Introduction
Hepatitis C virus (HCV) affects approximately
130–170 million people worldwide (1), however, no vaccines are available.
It is an important cause of chronic hepatitis, cirrhosis,
hepatocellular carcinoma (HCC), leading to a need for liver
transplantation (2,3). Treatment of chronic HCV is currently
based on the combination of pegylated interferon (IFN)-α and the
nucleotide analogue ribavirin, which is only effective in
approximately 50% of the patients, especially in HCV genotype 1
(4,5). HCV belongs to the Hepacivirus
genus within the Flaviviridae family, and is a
positive-stranded RNA virus with a genome of ∼9.6 kb. The HCV
genome contains a single open reading frame (ORF) encoding a large
polyprotein precursor of 3011 amino acids. The ORF is flanked by 5′
and 3′ untranslated regions. The precursor polyprotein is processed
by cellular and viral proteases into 10 proteins: structural (core,
E1 and E2), and non-structural proteins (p7, NS2, NS3, NS4A, NS4B,
NS5A and NS5B) (3,6). There are six major genotypes in HCV
classification (3). The major
prevalent type in Southern Taiwan is HCV 1b, which is the most
resistant type to interferon therapy (5,7).
Curcumin, derived from eastern traditional
medicines, Curcuma longa, has been found to have a variety
of beneficial properties, such as anti-inflammatory, antioxidant,
chemopreventive and chemotherapeutic activities (8,9).
Its multiple-target characteristics influence several activities of
intracellular molecules, including transcription nuclear factor-κB
(NF-κB), pro-inflammatory cyclooxygenase-2 and MAPK inhibitions, as
well as heme oxygenase-1 induction (9). In the antivirus bioactivity, certain
reports have indicated that curcumin showed anti-viral activity
against the human immunodeficiency (10,11), the coxsackie- (12) and the hepatitis B (HBV) viruses
(13). In the anti-HCV study, one
report showed that curcumin inhibited a lipogenic transcription
factor, sterol regulatory element binding protein-1
(SREBP-1)-induced HCV replication via the inhibition of the
PI3K-AKT pathway (14).
The catabolism of heme by heme oxygenase (HO)
resulted in the production of biliverdin, carbon monoxide and free
iron. HO-1, one of the phase II enzymes, is an enzyme in cells with
cytoprotective properties against oxidative damage (15) that has been reported to be induced
by the Nrf2 transcription factor (16). Curcumin-induced HO-1 expression
was first found in human endothelial cells (17), suggesting that a low dose of
curcumin induced HO-1 expression, which provided an intrinsic
antioxidant ability. Curcumin also induced HO-1 expression in
mesangial (18) and liver cells
(19–21), as well as in macrophages (22,23). The induction or overexpression of
HO-1 has been shown to interfere with the replication of certain
viruses, such as the human immunodeficiency virus (24), the HBV (25) and the HCV (26–28).
The properties of the transcription factor NF-κB are
extensively exploited in cells (29). In general, NF-κB is of great
importance in signal transduction pathways involved in chronic and
acute inflammatory diseases, as well as various types of cancer,
therefore, it is a good target for cancer prevention (30). Various reports have demonstrated
the correlation between curcumin and NF-κB. One of those reports
suggests the anti-inflammatory effect of curcumin, which suppresses
the ox-LDL-induced MCP-1 expression via the p38 MAPK and NF-κB
pathways in rat vascular smooth muscle cells (31). The anti-inflammatory effect of
curcumin has been reported to be due to the IκB/NF-κB system in rat
and human intestinal epithelial cells, including IEC-6, HT-29 and
Caco-2 cells (32). Curcumin has
also been found to have anti-metastatic properties via the
inhibition of NF-κB in the highly invasive and metastatic
MDA-MB-231 breast cancer cell line (33). Another signaling pathway,
Raf/MEK/extracellular signal-regulated kinases (ERK), is of crucial
importance in the regulation of cell growth, differentiation,
survival, as well as the transmission of oncogenic signals
(34). This pathway has also been
reported to be a target of curcumin. For example, curcumin
inhibited connective tissue growth factor gene expression by
suppressing ERK signaling in activated hepatic stellate cells
(35). Moreover, curcumin
inhibited phorbol myristate acetate-induced MCP-1 gene expression
by inhibiting ERK and NF-κB activities in U937 cells (36). However, the manner in which
curcumin affects the activities of NF-κB and ERK in HCV-infected
hepatoma cells has yet to be determine.
Only one study suggesting that curcumin inhibited
HCV replication by suppressing the AKT-SREBP-1 pathway is currently
available (14). In this study,
the correlation between curcumin-inhibited HCV replication, HO-1,
AKT, ERK and NF-κB molecules was examined.
Materials and methods
Cell culture and reagents
Huh7.5 cells expressing the HCV genotype 1b
subgenomic replicon (Con1/SG-Neo(I) hRlucFMDV2aUb) containing
Renilla luciferase reporter, kindly provided by Apath, were
cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10%
fetal bovine serum (FBS), 100 U/ml penicillin, 100 mg/ml
streptomycin and 0.5 mg/ml G418. The nuclear extraction kit was
purchased from Chemicon (Temecula, CA, USA). Curcumin (Acros
Organics, Geel, Belgium), LY294002, U0126 and Ro1069920 were
purchased from Tocris (Bristol, UK), and dissolved in dimethyl
sulfoxide (DMSO), then added into culture medium containing 0.1%
DMSO.
Cell viability assay
Cell viability was determined by colorimetric MTT
assay. Cells were cultured on 24-well plates at a density of
1×105 cells/well. After 24 h, the cells were incubated
with varying concentrations of curcumin or 0.1% DMSO for another 24
h. MTT was added to medium for 2 h, the medium was discarded and
DMSO was then added to dissolve the formazan product. Each well was
measured by light absorbance at 490 nm. The result was expressed as
a percentage, relative to the 0.1% DMSO-treated control group.
Luciferase reporter assay
Cells were subcultured at a density of
4×105 cells/well in 1 ml of culture medium in a 12-well
plastic dish for 6 h. Curcumin or DMSO was added to the medium for
24 h. The cells were lysed and cell lysates were prepared for a
Renilla luciferase assay (Promega, Madison, WI, USA) and
protein concentration assays, with Bio-Rad protein assay (Bio-Rad,
Hercules, CA, USA). The relative luciferase activities were
normalized to the same protein concentration.
