Introduction
Colorectal cancer is the most frequent malignancy of
the digestive tract, and one of the most common types of solid
organ cancer in both males and females in developed countries.
Worldwide, approximately 1 million cases are recorded annually, and
over half a million patients succumb to this disease each year
(1). Irinotecan (CPT-11) is a
topoisomerase I inhibitor widely used in the treatment of
colorectal tumors (2). However,
the use of CPT-11 in a clinical setting has been hampered by modest
efficacy and significant toxic side-effects (3). Thus, novel treatment strategies are
urgently needed to improve the clinical management of colorectal
cancer.
Tumor necrosis factor-related apoptosis-inducing
ligand (TRAIL), a tumor necrosis factor superfamily member has been
shown to induce apoptosis. TRAIL can bind to 5 different receptors:
TRAIL-R1 (death receptor 4, DR4), TRAIL-R2 (DR5), TRAIL-R3 (decoy
receptor, DcR1), TRAIL-R4 (DcR2) and osteoprotegerin (OPG). DR4 and
DR5 are the death receptors that signal for apoptosis, whereas
DcR1, DcR2 and OPG are considered antagonistic as they are unable
to induce such signaling due to the lack of intracellular death
domain or are secreted molecules (4). TRAIL triggers apoptosis through
binding to its receptors (DR4 and/or DR5) in a broad range of human
cancer cell lines while sparing most normal cell types (5). Thus, TRAIL is a promising cancer
therapeutic agent due to its tumor selectivity (6). Several studies have previously
demonstrated that many colon carcinoma cells were sensitive to
TRAIL both in vitro and in vivo except for HT-29
(7,8). They also found that CPT-11 enhanced
the antitumor effects of TRAIL on HT-29 colon carcinoma (9). However, the focus was mainly on the
TRAIL sensitive colon carcinoma cell lines such as COLO 205, SW948,
and HCT116, and the synergistic effects of TRAIL and CPT-11 on
HT-29 were not studied thoroughly (7). In this study, we investigated the
combined effects of TRAIL and CPT-11 at different doses and
explored the potential apoptosis mechanism on TRAIL-resistant HT-29
colon carcinoma cells in vitro and in vivo.
Materials and methods
Materials
Human HT-29 colon carcinoma cell line and TRAIL were
provided by DIAO Group (China). Female nude mice (6 weeks old) were
purchased from the animal experimental center of the Chinese
Academy of Science, Shanghai (China). CPT-11 was purchased from
Aventis (France). Sulforhodamine B (SRB) was supplied by Sigma
(USA). Roswell Park Memorial Institute (RPMI)-1640 medium and
trypsin were purchased from Gibco (USA). Mouse monoclonal antibody
for Bax was purchased from Santa Cruz Biotechnology, Inc., (USA).
Mouse monoclonal antibody for β-actin was purchased from Abcam
(UK). Mouse monoclonal antibody for caspase-9 was purchased from
Lab Vision (USA). LSAB kit was purchased from Dako (Japan). Goat
anti-mouse IgG-HRP was purchased from Rockland (USA). TRIzol kit
and apoptotic DNA ladder kit were purchased from Bioteke
Corporation (China). One-step RT-PCR kit was purchased from Takara
Biotechnology Co., Ltd., China. Polyvinylidene difluoride (PVDF)
membrane was purchased from Millipore. Enhanced chemiluminescence
(ECL) detection kit and X-ray film were purchased from Roche
(Switzerland). Annexin V-FITC apoptosis detection kit was purchased
from Jingmei Biotech Co., Ltd. (China).
Cell culture
The HT-29 colon carcinoma cell line was cultured in
RPMI-1640 medium supplemented with 10% fetal bovine serum plus
ampicillin and streptomycin routinely, and incubated in 5%
CO2 at 37°C.
TRAIL and CPT-11-mediated toxicity
Cell viability was measured by the SRB method
(10). Briefly, exponentially
growing tumor cells were seeded in a 96-well plate
(1×104/well). Cells were incubated without treatment for
24 and 72 h, respectively, in a negative control group 1 and 2.
