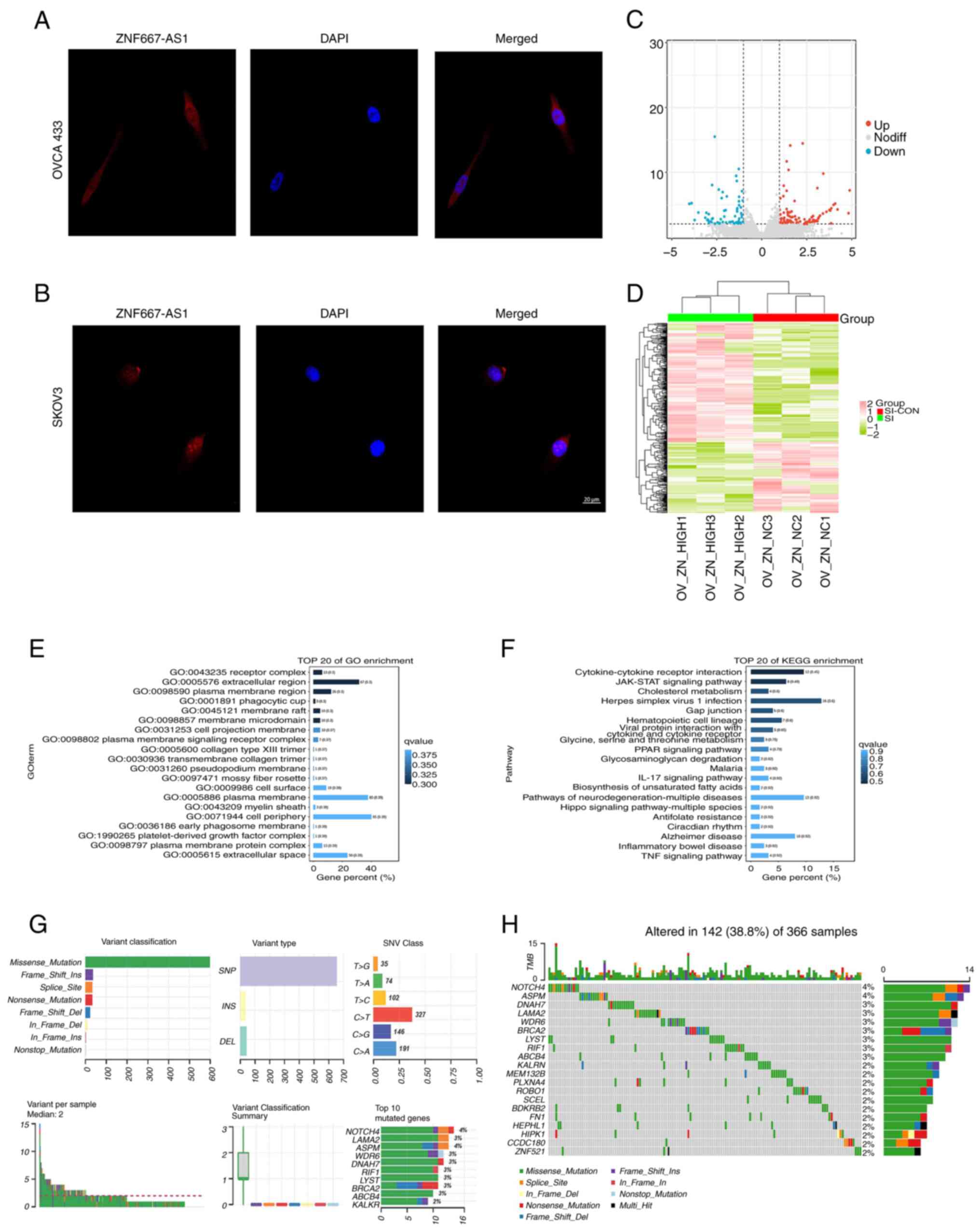
The present study subsequently analyzed the
correlation between lncRNA ZNF667-AS1 and various immune cell types
by using the SangerBox website. To investigate the infiltration
levels of immune cells, the present study employed several
algorithms, including TIMER, EPIC, IPS, MCPcounter, xCELL,
QUANTISEQ and CIBERSORT. These algorithms assess the composition
and activity of immune cells in the tumor microenvironment from
different perspectives. For example, TIMER focuses on estimating
tumor-infiltrating lymphocytes, whereas MCPcounter distinguishes
the abundance of different immune cell subpopulations. As shown in
Fig. 2, multiple algorithms
indicated that higher expression of lncRNA ZNF667-AS1 was
associated with increased immune cell infiltration in tumors such
as colon adenocarcinoma and thyroid carcinoma, whereas lower
expression was associated with decreased immune cell infiltration