Real-time RT-PCR analysis
Total RNA was isolated from Huh7.5 cells expressing
the HCV genotype 1b subgenomic replicon. Reverse transcription (RT)
was performed on 2 μg of total RNA by 1.5 μM random hexamer and
RevertAid™ reverse transcriptase (Fermentas, Glen Burnie, MD, USA).
Then, 1/20 volume of reaction mixture was used for quantitative
real-time PCR with HCV specific primers: 5′-AGCGTCTAGCCATGGCGT-3′
and 5′-GGTGTACTCACCGGTTCCG-3′, and GAPDH specific primers:
5′-CGGATTTGGTCGTATTGG-3′ and 5′-AGATGGT GATGGGATTTC-3′, as the
endogenous control. The quantitative real-time PCR was followed by
Maxima™ SYBR-Green qPCR Master Mix (Fermentas). Real-time PCR
reactions contained optimal volume of the reverse transcription
mixture, 600 nM each forward and reverse primer and 1X SYBR-Green
qPCR Master Mix in 25 μl. Reactions were incubated for 40 cycles in
an ABI GeneAmp® 7500 Sequence Detection System, with an
initial denaturization step at 95°C for 10 min, followed by 40
cycles of 95°C for 15 sec and 63°C for 1 min. PCR product
accumulation was monitored at several points during each cycle, by
measuring the increase in fluorescence. Gene expression changes
were assessed using the comparative Ct method. The relative amounts
of mRNA for HCV were optimized by subtracting the Ct values of HCV
from the Ct values of GAPDH mRNA (ΔCt). The ΔCt of the control
group was then subtracted from the ΔCt of the curcumin-treated
groups (ΔΔCt). Data were expressed as relative levels of HCV
RNA.
Western blotting
For western blotting, analytical 10% sodium dodecyl
sulfate (SDS)-polyacrylamide slab gel electrophoresis was
performed. Tissue extracts were prepared and a 30–60 μg aliquot of
protein extracts was analyzed. For immunoblotting, proteins in the
SDS-PAGE gels were transferred to a polyvinylidene difluoride
membrane using a trans-blot apparatus. Antibodies against HCV NS5A
and HCV NA5B (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA),
HO-1 (Assay Designs, Inc., Ann Arbor, MI, USA), pAKT (308) and pERK
(Santa Cruz Biotechnology, Inc.), NF-κB (Cell Signaling Technology,
Beverly, MA, USA), Sp1 (Millipore, Darmstadt, Germany), α-tubulin
(GeneTex, Inc., Irvine, CA, USA) and β-actin (Sigma-Aldrich, St.
Louis, MO, USA) were used as the primary antibodies. Mouse, rabbit
or goat IgG antibodies coupled with horseradish peroxidase were
used as the secondary antibodies. An enhanced chemiluminescence kit
and VL Chemi-Smart 3000 were used for detection, while the quantity
of each band was determined using MultiGauge software.
HO-1 knockdown by siRNA
Cells (3×106) were seeded in 10-cm dishes
for 6 h, then negative control small interfering (siRNA) (10 nM) or
HO-1 siRNA (10 nM) (Invitrogen) was transfected into cells using
the RNAiMAX Transfection Reagent (Invitrogen), according to the
manufacturer’s instructions. Subsequent to adding siRNA for 6 h,
the medium was changed to fresh condition medium for 18 h. Then the
transfected cells were then analyzed by western blotting.
Statistical analysis
Data were expressed as the mean ± SE. Statistical
evaluation was carried out by one-way ANOVA followed by Dunn’s
test. All statistics were calculated using SigmaStat version 3.5
(Systat Software). P<0.05 was considered to indicate a
statistically significant difference.
Results
Cytotoxicity of curcumin in Huh7.5 cells
expressing the HCV genotype 1b subgenomic replicon (Huh7.5-HCV
cells)
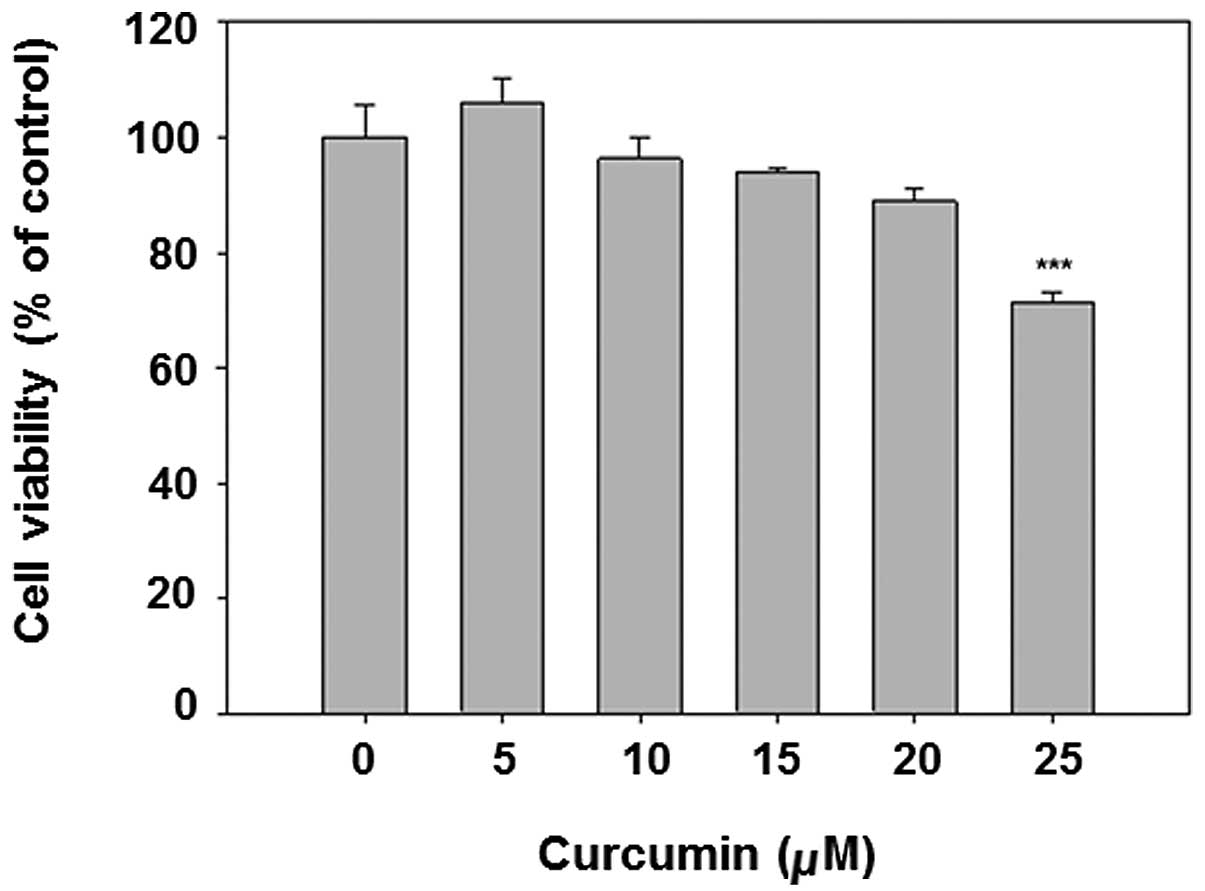
Curcumin is known to be an anticancer chemical at
high doses. To avoid the obvious cytotocicity in the subsequent
experiments, the MTT assay was applied for cytotoxicity analysis.
