Introduction
Hepatocellular carcinoma (HCC) can be a fatal, and
chronic infection caused by hepatitis B virus (HBV) and/or
hepatitis C virus (HCV) is usually related to hepatocarcinogenesis.
Globally, HBV carriers are estimated to total 3 million, and
approximately 1 million people suffer from HBV-related HCC
(1). HBV is an oncogenic virus
which integrates with the genome of hepatocytes. The quantity of
HBV-DNA is strongly associated with the generation of HCC (2,3).
In general, the diagnosis of HBV infection is based
on the detection of hepatitis B surface antigen (HBsAg) in the
serum. However, it was recently reported that HBV-DNA is
periodically detected in the serum or liver and is recognized as
occult HBV infection (4,5). In particular, HBcAb positivity is
thought to be a latent risk factor for hepatocarcinogenesis, since
the covalently closed circular DNA (cccDNA) of HBV is responsible
for the persistent HBV infection of hepatocytes in these cases
(6,7).
In order to examine the clinical characteristics of
HCC negative for HBsAg and the anti-hepatitis C virus antibody
(HCV-Ab) (NBNC-HCC) and HBV-DNA in relation to hepatocarcinogenesis
in NBNC-HCC, 32 cases of resected HCC were analyzed.
Materials and methods
Sample collection
Thirty-two cases of HCC (26 males and 6 females,
median age 65.7 years) that underwent surgical resection in Kobe
University and related hospitals were enrolled in this study.
Clinical characteristics including age, gender, biochemical data
and surrounding liver tissues were examined. Histological features
were graded according to the New Inuyama Classification (8).
Analysis of occult HBV infection
To examine occult HBV infection in HBsAg-negative
cases, DNA was extracted in 12 cases from surrounding liver tissues
using the QIAamp DNA Mini Kit (Qiagen, Tokyo, Japan). The detection
of HBV-DNA was carried out by employing PCR analysis using specific
primers (9,10). PCR products were directly sequenced
using an ABI PRISM® 3100 Avant Genetic Analyzer
(Applied Biosystems, Foster City, CA, USA), and nucleotide
alignment using 20 reference strains was performed with Clustal X
software. The HBV genotype was determined by phylogenetic analysis
using the Molecular Evolutionary Genetics Analysis 4 (MEGA4)
software program (available at http://www.megasoftware.net) (11).
Statistical analysis
Statistical analysis was carried out using JMP7
software (SAS Institute Japan, Co., Ltd.). P<0.05 was considered
significant.
Results
Clinical data
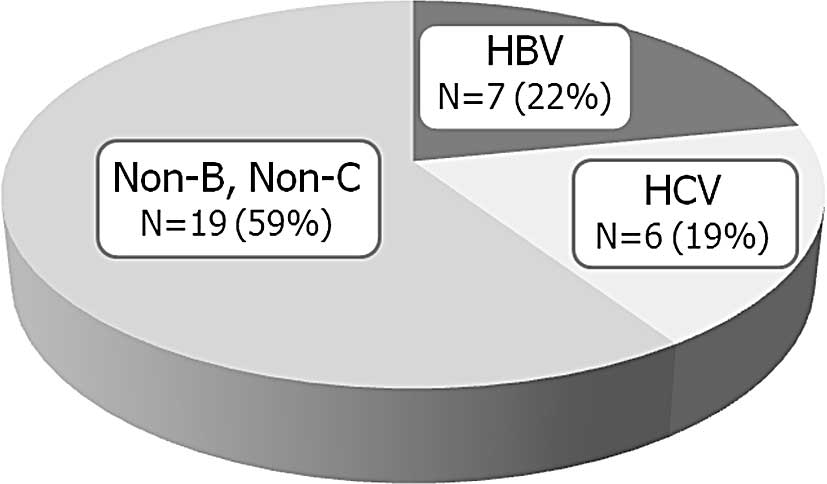
The etiology of the HCC cases is illustrated in
Fig. 1. Seven cases (22%) were
positive for HBsAg, 6 (19%) were positive for HCV-Ab and 19 (59%)
were negative for both HBsAg and HCV-Ab. Clinical data in the
hepatitis virus-related HCC (HV-HCC) cases were compared with those
in NBNC-HCC (Table I). The
activity and fibrosis scores of the background liver tissues in
HV-HCC were higher than those in NBNC-HCC. The average diameter of
the cancer in HV-HCC was smaller than that in NBNC-HCC.