Adherent cell cultures were fixed in situ by addition of 45
μl of cold 50% (w/v) trichloroacetic acid (TCA) and were kept for
60 min at 4°C. The supernatant was then discarded, and the plates
were washed 5 times with distilled water and air dried. In the
TRAIL or CPT-11 alone group (cells treated with TRAIL or CPT-11
alone), cells were incubated for 24 h, and then TRAIL or CPT-11 was
added to each well, respectively. The final concentrations of TRAIL
were 0.001, 0.01, 0.1, 1 and 10 μg/ml, respectively. The final
concentrations of CPT-11 were 0.1, 0.5, 1, 5 and 10 μg/ml,
respectively. The cytotoxic effect was evaluated 48 h after drug
challenge. Briefly, adherent cell cultures were fixed in
situ by addition of 45 μl of cold 50% (w/v) TCA and were kept
for 60 min at 4°C. The supernatant was then discarded, and the
plates were washed 5 times with distilled water and air dried. SRB
solution (0.4% w/v in 1% acetic acid) was added and the cells were
allowed to stain for 10 min at room temperature. Unbound SRB was
removed by washing 3 times with 1% acetic acid. The plates were
then air dried. Bound stain was dissolved with unbuffered 10 mM
Tris base [tris(hydroxymethyl)aminomethane]. The absorbance values
of the solution in each well were measured at 490 nm using a
microplate reader. Cell viability was measured by the formula: If
Ti ≥ Tz, cell viability (%) = [(Ti-Tz)/(C-Tz)]x100; If Ti < Tz,
cell viability (%) = [(Ti-Tz)/Tz]x100. (Ti, absorbance of the wells
treated with TRAIL or CPT-11; Tz, absorbance of the wells in
negative control 1; C, absorbance of the wells in negative control
2). All SRB experiments were performed in triplicate and repeated
at least 3 times.
The concentrations of reagents that induced a 50%
reduction in cell viability (IC50) were determined from
curves of reagent concentration vs. cell viability at 48 h of
incubation for the cell line analyzed. The sensitivity of cells to
the drug was evaluated by the value of IC50.
IC50 <10 μg/ml, indicated that the cells were
sensitive to the drug, while IC50 ≥10 μg/ml, indicated
that the cells were relatively resistant to the drug (11).
Combination of TRAIL and CPT-11-mediated
toxicity
According to the effect of TRAIL and CPT-11
treatment alone on cell viability, the concentrations of the
combination of TRAIL and CPT-11 were chosen as follows: TRAIL (0.1,
1 and 10 μg/ml, respectively), CPT-11 (1, 5 and 10 μg/ml,
respectively). Cell viability was then assessed by SRB assay.
Negative control group 1, negative control group 2, TRAIL alone
group, CPT-11 alone group and combination group (cells treated with
TRAIL and CPT-11) were designed for this experiment.
Evaluation of synergistic effect
The synergistic effect of the combination of TRAIL
and CPT-11 was analyzed by the Webb coefficient (12). Predicted cell viability (c) was
calculated according to the equation c = a x b/100, where a and b
indicated cell viability with single agents. Synergism in drug
interaction was indicated by observed cell viability of ≤70% of the
predicted cell viability (11).
According to the results of synergetic effect, the optimum
concentrations of 0.1 μg/ml TRAIL and 5 μg/ml CPT-11 were chosen
for later experiments in vitro.
DNA ladder assay
For detection of apoptosis by the DNA fragmentation
assay, 2×106 cells were plated into culture dishes 24 h
prior to drug treatment. Following treatment with TRAIL or CPT-11
for 48 h, HT-29 cells were washed with phosphate-buffered saline
(PBS) and harvested. Cells were then resuspended in 0.5 ml of lysis
buffer for 10 min at room temperature. After centrifugation at
12,000 rpm at room temperature for 5 min, the supernatant was
transferred to a new Eppendorf tube and equal volumes of
isopropanol were added, before centrifuging at 2,000 rpm at room
temperature for 30 sec. DNA was acquired by centrifugation of the
samples, washed, dried and dissolved in loading buffer, and
separated by electrophoresis during 90 min at 60 V on 2% agarose
gels, containing ethidium bromide (EB). Bands were visualised under
ultraviolet light.