The results show that curcumin dose-dependently decreased cell
viability (Fig. 1). The dose
<20 μM was selected for subsequent analysis, given that the
viability of 25 μM curcumin treatment is <80%.
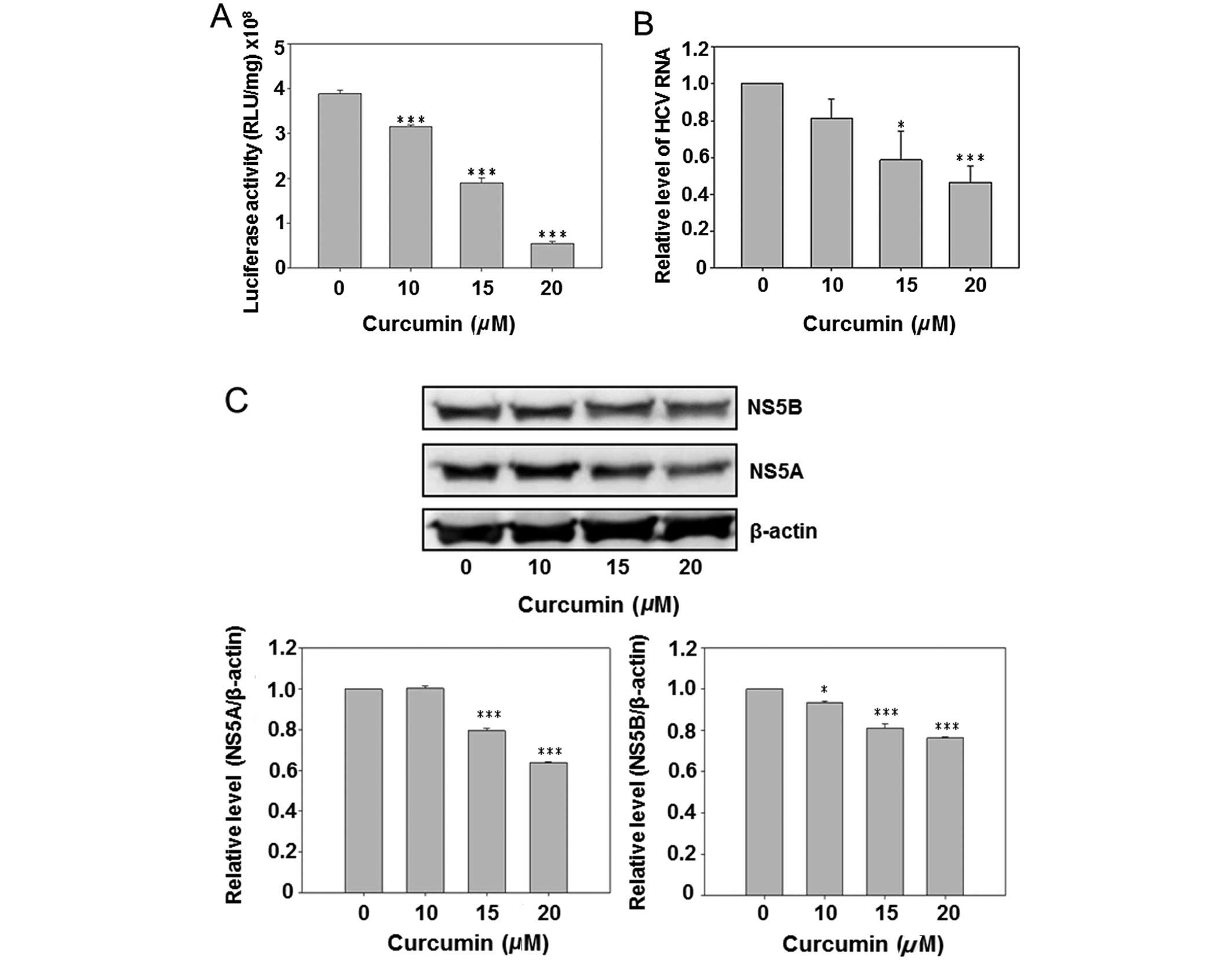
Curcumin reduced HCV replication and HCV
protein expression
Due to the presence of a luciferase reporter gene in
the HCV subgenomic replicon of Con1/SG-Neo(I)hRlucFMDV2aUb, the
culture medium luciferase activity was first analyzed subsequent to
curcumin treatment. The results show that curcumin dose-dependently
inhibited luciferase activity (Fig.
2A). However, the HCV RNA was also detected by real-time PCR.
Curcumin also reduced the intracellular HCV RNA expression in a
dose-dependent manner. Subsequent to curcumin treatment the
HCV-specific protein NS5A and NS5B were detected by western blot
analysis, indicating that curcumin dose-dependently inhibited
expression of the NS5A and NS5B. The above data suggest that
curcumin inhibited HCV replication in hepatoma cells.