in tumors such as brain lower grade glioma. These findings
indicated a potential role for this lncRNA in regulating the tumor
immune microenvironment. The present study further investigated the
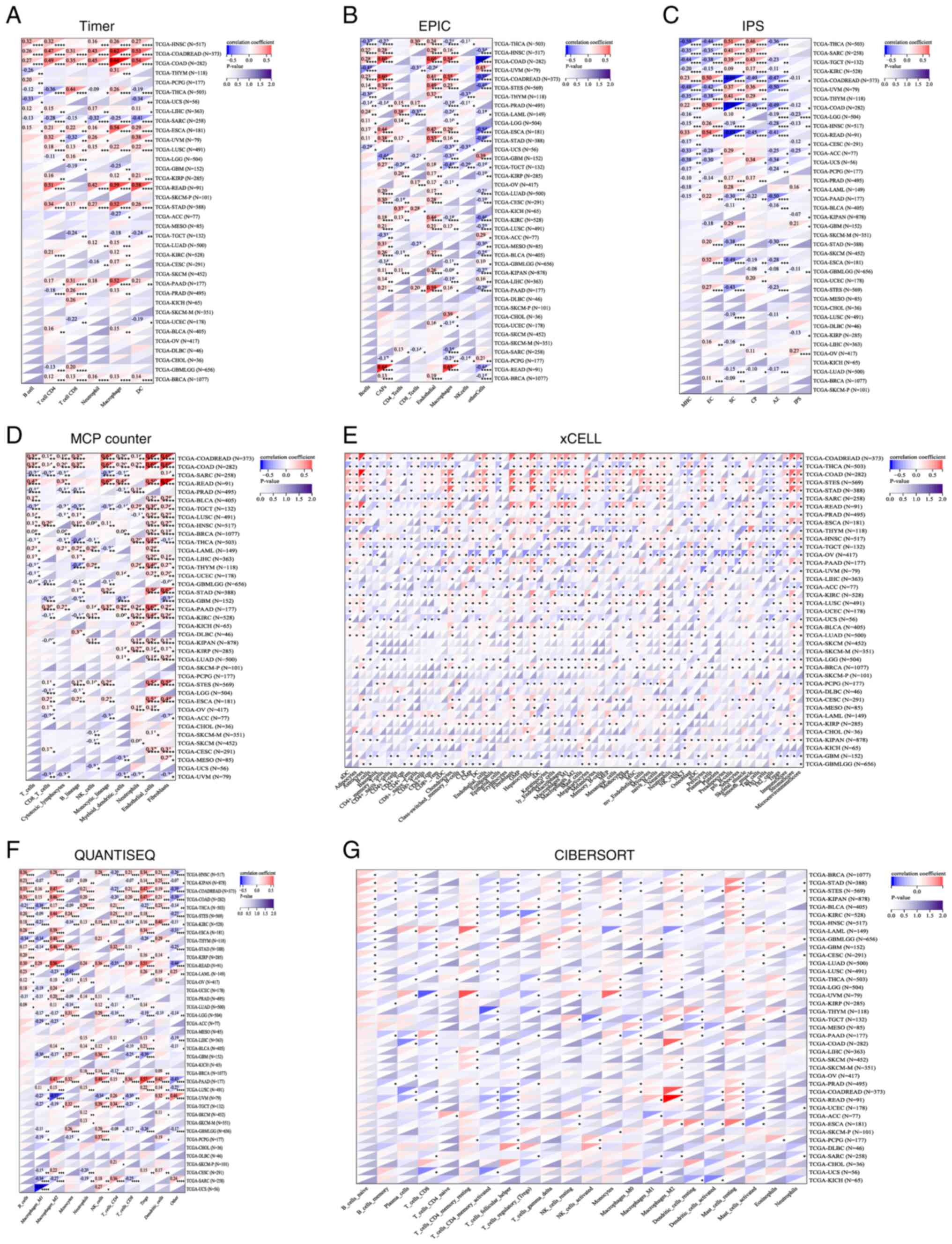
effect of genomic heterogeneity on the expression of lncRNA
ZNF667-AS1. The findings indicated that lncRNA ZNF667-AS1 had a
complex relationship with various metrics of genomic heterogeneity,
including mutations, copy number variations and chromosomal
structural variations in different types of cancer. Fig. 3 illustrated the intricate
correlations between lncRNA ZNF667-AS1 expression and these
heterogeneous parameters and highlights its potential role as a
marker of tumor evolution and treatment response.
 | Figure 2Relationship between lncRNA ZNF667-AS1
expression and pan-cancer immune cells: (A) Timer, (B) EPIC, (C)
IPS, (D) MCPcounter, (E) xCELL, (F) QUANTISEQ, (G) CIBERSORT.