 | Table I.Clinical characteristics of NBNC-HCC
and hepatitis virus-related HCC. |
Table I.
Clinical characteristics of NBNC-HCC
and hepatitis virus-related HCC.
| NBNC-HCC | HV-HCC | P-value |
|---|
| Cases (no.) | 19 | 13 | |
| Age (years) | 68.70±8.27 | 61.40±14.50 | 0.0794 |
| Tumor size (mm) | 74.70±36.20 | 48.80±29.60 | 0.0415a |
| Activity | 0.50±0.514 | 1.090±0.302 | 0.0019a |
| Fibrosis | 1.53±1.81 | 3.000±0.444 | 0.0061a |
| PLT
(×104/μl) | 20.40±9.80 | 15.90±8.01 | 0.1849 |
| ALT (IU/l) | 41.40±28.20 | 46.50±26.50 | 0.6151 |
| AFP (ng/ml) | 1,302±3,988 | 13,508±24,412 | 0.0984 |
| PIVKA-II
(mAU/ml) | 16,672±36,142 | 923±1,250 | 0.0738 |
Occult HBV infection in NBNC-HCC
To examine occult HBV infection, HBV-DNA was
assessed by PCR analysis from extracted liver tissue in NBNC-HCC.
The total characteristics of occult HBV were clinically compared
with those of non-occult HBV cases (Table II). No significant difference,
excluding the indocyanine green test, was detected between the two
groups.
 | Table II.Clinical data of NBNC-HCC between
occult HBV-infected cases and others. |
Table II.
Clinical data of NBNC-HCC between
occult HBV-infected cases and others.
| Occult HBV in
NBNC-HCC | Non-occult HBV in
NBNC-HCC | P-value |
|---|
| Cases (no.) | 7 | 12 | |
| Age (years) | 65.7±11.4 | 70.10±6.45 | 0.2925 |
| Tumor size (mm) | 84.7±46.3 | 70.2±10.2 | 0.4335 |
| Activity | 0.20±0.22 | 0.615±0.137 | 0.1284 |
| Fibrosis | 1.170±0.672 | 1.540±0.457 | 0.6530 |
| PLT
(×104/μl) | 24.40±3.91 | 18.50±2.68 | 0.2341 |
| ALT (IU/l) | 38.5±33.0 | 42.8±27.2 | 0.7690 |
| ICG-R 15% (%) | 11.30±1.62 | 17.80±2.01 | 0.0199a |
HBV-DNA in the pre-S/S region was amplified and
detected in 4 out of 7 cases (57%); the X region was detected in 4
cases (57%), and the core promoter/pre-core region was detected in
3 cases (43%). Mutation of codon 38 was detected in 1 case
(12,13). C1653T in the enhancer box α-region
was found in 3 out of 4 cases, and A1762T/G1764A in the basic core
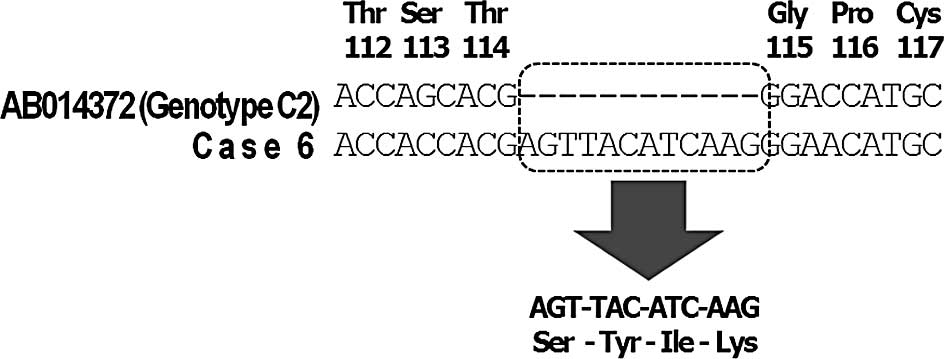
promoter region was detected in all 4 cases (Table III). A 12-nucleotide insertion
(4-amino acid insertion) in the S region was noted in 1 case
(Fig. 2). Phylogenetic analysis
showed that all 4 cases were grouped into genotype C (Fig. 3).