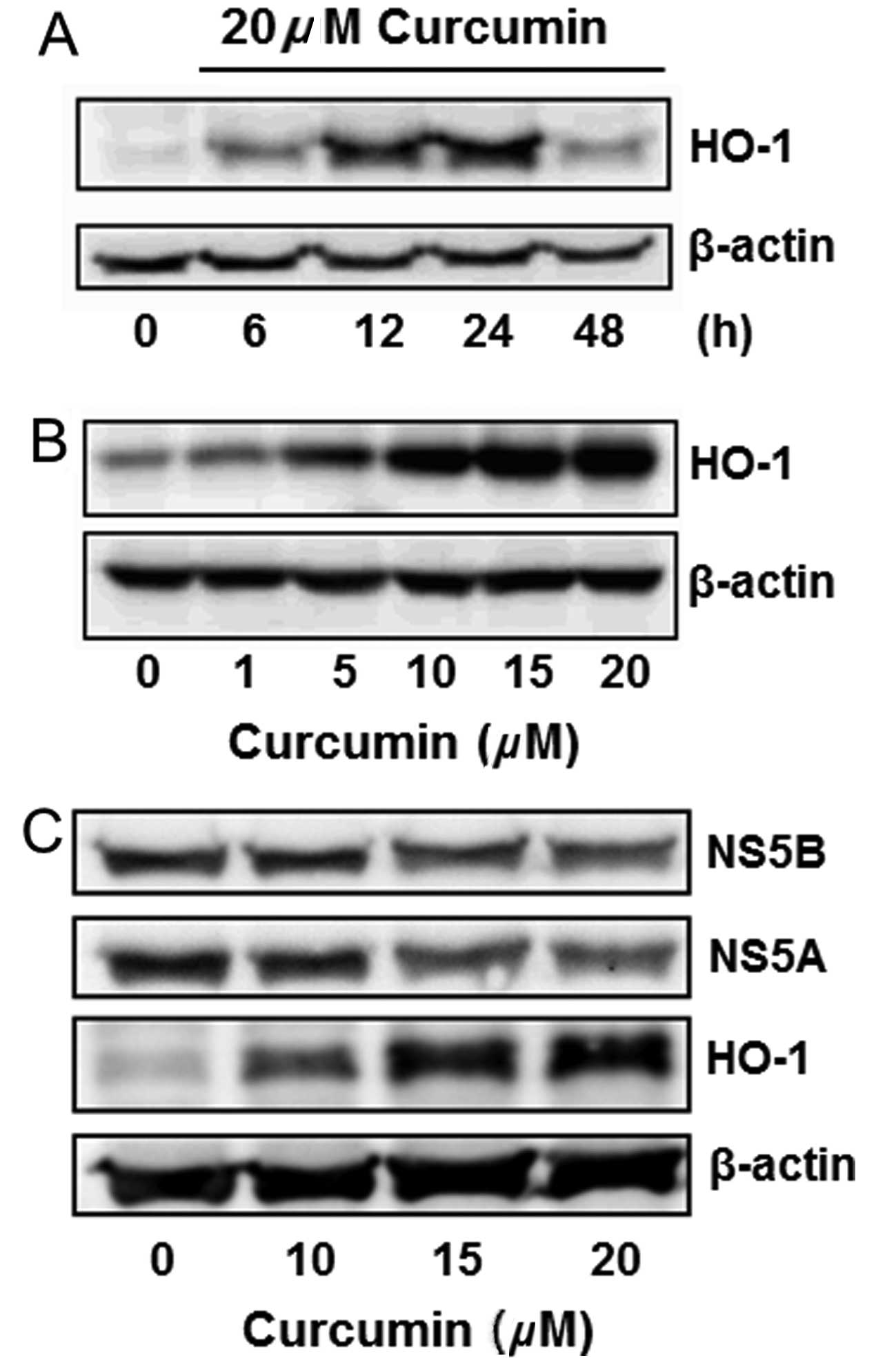
Curcumin induced HO-1 protein
expression
Curcumin is known to induce HO-1 expression in
various cells. This effect was analyzed in Huh7.5-HCV cells.
Curcumin slightly induced HO-1 expression in a 6-h treatment, while
significantly inducing it in 12 and 24 h. The HO-1 induction
declined after treatment for 48 h (Fig. 3A). Curcumin also induced HO-1
expression in a dose-dependent manner (Fig. 3B). The change of NS5A, NS5B and
HO-1 protein expressions was simultaneously detected by western
blot analysis, indicating that curcumin dose-dependently inhibited
the expression of NS5A and NS5B, while increasing the HO-1
expression (Fig. 3C).
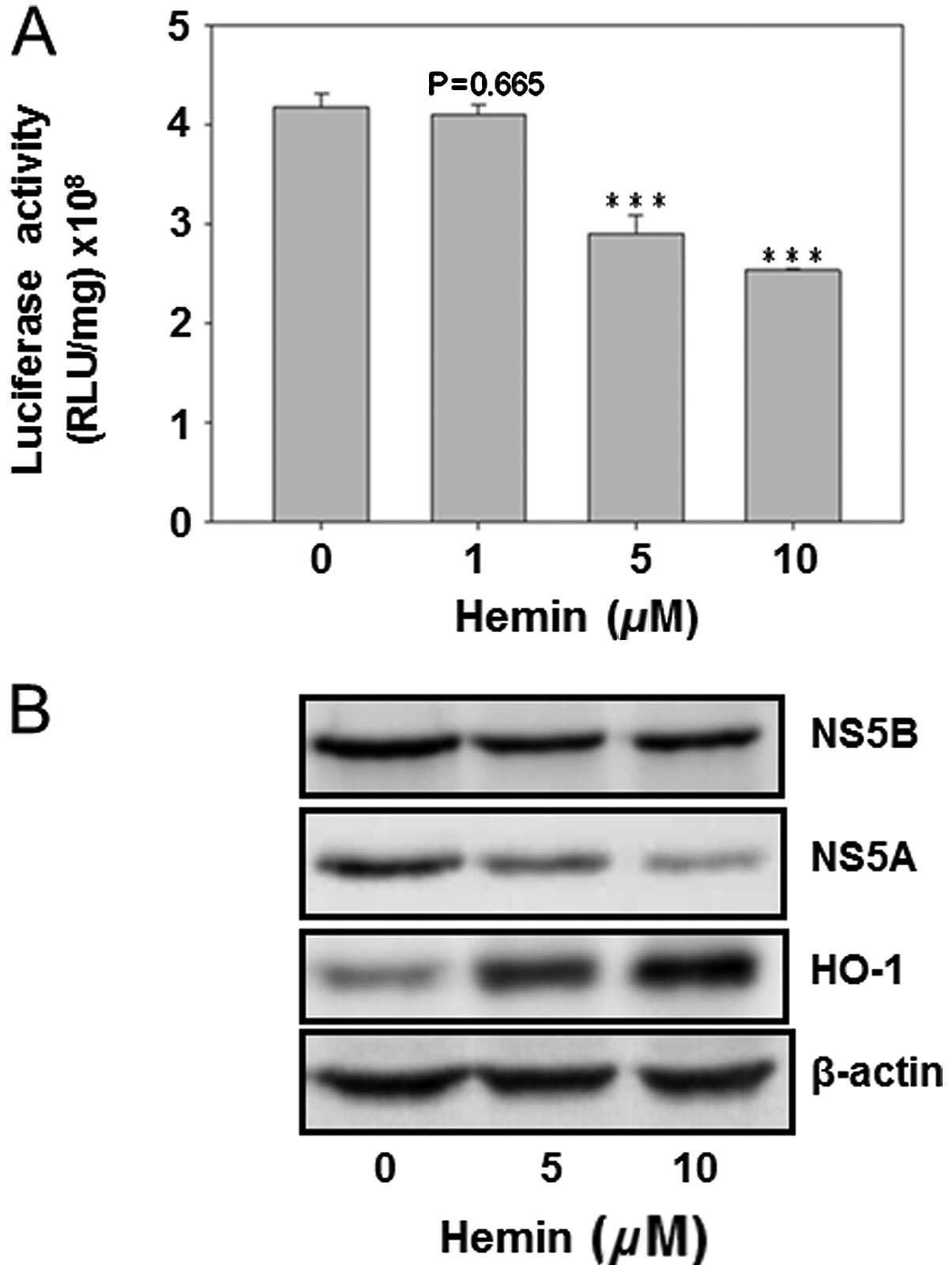
Hemin reduced HCV replication and the HCV
protein expression
The HO-1 inducer hemin was used to analyze its
effect on HCV replication as well as on the protein expression of
HCV NS5A and NS5B. The result showed that hemin dose-dependently
decreased HCV replication (Fig.