Flow cytometry analysis of apoptosis
To confirm that TRAIL- and CPT-11- mediated cell
death occurs by apoptosis, we employed flow cytometry and
determined the percentage of specific apoptotic cells. Apoptosis
was quantified using the Annexin V-FITC apoptosis detection kit
according to the manufacturer’s instructions. Briefly,
exponentially growing tumor cells were first digested with 0.25%
trypsin and counted. They were then diluted to a final
concentration of 1×105 cells/ml and inoculated into a
culture dish at 10 ml/dish. When 50–60% confluency was reached,
cells incubated with 5 μg/ml CPT-11 or 0.1 μg/ml TRAIL alone or
with the combination of both for 48 h were harvested by trypsin
release, washed twice with cold PBS, and permeabilized with 70%
ethanol in PBS for 30 min. Then they were resuspended in Annexin V
binding buffer. FITC-conjugated Annexin V (1 μg/ml) and propidium
iodide (50 μg/ml) were added to the cells and incubated for 15 min
at room temperature in the dark before flow cytometry analysis. The
samples were detected using an Elite-ESP flow cytometer
(Beckman-Coulter, USA). A minimum of 104 cells were
analyzed in each sample. All experiments were repeated at least 3
times.
Reverse transcription polymerase chain
reaction (RT-PCR)
Total-RNA was extracted from cells using TRIzol
reagent. The RNA was then quantified spectrophotometrically. RT-PCR
was run using One-Step RT-PCR kit. The primers for DR4, DR5 and
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were synthesized
by Shanghai Genebase Gene-Tech Co., Ltd. (China). GAPDH was used as
internal control primer. The upstream primers of DR4, DR5 and GAPDH
were, 5′-CTGAGCAACGCAGACTCGCTGTCCAC-3′, 5′-GCCTC
ATGGACAATGAGATAAAGGTGGCT-3′ and 5′-ACCAC AGTCCATGCCATCAC-3′,
respectively. The downstream primers of DR4, DR5 and GAPDH were,
5′-ACAGCATCAG AGTCTCAGTGGGGTCAGC-3′, 5′-CCAAATCTCAAAGTA
CGCACAAACGG-3′ and 5′-TCCACCACCCTGTTGCTG TA-3′, respectively. The
PCR products were a 202-bp fragment for DR4, a 220-bp fragment for
DR5, and a 485-bp fragment for GAPDH, respectively.
PCR was performed in a 25 μl reaction volume. PCR
cycling conditions of DR4 were as follows: an initial denaturing
step at 94°C for 2 min, 30 cycles repeating a denaturation step of
94°C for 10 sec, followed by an optimized annealing temperature of
59°C for 10 sec, and a final elongation step at 72°C for 20 sec.
PCR cycling conditions of DR5 were as follows: an initial
denaturing step at 95°C for 2 min, 30 cycles repeating 94°C for 10
sec, 52°C for 10 sec and 72°C for 20 sec.
Amplified products of DR4, DR5 and GAPDH were
separated by 1.5% agarose gel electrophoresis in EB stained and
viewed under ultraviolet light. The electrophoresis images were
obtained and the DR4, DR5 and GAPDH band integrated optical density
values were analyzed using the Bio-Rad image system.
Semiquantitative analysis of DR4 and DR5 mRNA was performed by
comparison to GAPDH.
Western blot analysis
Cells incubated with 5 μg/ml CPT-11 or 0.1 μg/ml
TRAIL alone or in combination for 48 h were lysed in lysis buffer.
Protein content of the supernatant was measured using the
bicinchoninic acid (BCA) method. Cell lysate protein (25 μg) was
separated by SDS-PAGE using a Trisglycine system and then gels were
electroblotted onto PVDF membranes for 1.5 h. The membranes were
then incubated with 5% non-fat dry milk in PBS for 1 h for blocking
nonspecific binding sites, and then incubated with the appropriate
primary antibody concentration (1:400 dilution for caspase-9 and
Bax, and 1:2,000 for β-actin) for 1 h at 37°C in 5% non-fat dry
milk. Membranes were subsequently rinsed in PBS, and then incubated
for 1 h at 37°C with secondary antibody (horseradish peroxidase
conjugated anti-mouse IgG at 1:2,000 dilution). Following
incubation, membranes were rinsed and blots were visualized by
incubation with ECL detection reagents. Signal density was obtained
by scanning exposed X-ray films on a Bio-Rad imaging system.
Normalized density was obtained by dividing the rough density
values of a sample band over loading control band (β-actin).