 | Table III.Mutation/variation of occult HBV
infection. |
Table III.
Mutation/variation of occult HBV
infection.
| Case no. | HBsAg | Pre-S/S | X region
| BCP region
|
|---|
| C1653T | T1753V | A1762T/G1764A |
|---|
| 2 | ND | ND | T | T | T/A |
| 3 | ND | ND | T | T | T/A |
| 5 | + | + | T | T | T/A |
| 6 | + | 4-AA insertion | ND | ND | ND |
| 11 | − | + | ND | ND | ND |
| 12 | ND | + | ND | ND | ND |
| 16 | + | ND | C | T | ND |
HBcAb was serologically examined in 12 out of 19
NBNC-HCC cases. Five out of 12 cases (42%) were positive for HBcAb,
which were thought to have been previously infected and resolved
naturally.
Discussion
Liver resection for HCC is a potentially curative
treatment. However, it has been reported that resection is possible
in only 30–35% of the HCC cases evaluated for surgical therapy due
to co-existing cirrhosis or the multicentric generation of HCC
(14). NBNC-HCC is estimated to
comprise 10% of HCC in Japanese patients. In this study, 19 out of
32 resected HCC cases (56%) were diagnosed as NBNC-HCC. It has been
suggested that cases of hepatic fibrosis of HCC with hepatitis
viral infection are usually more severe than those of NBNC-HCC. In
this study, HCC of more than 5 cm in diameter was present in 13 out
of 19 cases (68%) of NBNC-HCC. In addition, the average diameter of
NBNC-HCC was larger than that of HV-HCC. It is thought that HV-HCC
is diagnosed at an early stage, since patients with chronic
infection of hepatitis virus are regularly examined. It was
recently reported that the prevalence of non-alcoholic
steatohepatitis is increasing among NBNC-HCC cases (15). In this study, 5 cases (26%) showed
impaired glucose tolerance, and 3 cases (16%) were obese. Thus our
findings indicated that abnormal glucose tolerance was associated
with hepatocarcinogenesis in this study.
Occult HBV infection is diagnosed when HBsAg in
serum is negative and HBV-DNA in serum or liver is positive,
regardless of the positivity for anti-HBc. It is still
controversial whether or not occult HBV infection is associated
with liver damage. Althought it was reported that occult HBV
infection does not progress to severe liver disease (16), another study showed that occult HBV
infection caused liver disease progression in cases of chronic
infection of HCV (17,18). In this study, the activity and
fibrosis scores in the HCC cases with occult HBV infection were
lower than those in HV-HCC. Thus, our data indicated that occult
HBV infection is not related to the progression of liver disease.
As for hepatocarcinogenesis, it is also controversial whether or
not occult HBV infection is associated with HCC. Although it was
reported that occult HBV infection is an independent risk factor
for carcinogenesis in patients with HCV (19,20),
another study showed that occult HBV infection was not related to
hepatocarcinogenesis (21).
Several studies have demonstrated that the basic
core promoter region in the HBV genome is associated with
hepatocarcinogenesis (22,23). In Japan, it was reported that
C1653T and T1753V are associated with HCC (24,25).