4A). Furthermore, curcumin inhibited the protein expression of
NS5A and NS5B, while enhancing the HO-1 protein expression. This
finding suggested that HO-1 protein inhibited HCV replication in
Huh7.5-HCV cells (Fig. 4).
HO-1 knockdown partially reversed the
curcumin-reduced viral protein expression
In order to prove the direct relationship between
curcumin-induced HO-1 and curcumin-inhibited HCV replication, the
HO-1 specific siRNA was used for analysis. HO-1 siRNA significantly
inhibited basal and curcumin-induced HO-1 expression (Fig. 5A). HO-1 knockdown slightly
increased the NS5A and NS5B protein expressions in the basal
condition. At the same time, it partially but significantly
reversed the curcumin-inhibited the expression of NS5A and NS5B,
suggesting that curcumin-induced HO-1 was involved in
curcumin-inhibited HCV replication, while having additional
mechanisms regarding the anti-HCV effect of curcumin.
 | Figure 5.The role of HO-1, AKT, ERK and NF-κB
on curcumin-inhibited HCV protein expression is shown. (A)
Knockdown of HO-1 partially reversed curcumin-inhibited HCV protein
expression. Cells (3×106) were seeded in a 10-cm dish
for 6 h, and negative control small interfering (siRNA) (10 nM) or
HO-1 siRNA (10 nM) was transfected into cells. Subsequent to a 6-h
addition of siRNA, the medium was changed to fresh condition medium
for 18 h, and the transfected cells were analyzed by western
blotting (*P<0.05 and ***P<0.001, in 2 groups,
respectively). (B) Curcumin inhibited AKT, ERK and NF-κB. Cells
were subcultured at a density of 1.5×106 cells in 8 ml
of culture medium in a 10-cm plastic dish for 6 h. Curcumin or DMSO
was added to the medium for 24 h. Total cell lysates (up) or
cytosol-nuclear fraction (down) were isolated by western blot
analysis. Sp1 is a dominant nuclear protein and α-tubulin is a
cytosolic protein. (C) Effect of AKT, ERK and NF-κB inhibitors on
the HCV protein expression is shown. Cells were subcultured at a
density of 1.5×106 cells in 8 ml of culture medium in a
10-cm plastic dish for 6 h. Chemical (LY, LY294002; U0, U0126; Ro,
Ro1069920) or DMSO was added to the medium for 24 h. Total cell
lysates were isolated for western blot analysis.
(***P<0.001 compared to control). The experiments
were repeated three times. |
Effect of the PI3K-AKT, MEK-ERK and NF-κB
pathways on curcumin-inhibited HCV replication
Fig. 5A shows that
HO-1 is partially involved in curcumin-inhibited HCV replication.
Additional signaling pathways affected by curcumin were analyzed,
demonstrating that curcumin inhibited the protein phosphorylation
of ERK and AKT, as well as the cytoplasmic protein expression of
NF-κB (Fig. 5B). Therefore, the
specific inhibitors of PI3K-AKT (LY294002), MEK-ERK (U0126) and
NF-κB (Ro 106-9920) were used to identify the role of AKT, ERK and
NF-κB in the HCV protein expression. Fig. 5C shows that curcumin was the only
chemical to induce the HO-1 expression. Of the three inhibitors,
only PI3K-AKT LY294002 slightly inhibited the HCV protein
expression, while MEK-ERK U0126 and NF-κB inhibitors Ro 1069920 had
a slight effect on increasing the HCV protein expression,
suggesting that curcumin-inhibited HCV replication was also
partially mediated via PI3K-AKT inhibition.
Discussion
Curcumin is a common chemical ingredient of curry.
It has, however, been studied in clinical trials regarding its
applicability in treating patients suffering from pancreatic and
colon cancer, as well as multiple myeloma (37). In Taiwan, several doctors of
traditional Chinese medicine consider curcumin to be beneficial for
patients suffering from hepatitis. The results of this study
demonstrate that curcumin inhibits HCV replication in cellular
analysis, and its mechanism partially occurs through HO-1 induction
and PI3K-AKT inhibition.
HO-1, a curcumin-induced gene, is thought to be a
potential therapeutic protein for the re-establishment of
homeostasis in several pathologic conditions (38) and is also involved in inhibiting
HCV replication (28). The HO-1
products biliverdin and iron contribute to certain anti-HCV
mechanisms of HO-1 (26,39,40). In this study, HO-1 knockdown
partially reversed curcumin-inhibited HCV replication, supporting
the evidence for the anti-HCV effect of HO-1. Since HO-1 is induced
by ROS or certain electrophiles, ROS has also been reported to
inhibit HCV replication (41,42). Arsenic trioxide-inhibited HCV
replication is also suggested to be mediated through the induction
of oxidative stress (43). HO-1,
an oxidative stress-induced gene, may be involved in the
ROS-inhibited HCV replication.
As a downstream kinase of PI3K, AKT is an important
molecule in regulating a wide range of signaling pathways (44). In HCV-infected cells, the PI3K-AKT
signaling pathway is involved in certain pathological mechanisms.