*P<0.05, **P<0.01,
***P<0.001. lncRNA, long noncoding RNA; IPS, immune
score. |
 | Figure 3Relationship between lncRNA ZNF667-AS1
expression and pan-cancer various heterogeneity metrics: (A) TMB,
(B) MATH, (C) MSI, (D) NEO, (E) PURITY, (F) PLOIDY, (G) HRD, (H)
LOH. lncRNA, long noncoding RNA; TMB, tumor mutation burden; MATH,
variant-allele tumor heterogeneity; MSI,microsatellite instability;
NEO, neopeptide; HRD, homologous recombination deficiency; LOH,
loss of heterozygosity. |
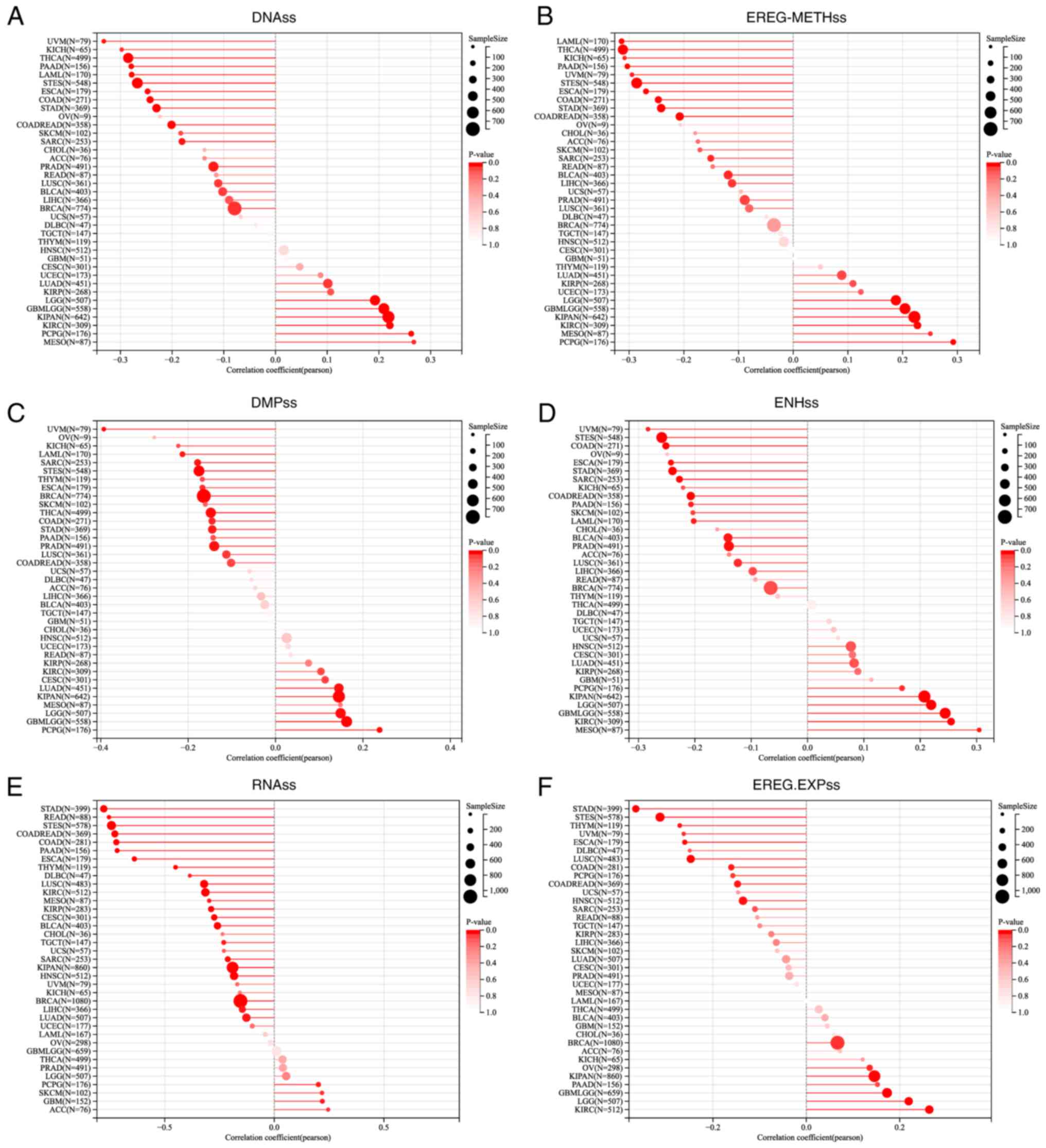
Tumor stemness, a key factor in cancer biology,
refers to the ability of tumor cells or tissues to maintain or
acquire stem cell characteristics. This information is crucial for
understanding tumor biology, predicting disease prognosis and
developing new anticancer treatments. As shown in Fig. 4, in most types of cancer, lncRNA
ZNF667-AS1 expression was negatively associated with tumor stemness
and this relationship was confirmed by several methods, including
DNA, EREG-METH, DMPs, ENHs, RNAss and EREG-EXPs. These results
showed that lncRNA ZNF667-AS1 may prevent tumor cells from
acquiring stem-like properties.