Although T1753V was detected in only 1 case, C1653T and
A1762T/G1764A were relatively common in this study. It was also
reported that a mutation of the X region is associated with HCC
(12,13), but the codon 38 mutation was
detected in only 1 case and no mutation was found in codon 31 in
the present study. It is probable that the mutation/variation in
occult HBV infection in relation to HCC is different from that in
HBsAg-positive cases. A recent study showed that virological
factors of HBV related to HCC are different between occult
HBV-infected and HBsAg-positive patients, and G1721A, M1I and Q2K
in the pre-S2 gene might be useful viral markers for HCC in occult
HBV carriers (24).
In this study, a 12-nucleotide insertion (4-amino
acid insertion) between nt 114 and 115 in the S region was found in
1 case (Fig. 2). We considered
that the α-loop region in the S region was important for the
conformation of the S antigen, and amino acid substitutions at 122,
123, 126, 141 and 145 were found in the vaccine-escaping mutant. It
was possible that the insertion caused the conformational change to
the S antigen, and this resulted in the serological HBsAg
negativity in this case. It has also been reported that several
insertions in the HBx and Pre-S2 regions are associated with HCC
(25,26), but there were no such mutations
identified in this study.
In the present study, phylogenetic analysis revealed
that all cases of occult HBV infection were grouped into genotype
C. Since HBV genotype C is the most prevalent in Japanese patients,
it is reasonable that occult HBV in Japan is derived from genotype
C.
In conclusion, HBV-DNA was frequently found in the
NBNC-HCC tissues. No significant difference was noted between
occult HBV and other forms of infection among the NBNC-HCC cases.
Therefore, further investigation is necessary to assess whether
occult HBV infection is related to hepatocarcinogenesis.
Acknowledgements
This study was supported by a
grant-in-aid through the Program of Founding Research Centers for
Emerging and Reemerging Infectious Diseases, the Ministry of
Education, Culture, Sports, Science and Technology (MEXT),
Japan.
References
|
1.
|
Lee WM: Hepatitis B virus infection. N
Engl J Med. 337:1733–1743. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Tang B, Kruger WD, Chen G, et al:
Hepatitis B viremia is associated with increased risk of
hepatocellular carcinoma in chronic carriers. J Med Virol.
72:35–40. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Harris RA, Chen G, Lin WY, Shen FM, London
WT and Evans AA: Spontaneous clearance of high-titer serum HBV DNA
and risk of hepatocellular carcinoma in a Chinese population.
Cancer Causes Control. 14:995–1000. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Shih LN, Sheu JC, Wang JT, et al: Serum
hepatitis B virus DNA in healthy HBsAg-negative Chinese adults
evaluated by polymerase chain reaction. J Med Virol. 32:257–260.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Marusawa H, Uemoto S, Hijikata M, et al:
Latent hepatitis B virus infection in healthy individuals with
antibodies to hepatitis B core antigen. Hepatology. 31:488–495.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Tuttleman JS, Pourcel C and Summers J:
Formation of the pool of covalently closed circular viral DNA in
hepadnavirus-infected cells. Cell. 47:451–460. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Seeger C and Mason WS: Hepatitis B virus
biology. Microbiol Mol Biol Rev. 64:51–68. 2000. View Article : Google Scholar
|
|
8.
|
Ichida F, Tsuji T, Omata M, et al: New
Inuyama Classification; new criteria for histological assessment of
chronic hepatitis. Int Hepatol Commun. 6:112–119. 1996. View Article : Google Scholar
|
|
9.
|
Sugauchi F, Mizokami M, Orito E, et al: A
novel variant genotype C of hepatitis B virus identified in
isolates from Australian Aborigines: complete genome sequence and
phylogenetic relatedness. J Gen Virol. 82:883–892. 2001.
|
|
10.