For example, the activities of PI3K, AKT and their downstream
target mTOR are increased in the HCV-replicating cells (45). HCV NS5A binds to PI3K, while
enhancing the phosphotransferase activity of the catalytic domain
(46). The HCV-activated PI3K-AKT
contributes to cell survival enhancement. In addition to cell
survival, AKT leads to the protein accumulation of SREBP-1, an
important transcription factor regulating genes involved in fatty
acid and cholesterol synthesis (47). HCV NS4B has been found to enhance
the protein expression levels of SREBPs and fatty acid synthase
through PI3K activity, subsequently inducing a lipid accumulation
in hepatoma cells (48).
Therefore, inhibition of the PI3K-SREBP signaling pathway should
decrease the HCV-induced HCC development and the cellular fatty
acid level. Curcumin has been reported to inhibit HCV replication
via suppression of the AKT-SREBP-1 pathway (14). In the present study, data also
demonstrated that curcumin-inhibited PI3K-AKT was slightly involved
in the anti-HCV activity of curcumin.
Activation of the MEK-ERK signal cascade enhances
the replication of viruses, such as the human immunodeficiency
(49), the influenza (50), the corona- (51) and the herpes simplex viruses
(52). By contrast, in the case
of HBV, activation of MEK-ERK signaling led to the inhibition of
HBV replication (53). In the HCV
study, interleukin-1 has been reported to have the potential to
effectively inhibit HCV replication and protein expression by
activating the ERK signaling pathway (54). HCV IRES-dependent protein
synthesis was enhanced by MEK-ERK inhibitor PD98059 (55). Another report also suggests that
inhibition of MEK-ERK signaling leads to the upregulation of HCV
replication and protein production (56). Consistent with the results of the
present study, those findings confirm that the curcumin-inhibited
MEK-ERK signaling pathway contributes to the increase of HCV
replication.
NF-κB, one of the major signaling transduction
molecules activated in response to oxidative stress, is able to
modulate the transcription of a large number of downstream genes.
The HCV core protein has been shown to activate NF-κB, inducing
resistance to TNF-α-induced apoptosis in hepatoma cells (57). HCV NS2 activates the IL-8 gene
expression by activating the NF-κB pathway in HepG2 cells (58). In the infectious JFH1 model, HCV
is suggested to enhance hepatic fibrosis progression through the
induction of TGF-β1, mediated by a ROS-induced and NF-κB-dependent
pathway (59). These evidences
indicate that the activation of NF-κB by HCV induces hepatic
disease progression. In this study, the NF-κB expression is
abundant in the cytoplasm of Huh7.5 cells, expressing the HCV
genotype 1b subgenomic replicon (Fig.
5B). The absence of NF-κB nuclear translocation indicates that
NF-κB is not likely to participate in the mechanism of
hepatocarcinogenesis in this cell line. The absense of complete HCV
core and HCV NS2 sequences in the subgenomic replicon used in this
study, is likely to be the reason for the absence of NF-κB nuclear
translocation. Therefore, it is likely to contribute to the
inability of the NF-κB inhibitor to suppress the HCV protein
expression in this cell line. In fact, the genomic variation of HCV
core protein generates a distinct functional regulation of NF-κB,
which may inhibit or activate NF-κB activity (60).
In certain reports, the inhibition of NF-κB shows
anti-HCV activity: for example, the Acacia confusa (61) and San-Huang-Xie-Xin-Tang extracts
(62) suppress HCV replication
associated with NF-κB inhibition. In the present study,
curcumin-inhibited NF-κB does not have any benefit in anti-HCV
activity. Thus, the presence or absence of the inhibition of NF-κB
in anti-HCV therapy is likely to depend on the activation status of
NF-κB, although additional investigations are required on the
subject.
In conclusion, this study proved that curcumin
inhibits HCV replication through the induction of the HO-1
expression and the inhibition of the PI3K-AKT signaling pathway.
However, the curcumin-inhibited MEK-ERK mechanism contributes
negatively to its anti-HCV activity.
Acknowledgements
This study was financed by grants from
the National Science Council (NSC98-2320-B-415-002-MY3) and from
the Chiayi Christian Hospital, Taiwan.
References
|
1.
|
D LavanchyThe global burden of hepatitis
CLiver Int29Suppl 1S74S81200910.1111/j.1478-3231.2008.01934.x
|
|
2.
|
N BostanT MahmoodAn overview about
hepatitis C: a devastating virusCrit Rev
Microbiol3691133201010.3109/1040841090335745520345213
|
|
3.
|
D MoradpourF PeninCM RiceReplication of
hepatitis C virusNat Rev
Microbiol5453463200710.1038/nrmicro1645
|
|
4.
|
JJ FeldJH HoofnagleMechanism of action of
interferon and ribavirin in treatment of hepatitis
CNature436967972200510.1038/nature0408216107837
|
|
5.
|
S MunirS SaleemM IdreesHepatitis C
treatment: current and future perspectivesVirol
J7296201010.1186/1743-422X-7-29621040548
|
|
6.
|
BD LindenbachCM RiceUnravelling hepatitis
C virus replication from genome to
functionNature436933938200510.1038/nature0407716107832
|
|
7.
|
CM LeeCH HungSN LuViral etiology of
hepatocellular carcinoma and HCV genotypes in
TaiwanIntervirology497681200610.1159/00008726716166793
|
|
8.
|
H HatcherR PlanalpJ ChoFM TortiSV
TortiCurcumin: from ancient medicine to current clinical trialsCell
Mol Life Sci6516311652200810.1007/s00018-008-7452-418324353
|
|
9.
|
A GoelAB KunnumakkaraBB AggarwalCurcumin
as ‘Curecumin’: from kitchen to clinicBiochem
Pharmacol757878092008
|
|
10.
|
CJ LiLJ ZhangBJ DezubeCS CrumpackerAB
PardeeThree inhibitors of type 1 human immunodeficiency virus long
terminal repeat-directed gene expression and virus replicationProc
Natl Acad Sci USA9018391842199310.1073/pnas.90.5.18398446597
|
|
11.
|
A MazumderK RaghavanJ WeinsteinKW KohnY
PommierInhibition of human immunodeficiency virus type-1 integrase
by curcuminBiochem
Pharmacol4911651170199510.1016/0006-2952(95)98514-A7748198
|
|
12.