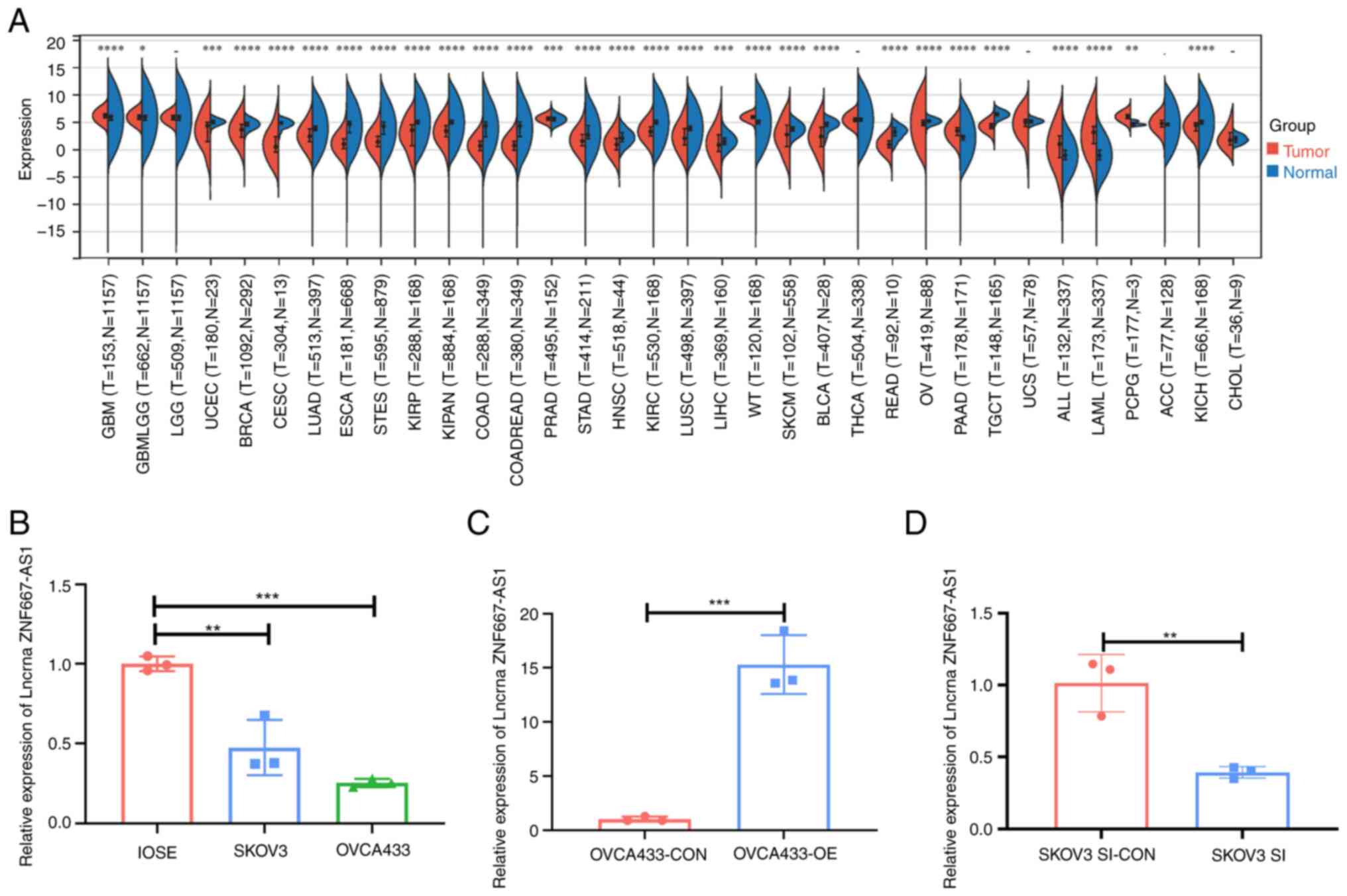
Expression of the lncRNA ZNF667-AS1 is
downregulated in human OC tissues
Analysis of TCGA data using the SangerBox platform
revealed downregulation of lncRNA ZNF667-AS1 across multiple types
of cancer, including OC (Fig. 5A).
The present study validated this finding in ovarian cell lines;
lower ZNF667-AS1 levels occurred in the SKOV3 and OVCA433 cancer
lines than in normal IOSE80 cells. ZNF667-AS1 expression was
particularly diminished in OVCA433 cells (Fig. 5B).
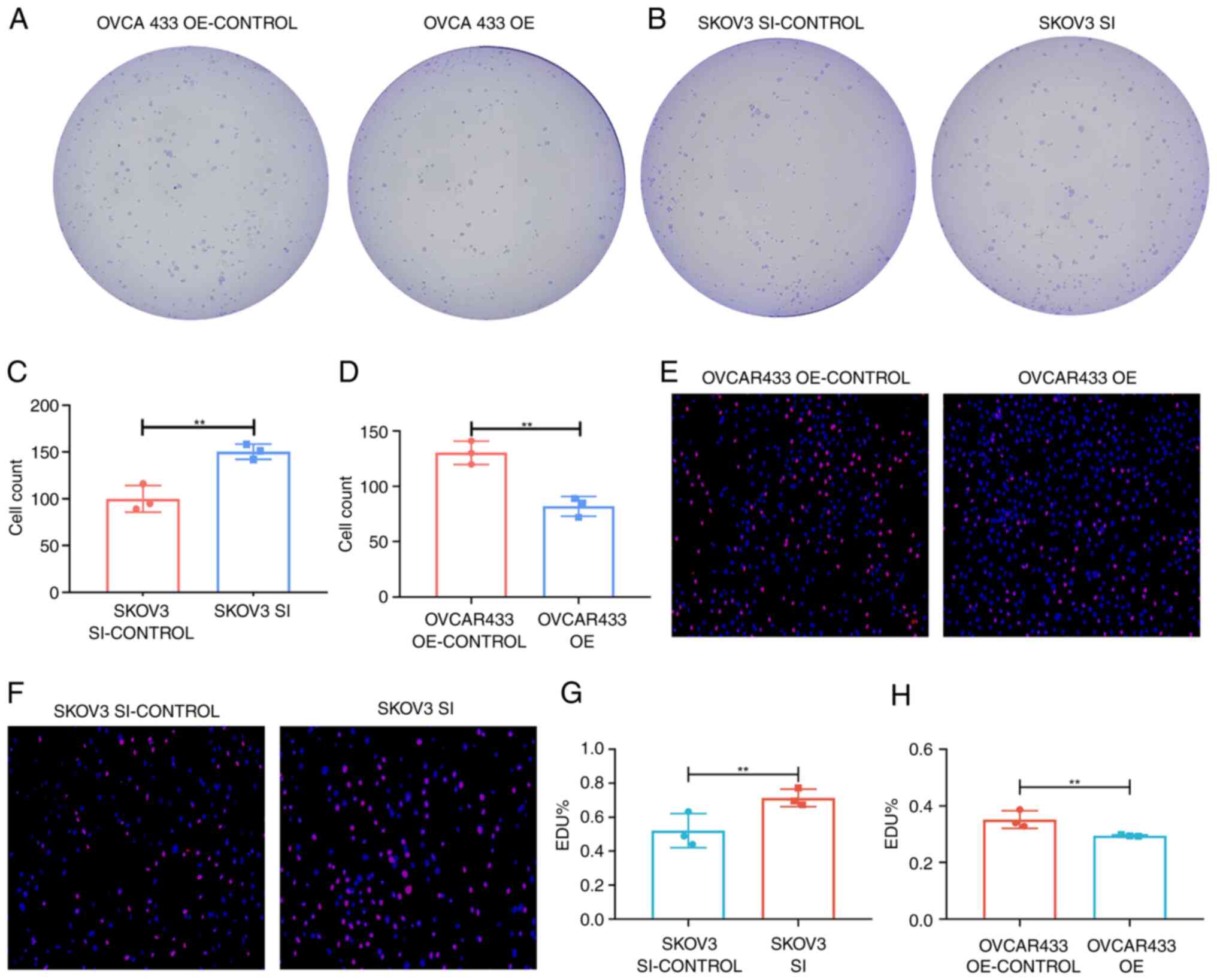
lncRNA ZNF667-AS1 mediates cell
proliferation in vitro
To elucidate the functional role of ZNF667-AS1 in
OC, the present study generated ZNF667-AS1-overexpressing OVCA433
cells and ZNF667-AS1-knockdown SKOV3 cells via plasmid transfection
and siRNA strategies, respectively. Successful overexpression and
knockdown were confirmed by RT-qPCR (Fig. 5C and D). A colony formation assay revealed
significantly reduced proliferation in ZNF667-AS1-overexpressing
OVCA433 cells compared with controls. Conversely, ZNF667-AS1
knockdown enhanced SKOV3 cell proliferation (Fig. 6A-D). The EdU experiment showed
consistent results (Fig. 6E-H). In
addition, western blotting revealed that overexpression of
ZNF667-AS inhibited the expression of the proliferation marker PCNA
(Fig. 7E).
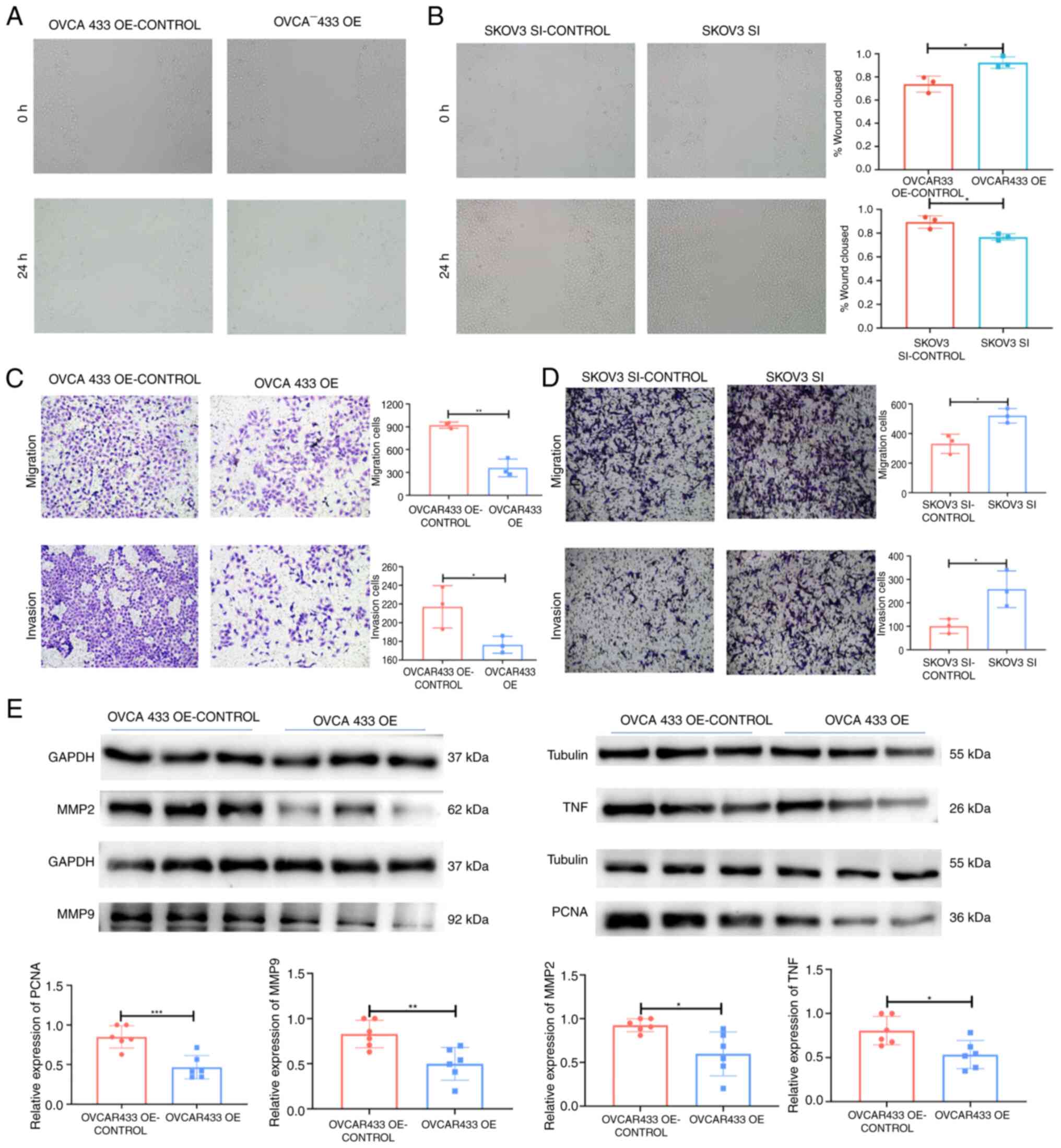 | Figure 7lncRNA ZNF667-AS1 inhibited OC cell
migration, invasion in vitro and western blotting
validation. (A and B) Transwell assay showing that upregulation of
ZNF667-AS1 suppressed cell migration (P<0.01) and invasion
(P<0.05) in the OVCA433 cell line and that knockdown of TPTEP1