|
Sugauchi F, Ohno T, Orito E, et al:
Influence of hepatitis B virus genotypes on the development of preS
deletions and advanced liver disease. J Med Virol. 70:537–544.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Tamura K, Dudley J, Nei M and Kumar S:
MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software
version 4.0. Mol Biol Evol. 24:1596–1599. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Yeh CT, Shen CH, Tai DI, Chu CM and Liaw
YF: Identification and characterization of a prevalent hepatitis B
virus X protein mutant in Taiwanese patients with hepatocellular
carcinoma. Oncogene. 19:5213–5220. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Muroyama R, Kato N, Yoshida H, et al:
Nucleotide change of codon 38 in the X gene of hepatitis B virus
genotype C is associated with an increased risk of hepatocellular
carcinoma. J Hepatol. 45:805–812. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Sotiropoulos GC, Lang H, Frilling A, et
al: Resectability of hepatocellular carcinoma: evaluation of 333
consecutive cases at a single hepatobiliary specialty center and
systematic review of the literature. Hepatogastroenterology.
53:322–329. 2006.
|
|
15.
|
Hatanaka K, Kudo M, Fukunaga T, et al:
Clinical characteristics of NonBNonC-HCC: comparison with HBV and
HCV-related HCC. Intervirology. 50:24–31. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Kao JH, Chen PJ, Lai MY and Chen DS:
Occult hepatitis B virus infection and clinical outcomes of
patients with chronic hepatitis C. J Clin Microbiol. 40:4068–4071.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Mrani S, Chemin I, Menouar K, et al:
Occult HBV infection may represent a major risk factor of
non-response to antiviral therapy of chronic hepatitis C. J Med
Virol. 79:1075–1081. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Tamori A, Nishiguchi S, Kubo S, et al:
Possible contribution to hepatocarcinogenesis of X transcript of
hepatitis B virus in Japanese patients with hepatitis C virus.
Hepatology. 29:1429–1434. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Pollicino T, Squadrito G, Cerenzia G, et
al: Hepatitis B virus maintains its pro-oncogenic properties in the
case of occult HBV infection. Gastroenterology. 126:102–110. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Tamori A, Hayashi T, Shinzaki M, et al:
Frequent detection of hepatitis B virus DNA in hepatocellular
carcinoma of patients with sustained virologic response for
hepatitis C virus. J Med Virol. 81:1009–1014. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Kusakabe A, Tanaka Y, Orito E, et al: A
weak association between occult HBV infection and non-B non-C
hepatocellular carcinoma in Japan. J Gastroenterol. 42:298–305.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Kim JK, Chang HY, Lee JM, et al: Specific
mutations in the enhancer II/core promoter/precore regions of
hepatitis B virus subgenotype C2 in Korean patients with
hepatocellular carcinoma. J Med Virol. 81:1002–1008. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Kao JH, Chen PJ, Lai MY and Chen DS: Basal
core promoter mutations of hepatitis B virus increase the risk of
hepatocellular carcinoma in hepatitis B carriers. Gastroenterology.
124:327–334. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Shinkai N, Tanaka Y, Ito K, et al:
Influence of hepatitis B virus X and core promoter mutations on
hepatocellular carcinoma among patients infected with subgenotype
C2. J Clin Microbiol. 45:3191–3197. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Tanaka Y, Mukaide M, Orito E, et al:
Specific mutations in enhancer II/core promoter of hepatitis B
virus subgenotypes C1/C2 increase the risk of hepatocellular
carcinoma. J Hepatol. 45:646–653. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Chen CH, Changchien CS, Lee CM, et al: A
study on sequence variations in pre-S/surface, X and enhancer
II/core promoter/precore regions of occult hepatitis B virus in
non-B, non-C hepatocellular carcinoma patients in Taiwan. Int J
Cancer. 125:621–629. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Tabor E: Pathogenesis of hepatitis B
virus-associated hepatocellular carcinoma. Hepatol Res. 37:110–114.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Lupberger J and Hildt E: Hepatitis B
virus-induced oncogenesis. World J Gastroenterol. 13:74–81. 2007.
View Article : Google Scholar
|