Combined effects on HT-29 xenograft on
nude mice
All animal procedures were approved by the Animal
Care and Scientific Committee of Sichuan University. Thirty-six
HT-29 bearing nude mice were divided into 6 groups (6 mice/group);
the control group, treated with normal saline; the TRAIL-5 mg/kg
group, treated with TRAIL with a concentration of 5 mg/mice weight
(kg); the TRAIL-15 mg/kg group, treated with TRAIL with a
concentration of 15 mg/mice weight (kg); the CPT-11-12.5 mg/kg
group, treated with CPT-11 with a concentration of 12.5 mg/mice
weight (kg); the TRAIL-5 mg/kg + CPT-11-12.5 mg/kg group, treated
with TRAIL with a concentration of 5 mg/mice weight (kg) and CPT-11
with a concentration of 12.5 mg/mice weight (kg); the TRAIL-15
mg/kg + CPT-11-12.5 mg/kg group, treated with TRAIL with a
concentration of 15 mg/mice weight (kg) and CPT-11 with a
concentration of 12.5 mg/mice weight (kg). Saline was injected
through the tail vein 15 times in 15 days; TRAIL was injected
through the tail vein 5 times in 5 days, and CPT-11 was injected
into the peritoneal cavity 6 times in 6 days. The length, width,
and weight of the tumor were measured using a slide caliper every 5
days. Tumor volume (TV) was estimated using the formula: TV
(mm3) = (width2 x length)/2. Inhibitive rate
of tumor was calculated using the formula: inhibitive rate of tumor
(%) = (1 - average weight in treated group/average weight in
control) x 100% (13).
Statistical analysis
Results were expressed as the mean ± standard
deviation (SD) with the exception of the tumor inhibitive rate.
Statistical comparisons of mean values were analyzed by one-way
ANOVA, followed by the Student’s t-test. Data of tumor inhibitive
rate were analyzed using the Chi-square test. Linear correlation
between cell viability and concentrations of CPT-11 or TRAIL was
analyzed using SPSS 13.0 software. Synergetic effect was evaluated
using the Webb coefficient. All P-values were 2-sided and P<0.05
was considered to indicate statistically significant
differences.
Results
SRB assay
Results of the SRB assay are shown in Fig. 1 and demonstrate that TRAIL induced
limited cell death, and there was significant negative correlation
between cell viability and TRAIL concentration (r=−0.779,
P<0.05). IC50 >10 μg/ml suggesting that HT-29
cells were relatively resistant to TRAIL. The cell viability of
HT-29 cells treated with 10 μg/ml TRAIL was significantly higher
than in 0, 0.01, 0.01, 0.1 and 1 μg/ml TRAIL groups (Fig. 1A). CPT-11 induced cell death in a
dose-dependent manner (r=−0.863, P<0.05). IC50 was
<10 μg/ml suggesting that HT-29 cells were sensitive to CPT-11.
The cell viability of HT-29 treated with 1 μg/ml CPT-11 was
significantly higher than in 0, 0.1 and 0.5 μg/ml CPT-11 groups,
and the cell viability of HT-29 cells treated with 10 μg/ml CPT-11
was higher than in 0, 0.1, 0.5, 1 and 5 μg/ml CPT-11 groups
(Fig. 1B). Webb coefficient
analysis showed that only 0.1 μg/ml TRAIL combined with 5 μg/ml
CPT-11 had a significant synergistic effect (P<0.05) (Fig. 1C and Table I).