promoted cell migration (P<0.05) and invasion (P<0.05) in the
SKOV3 cell line (magnification, x100). (C and D) Wound-healing
assay showing that upregulation of ZNF667-AS1 suppressed cell
migration in the OVCA433 cell line (P<0.05) and that knockdown
of ZNF667-AS1 promoted cell migration in the SKOV3 cell line
(P<0.05) (magnification, x100). (E) western blotting showed the
association between ZNF667-AS1 expression level and tumor indexes
of MMP2, MMP9, PCNA and TNF. *P<0.05,
**P<0.01, ***P<0.001. lncRNA, long
noncoding RNA; OC, ovarian cancer; PCNA, proliferating cell nuclear
antigen. |
Alterations in the expression of
lncRNA ZNF667-AS1 affect OC cell migration and invasion in
vitro
The present study further examined the effects of
ZNF667-AS1 on OC cell motility via Transwell and wound healing
assays. Wounds were introduced into layers of SKOV3-SI, OVCA433-OE
and corresponding control cells by scratching and the cells were
subsequently cultured for 24 h. Overexpression of the lncRNA
ZNF667-AS1 significantly decreased the spreading potential of
OVCA433 cells, whereas downregulation of the lncRNA ZNF667-AS1
significantly increased the spreading potential of SKOV3 cells
(Fig. 7A and B). The Transwell assay revealed a similar
trend with respect to the invasion and migration abilities of SKOV3
and OVCA433 cells (Fig. 7C and
D). In addition, overexpression of
ZNF667-AS inhibited the expression of the migration and invasion
markers MMP2 and MMP9 (Fig.
7E).
lncRNA ZNF667-AS1 mediates the TNF
signaling pathway to affect ovarian cancer functions
The FISH results (Fig.
8A and B) revealed that lncRNA
ZNF667-AS1 was mainly expressed in the nuclei of SKOV3 and OVCA433