|
X SiY WangJ WongJ ZhangBM McManusH
LuoDysregulation of the ubiquitin-proteasome system by curcumin
suppresses coxsackievirus B3 replicationJ
Virol8131423150200710.1128/JVI.02028-0617229707
|
|
13.
|
MM RechtmanO Har-NoyI Bar-YishayCurcumin
inhibits hepatitis B virus via down-regulation of the metabolic
coactivator PGC-1alphaFEBS
Lett58424852490201010.1016/j.febslet.2010.04.06720434445
|
|
14.
|
K KimKH KimHY KimHK ChoN SakamotoJ
CheongCurcumin inhibits hepatitis C virus replication via
suppressing the Akt-SREBP-1 pathwayFEBS
Lett584707712201010.1016/j.febslet.2009.12.01920026048
|
|
15.
|
LE OtterbeinMP SoaresK YamashitaFH
BachHeme oxygenase-1: unleashing the protective properties of
hemeTrends
Immunol24449455200310.1016/S1471-4906(03)00181-912909459
|
|
16.
|
TO KhorMT HuangKH KwonJY ChanBS ReddyAN
KongNrf2-deficient mice have an increased susceptibility to dextran
sulfate sodium-induced colitisCancer
Res661158011584200610.1158/0008-5472.CAN-06-356217178849
|
|
17.
|
R MotterliniR ForestiR BassiCJ
GreenCurcumin, anti-oxidant and anti-inflammatory agent, induces
heme oxygenase-1 and protects endothelial cells against oxidative
stressFree Radic Biol
Med2813031312200010.1016/S0891-5849(00)00294-X10889462
|
|
18.
|
J GaedekeNA NobleWA BorderCurcumin blocks
fibrosis in anti-Thy 1 glomerulonephritis through up-regulation of
heme oxygenase 1Kidney
Int6820422049200510.1111/j.1523-1755.2005.00658.x16221204
|
|
19.
|
SJ McNallyEM HarrisonJA RossOJ GardenSJ
WigmoreCurcumin induces heme oxygenase-1 in hepatocytes and is
protective in simulated cold preservation and warm reperfusion
injuryTransplantation81623626200610.1097/01.tp.0000184635.62570.1316495813
|
|
20.
|
W BaoK LiS RongCurcumin alleviates
ethanol-induced hepatocytes oxidative damage involving heme
oxygenase-1 inductionJ
Ethnopharmacol128549553201010.1016/j.jep.2010.01.02920080166
|
|
21.
|
EO FarombiS ShrotriyaHK NaSH KimYJ
SurhCurcumin attenuates dimethylnitrosamine-induced liver injury in
rats through Nrf2-mediated induction of heme oxygenase-1Food Chem
Toxicol4612791287200810.1016/j.fct.2007.09.09518006204
|
|
22.
|
KM KimHO PaeM ZhungInvolvement of
anti-inflammatory heme oxygenase-1 in the inhibitory effect of
curcumin on the expression of pro-inflammatory inducible nitric
oxide synthase in RAW264.7 macrophagesBiomed
Pharmacother62630636200810.1016/j.biopha.2008.01.00818325727
|
|
23.
|
HY HsuLC ChuKF HuaLK ChaoHeme oxygenase-1
mediates the anti-inflammatory effect of Curcumin within
LPS-stimulated human monocytesJ Cell
Physiol215603612200810.1002/jcp.2120618357586
|
|
24.
|
K DevadasS DhawanHemin activation
ameliorates HIV-1 infection via heme oxygenase-1 inductionJ
Immunol17642524257200610.4049/jimmunol.176.7.425216547262
|
|
25.
|
U ProtzerS SeyfriedM QuasdorffAntiviral
activity and hepatoprotection by heme oxygenase-1 in hepatitis B
virus
infectionGastroenterology13311561165200710.1053/j.gastro.2007.07.02117919491
|
|
26.
|
E LehmannWH El-TantawyM OckerThe heme
oxygenase 1 product biliverdin interferes with hepatitis C virus
replication by increasing antiviral interferon
responseHepatology51398404201010.1002/hep.2333920044809
|
|
27.
|
Y ShanJ ZhengRW LambrechtHL
BonkovskyReciprocal effects of micro-RNA-122 on expression of heme
oxygenase-1 and hepatitis C virus genes in human
hepatocytesGastroenterology13311661174200710.1053/j.gastro.2007.08.00217919492
|
|
28.
|
Z ZhuAT WilsonMM MathahsHeme oxygenase-1
suppresses hepatitis C virus replication and increases resistance
of hepatocytes to oxidant
injuryHepatology4814301439200810.1002/hep.2249118972446
|
|
29.
|
V TergaonkarNFkappaB pathway: a good
signaling paradigm and therapeutic targetInt J Biochem Cell
Biol3816471653200610.1016/j.biocel.2006.03.02316766221
|
|
30.
|
S LuqmanJM PezzutoNFkappaB: a promising
target for natural products in cancer chemopreventionPhytother
Res24949963201020577970
|
|
31.
|
Y ZhongT LiuZ GuoCurcumin inhibits
ox-LDL-induced MCP-1 expression by suppressing the p38MAPK and
NF-kappaB pathways in rat vascular smooth muscle cellsInflamm
Res616167201210.1007/s00011-011-0389-322005927
|
|
32.
|
C JobinCA BradhamMP RussoCurcumin blocks
cytokine-mediated NF-kappa B activation and proinflammatory gene
expression by inhibiting inhibitory factor I-kappa B kinase
activityJ Immunol16334743483199910477620
|
|
33.
|
AC BhartiN DonatoS SinghBB
AggarwalCurcumin (diferuloylmethane) down-regulates the
constitutive activation of nuclear factor-kappa B and IkappaBalpha
kinase in human multiple myeloma cells, leading to suppression of
proliferation and induction of
apoptosisBlood10110531062200310.1182/blood-2002-05-1320
|
|
34.
|
GL JohnsonR LapadatMitogen-activated
protein kinase pathways mediated by ERK, JNK, and p38 protein
kinasesScience29819111912200210.1126/science.107268212471242
|
|
35.
|
A ChenS ZhengCurcumin inhibits connective
tissue growth factor gene expression in activated hepatic stellate
cells in vitro by blocking NF-kappaB and ERK signallingBr J
Pharmacol153557567200810.1038/sj.bjp.070754217965732
|
|
36.
|
JH LimTK KwonCurcumin inhibits phorbol
myristate acetate (PMA)-induced MCP-1 expression by inhibiting ERK
and NF-kappaB transcriptional activityFood Chem
Toxicol484752201010.1016/j.fct.2009.09.01319766691
|
|
37.