cells. To elucidate the molecular mechanisms by which ZNF667-AS1
functions in OC, RNA-seq was performed on ZNF667-AS1-overexpressing
and control OVCA433 cells. Differentially expressed genes were
identified using volcano plot and heatmap analyses (Fig. 8C and D). KEGG pathway enrichment analysis
revealed significant enrichment of differentially expressed genes
in the TNF pathway (Fig. 8E and
F). TNF-α is expressed mainly on
the cell membrane, but its signaling process involves gene
expression regulation in the nucleus (19) and ZNF667-AS1 is expressed mainly in
the nucleus. Western blotting confirmed that overexpression of
ZNF667-AS1 decreased the expression of TNF (Fig. 7E).
Missense mutations were the predominant type among
the differentially expressed genes, with NOTCH4 being the most
frequently mutated gene in OC (Fig.
8G). These results suggested that ZNF667-AS1 was a key
regulator of the TNF signaling cascade in OC.
Discussion
The advent of targeted cancer therapies has
revolutionized cancer treatment by selectively modulating
deregulated oncogenic signaling pathways to improve patient
survival. While progress has been made in identifying OC biomarkers
and in targeted therapeutics, further research is necessary to
develop more effective diagnostic, prognostic and treatment
strategies. Identifying key OC biomarkers is a critical step
towards improving clinical outcomes.
A growing body of evidence shows that lncRNA
expression is closely associated with cancer pathogenesis,
metastasis and clinical outcomes (20-22).
By modulating gene expression programs and key oncogenic signaling
cascades, such as the PI3K/AKT pathway, lncRNAs can act as tumor
suppressors or oncogenes (23,24).
Studies have reported frequent downregulation of the lncRNA
ZNF667-AS1in various types of malignancy, which suggests a
potential tumor-suppressive role (13,25).
Silencing of ZNF667-AS1 occurs early in carcinogenesis and is
sustained throughout cancer progression to invasive and metastatic
disease (26). Furthermore, low
ZNF667-AS1 expression is significantly associated with an advanced
tumor stage and poor prognosis in several types of cancer (27).
The pan-cancer analysis of the present study
revealed widespread downregulation of ZNF667-AS1 in nearly half of
the types of cancer examined. Notably, the prognosis for certain
solid tumors, such as OC, showed improved prognosis when ZNF667-AS1
was expressed. Along with the associations between ZNF667-AS1 and
genomic heterogeneity and stemness, these data highlighted its
potential as a pan-cancer prognostic marker.
Functionally, The present study found that
overexpression of ZNF667-AS1 significantly inhibited OC cell
proliferation, migration and invasion in vitro, whereas
knockdown of ZNF667-AS1 promoted these oncogenic properties. These
complementary results demonstrated that ZNF667-AS1 is a potential
therapeutic target in OC. Additionally, protein markers for
proliferation, invasion and migration, including PCNA, MMP2 and
MMP9, tended to decrease following ZNF667-AS1 overexpression.
TNF-α is a crucial cytokine and TNF-α can kill
target cells and regulate adaptive immunity to protect the body
(28). The upregulation of TNF-α,
a proinflammatory cytokine frequently detected in various
malignancies, is associated with an increased incidence of OC
(29). Additionally, IL-15
agonists (such as ALT-803) can upregulate TNF-α expression and
restore natural killer cell function in the ascites of OC patients
(30). In the present study,
pathway enrichment analysis revealed that TNF signaling pathway was
one of the top pathways modulated by ZNF667-AS1 in OC, as validated
by western blotting. These results suggested that ZNF667-AS1 may
regulate ovarian cancer progression by interacting with the TNF
signaling pathway.
The present study had certain limitations, including
a reliance on in vitro experiments alone. Future in
vivo studies are warranted to validate the tumor-suppressive
functions of ZNF667-AS1 in OC models. Additionally, further
research is needed to delineate the specific mechanisms by which
ZNF667-AS1 interacts with and regulates TNF signaling in OC.
The present study revealed that ZNF667-AS1 was an
important regulator of OC progression, potentially by modulating
TNF signaling. These findings shed light on the functional and
mechanistic roles of lncRNA ZNF667-AS1 in OC pathogenesis and may
facilitate the development of novel diagnostic, prognostic, or
therapeutic approaches that leverage ZNF667-AS1 in the clinical
management of ovarian cancer.
Supplementary Material
Primer sequence and plasmid
sequence
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
FFW and TWG performed the study and drafted the
article. FFW, TCZ and XYM conducted cell culture, data analysis and
interpretation. FFW and TWG contributed to the study design. All
authors discussed the results and agreed to be accountable for all
aspects of the work. All authors read and approved the final
version of the manuscript. TG and FW confirm the authenticity of
all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Corpus CU and Vulva UOV: Gynecologic
Cancer Incidence, United States-2012-2016; Centers for Disease
Control and Prevention, US Department of Health and Human Services:
Atlanta, GA, USA, 2019.
|
|
2
|
Dellino M, Cascardi E, Laganà AS, Di Vagno
G, Malvasi A, Zaccaro R, Maggipinto K, Cazzato G, Scacco S, Tinelli
R, et al: Lactobacillus crispatus M247 oral administration: Is it
really an effective strategy in the management of
papillomavirus-infected women? Infect Agent Cancer.
17(53)2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36
Cancers in 185 Countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Viviani S, Dellino M, Ramadan S, Peracchio
C, Marcheselli L, Minoia C and Guarini A: Fertility preservation
strategies for patients with lymphoma: A real-world practice survey
among Fondazione Italiana Linfomi centers. Tumori. 108:572–577.
2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Chen Q, Zhou L, Ma D, Hou J, Lin Y, Wu J
and Tao M: LncRNA GAS6-AS1 facilitates tumorigenesis and metastasis
of colorectal cancer by regulating TRIM14 through
miR-370-3p/miR-1296-5p and FUS. J Transl Med.
20(356)2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A Long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Tan DSW, Chong FT, Leong HS, Toh SY, Lau
DP, Kwang XL, Zhang X, Sundaram GM, Tan GS, Chang MM, et al: Long
noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor
addiction and modulates treatment response in squamous cell
carcinoma. Nat Med. 23:1167–1175. 2017.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Xu Y, Ge Z, Zhang E, Zuo Q, Huang S, Yang
N, Wu D, Zhang Y, Chen Y, Xu H, et al: The lncRNA TUG1 modulates
proliferation in trophoblast cells via epigenetic suppression of
RND3. Cell Death Dis. 8:e3104. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lin H, Xu X, Chen K, Fu Z, Wang S, Chen Y,
Zhang H, Niu Y, Chen H, Yu H, et al: LncRNA CASC15, MiR-23b cluster
and SMAD3 form a novel positive feedback loop to promote
epithelial-mesenchymal transition and metastasis in ovarian cancer.