|
A ShehzadF WahidYS LeeCurcumin in cancer
chemo-prevention: molecular targets, pharmacokinetics,
bioavailability, and clinical trialsArch
Pharm343489499201010.1002/ardp.20090031920726007
|
|
38.
|
MP SoaresFH BachHeme oxygenase-1: from
biology to therapeutic potentialTrends Mol
Med155058200910.1016/j.molmed.2008.12.00419162549
|
|
39.
|
Z ZhuAT WilsonBA LuxonBiliverdin inhibits
hepatitis C virus nonstructural 3/4A protease activity: mechanism
for the antiviral effects of heme
oxygenase?Hepatology5218971905201010.1002/hep.2392121105106
|
|
40.
|
C FillebeenK PantopoulosIron inhibits
replication of infectious hepatitis C virus in permissive Huh7.5.1
cellsJ Hepatol53995999201010.1016/j.jhep.2010.04.04420813419
|
|
41.
|
J ChoiKJ LeeY ZhengAK YamagaMM LaiJH
OuReactive oxygen species suppress hepatitis C virus RNA
replication in human hepatoma
cellsHepatology398189200410.1002/hep.2000114752826
|
|
42.
|
M YanoM IkedaK AbeOxidative stress induces
anti-hepatitis C virus status via the activation of extracellular
signal-regulated
kinaseHepatology50678688200910.1002/hep.2302619492433
|
|
43.
|
M KurokiY AriumiM IkedaH DansakoT WakitaN
KatoArsenic trioxide inhibits hepatitis C virus RNA replication
through modulation of the glutathione redox system and oxidative
stressJ Virol8323382348200910.1128/JVI.01840-0819109388
|
|
44.
|
DP BrazilZZ YangBA HemmingsAdvances in
protein kinase B signalling: AKTion on multiple frontsTrends
Biochem Sci29233242200410.1016/j.tibs.2004.03.00615130559
|
|
45.
|
P MannovaL BerettaActivation of the
N-Ras-PI3K-Akt-mTOR pathway by hepatitis C virus: control of cell
survival and viral replicationJ
Virol7987428749200510.1128/JVI.79.14.8742-8749.200515994768
|
|
46.
|
A StreetA MacdonaldK CrowderM HarrisThe
Hepatitis C virus NS5A protein activates a phosphoinositide
3-kinase-dependent survival signaling cascadeJ Biol
Chem2791223212241200410.1074/jbc.M31224520014709551
|
|
47.
|
T PorstmannB GriffithsYL ChungPKB/Akt
induces transcription of enzymes involved in cholesterol and fatty
acid biosynthesis via activation of
SREBPOncogene2464656481200516007182
|
|
48.
|
CY ParkHJ JunT WakitaJH CheongSB
HwangHepatitis C virus nonstructural 4B protein modulates sterol
regulatory element-binding protein signaling via the AKT pathwayJ
Biol Chem28492379246200910.1074/jbc.M80877320019204002
|
|
49.
|
D Yangxand GabuzdaRegulation of human
immunodeficiency virus type 1 infectivity by the ERK
mitogen-activated protein kinase signaling pathwayJ
Virol7334603466199910074203
|
|
50.
|
S PleschkaT WolffC EhrhardtInfluenza virus
propagation is impaired by inhibition of the Raf/MEK/ERK signalling
cascadeNat Cell Biol3301305200110.1038/3506009811231581
|
|
51.
|
Y CaiY LiuX ZhangSuppression of
coronavirus replication by inhibition of the MEK signaling pathwayJ
Virol81446456200710.1128/JVI.01705-0617079328
|
|
52.
|
KD SmithJJ MezhirK BickenbachActivated MEK
suppresses activation of PKR and enables efficient replication and
in vivo oncolysis by Deltagamma(1)34.5 mutants of herpes simplex
virus 1J
Virol8011101120200610.1128/JVI.80.3.1110-1120.200616414988
|
|
53.
|
Y ZhengJ LiDL JohnsonJH OuRegulation of
hepatitis B virus replication by the ras-mitogen-activated protein
kinase signaling pathwayJ
Virol7777077712200310.1128/JVI.77.14.7707-7712.200312829809
|
|
54.
|
H ZhuC LiuInterleukin-1 inhibits hepatitis
C virus subgenomic RNA replication by activation of extracellular
regulated kinase pathwayJ
Virol7754935498200310.1128/JVI.77.9.5493-5498.200312692250
|
|
55.
|
T MurataM HijikataK ShimotohnoEnhancement
of internal ribosome entry site-mediated translation and
replication of hepatitis C virus by
PD98059Virology340105115200510.1016/j.virol.2005.06.01516005928
|
|
56.
|
J NdjomouIW ParkY LiuLD MayoJJ
HeUp-regulation of hepatitis C virus replication and production by
inhibition of MEK/ERK signalingPLoS
One4e7498200910.1371/journal.pone.000749819834602
|
|
57.
|
DI TaiSL TsaiYM ChenActivation of nuclear
factor kappaB in hepatitis C virus infection: implications for
pathogenesis and
hepatocarcinogenesisHepatology31656664200010.1002/hep.51031031610706556
|
|
58.
|
JK OemC Jackel-CramYP LiHepatitis C virus
non-structural protein-2 activates CXCL-8 transcription through
NF-kappaBArch
Virol153293301200810.1007/s00705-007-1103-118074095
|
|
59.
|
W LinWL TsaiRX ShaoHepatitis C virus
regulates transforming growth factor beta1 production through the
generation of reactive oxygen species in a nuclear factor
kappaB-dependent
mannerGastroenterology138250925182518.e1201010.1053/j.gastro.2010.03.008
|
|
60.
|
RB RayR SteeleA BasuDistinct functional
role of hepatitis C virus core protein on NF-kappaB regulation is
linked to genomic variationVirus
Res872129200210.1016/S0168-1702(02)00046-112135786
|
|
61.
|
JC LeeWC ChenSF WuAnti-hepatitis C virus
activity of Acacia confusa extract via suppressing
cyclooxygenase-2Antiviral
Res893542201110.1016/j.antiviral.2010.11.00321075144
|
|
62.
|
JC LeeCK TsengSF WuFR ChangCC ChiuYC
WuSan-Huang-Xie-Xin-Tang extract suppresses hepatitis C virus
replication and virus-induced cyclooxygenase-2 expressionJ Viral
Hepat18e315e324201110.1111/j.1365-2893.2010.01424.x21692943
|