Int J Biol Sci. 18:1989–2002. 2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Cao HL, Liu ZJ, Huang PL, Yue YL and Xi
JN: lncRNA-RMRP promotes proliferation, migration and invasion of
bladder cancer via miR-206. Eur Rev Med Pharmacol Sci.
23:1012–1021. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kong X, Duan Y, Sang Y, Li Y, Zhang H,
Liang Y, Liu Y, Zhang N and Yang Q: LncRNA-CDC6 promotes breast
cancer progression and function as ceRNA to target CDC6 by sponging
microRNA-215. J Cell Physiol. 234:9105–9117. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Xiao Z, Liu Y, Zhao J, Li L, Hu L, Lu Q,
Zeng Z, Liu X, Huang D, Yang W and Xu Q: Long noncoding RNA
LINC01123 promotes the proliferation and invasion of hepatocellular
carcinoma cells by modulating the miR-34a-5p/TUFT1 axis. Int J Biol
Sci. 16:2296–2305. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Vrba L, Garbe JC, Stampfer MR and Futscher
BW: A lincRNA connected to cell mortality and
epigenetically-silenced in most common human cancers. Epigenetics.
10:1074–1083. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Chen X, Huang Y, Shi D, Nie C, Luo Y, Guo
L, Zou Y and Xie C: LncRNA ZNF667-AS1 Promotes ABLIM1 Expression by
Adsorbing microRNA-1290 to Suppress Nasopharyngeal Carcinoma Cell
Progression. Onco Targets Ther. 13:4397–4409. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yang H, Cai MY, Rong H, Ma LR and Xu YL:
ZNF667-AS1, a positively regulating MEGF10, inhibits the
progression of uveal melanoma by modulating cellular
aggressiveness. J Biochem Mol Toxicol. 35(e22732)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wang D, Wang Y, Zou X, Shi Y, Liu Q, Huyan
T, Su J, Wang Q, Zhang F, Li X and Tie L: FOXO1 inhibition prevents
renal ischemia-reperfusion injury via cAMP-response element binding
protein/PPAR-γ coactivator-1α-mediated mitochondrial biogenesis. Br
J Pharmacol. 177:432–448. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Feng Y, Zhang Z, Yang H, Miao F, Li Y,
Zhang M, Cao Y and Li M: The lncRNA TPTEP1 suppresses PI3K/AKT
signalling and inhibits ovarian cancer progression by interacting
with PTBP1. J Cell Mol Med. 28(e70106)2024.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Huang Z, Senocak F, Jayaraman A and Hahn
J: Integrated modeling and experimental approach for determining
transcription factor profiles from fluorescent reporter data. BMC
Syst Biol. 2(64)2008.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Cervena K, Vodenkova S and Vymetalkova V:
MALAT1 in colorectal cancer: Its implication as a diagnostic,
prognostic and predictive biomarker. Gene.
843(146791)2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Qiu MT, Hu JW, Yin R and Xu L: Long
noncoding RNA: An emerging paradigm of cancer research. Tumour
Biol. 34:613–620. 2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv
XW and Li J: Long noncoding RNAs: Novel insights into hepatocelluar
carcinoma. Cancer Lett. 344:20–27. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Katsushima K, Natsume A, Ohka F, Shinjo K,
Hatanaka A, Ichimura N, Sato S, Takahashi S, Kimura H, Totoki Y, et
al: Targeting the Notch-regulated non-coding RNA TUG1 for glioma
treatment. Nat Commun. 7(13616)2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Huang Y, Zhang J, Hou L, Wang G, Liu H,
Zhang R, Chen X and Zhu J: LncRNA AK023391 promotes tumorigenesis
and invasion of gastric cancer through activation of the PI3K/Akt
signaling pathway. J Exp Clin Cancer Res. 36(194)2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhao LP, Li RH, Han DM, Zhang XQ, Nian GX,
Wu MX, Feng Y, Zhang L and Sun ZG: Independent prognostic Factor of
low-expressed LncRNA ZNF667-AS1 for cervical cancer and inhibitory
function on the proliferation of cervical cancer. Eur Rev Med
Pharmacol Sci. 21:5353–5360. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Vrba L and Futscher BW: Epigenetic
silencing of MORT is an early event in cancer and is associated
with luminal, receptor positive breast tumor subtypes. J Br Cancer.
20(198)2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Vrba L and Futscher BW: Epigenetic
silencing of lncRNA MORT in 16 TCGA cancer types. F1000Research.
7(211)2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Liu M, Yi Y and Zhao M: Effect of
dexmedetomidine anesthesia on perioperative levels of TNF-α and
IL-6 in patients with ovarian cancer. Oncol Lett. 17:5517–5522.
2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Gupta M, Babic A, Beck AH and Terry K:
TNF-α expression, risk factors and inflammatory exposures in
ovarian cancer: Evidence for an inflammatory pathway of ovarian
carcinogenesis? Hum Pathol. 54:82–91. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Felices M, Chu S, Kodal B, Bendzick L,
Ryan C, Lenvik AJ, Boylan KLM, Wong HC, Skubitz APN, Miller JS and
Geller MA: IL-15 super-agonist (ALT-803) enhances natural killer
(NK) cell function against ovarian cancer. Gynecol Oncol.
145:453–456. 2017.PubMed/NCBI View Article : Google Scholar
|