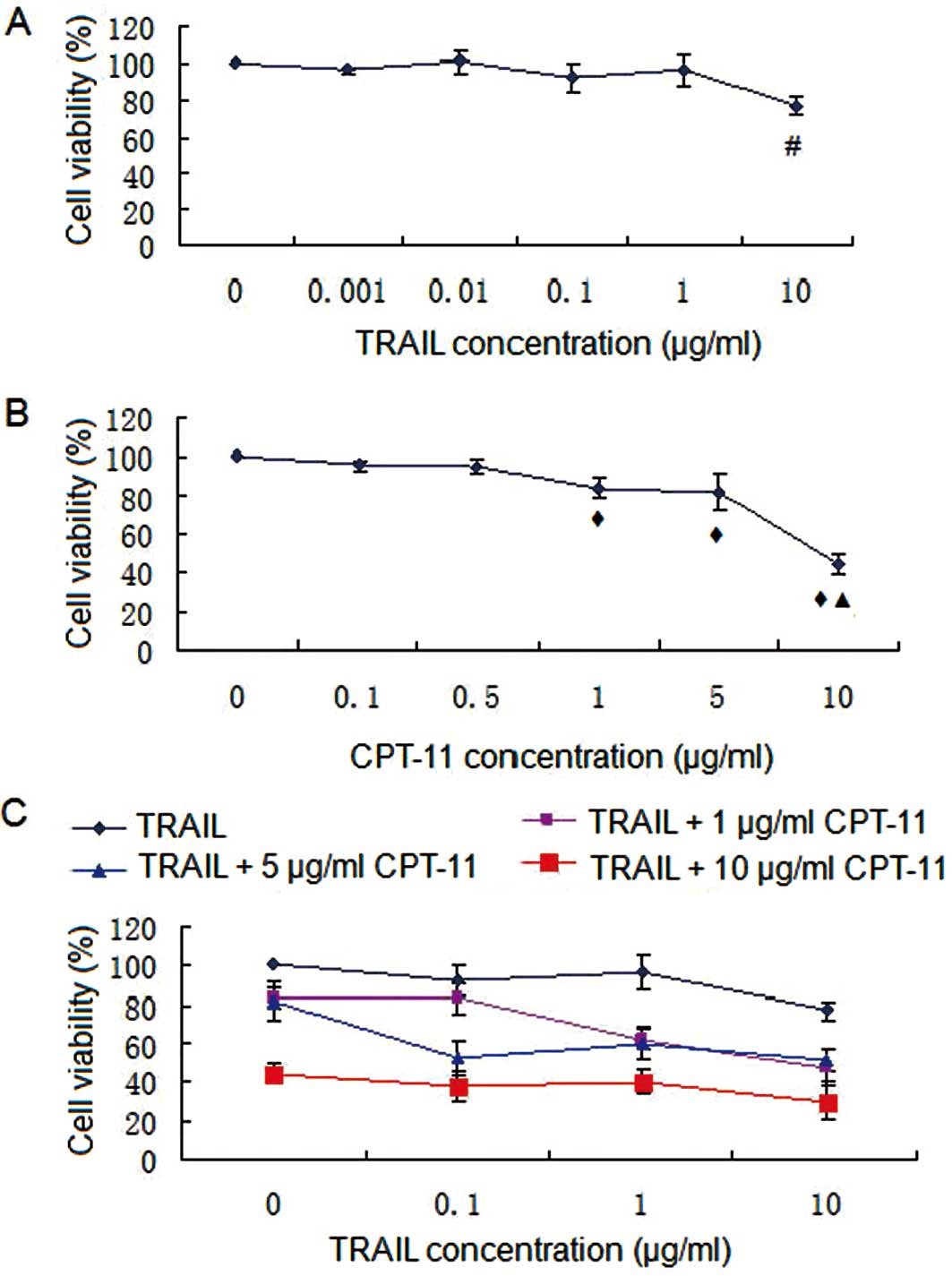 | Figure 1.SRB assay. (A) TRAIL treatment; (B)
CPT-11 treatment; (C) combination treatment. #P<0.05
compared with 0, 0.01, 0.01, 0.1 and 1 μg/ml TRAIL;
◆P<0.05 compared with 0, 0.1 and 0.5 μg/ml CPT-11;
▲P<0.05 compared with 0, 0.1, 0.5, 1 and 5 μg/ml
CPT-11. TRAIL induced limited cell death, and there was significant
negative correlation between cell viability and TRAIL concentration
(r=−0.779, P<0.05). IC50 was >10 μg/ml suggesting
that HT-29 cells were relatively resistant to TRAIL. CPT-11 induced
cell death in a dose-dependent manner (r=−0.863, P<0.05).
IC50 was <10 μg/ml suggesting that HT-29 cells were
sensitive to CPT-11. |
 | Table I.Analysis of the synergetic effect of
TRAIL and CPT-11 on the cell viability of HT-29 cells. |
Table I.
Analysis of the synergetic effect of
TRAIL and CPT-11 on the cell viability of HT-29 cells.
| Group | Concentration
(μg/ml) | Observed cell
viability (%) | 70% estimated cell
viability (70% c) | P-value |
|---|
| T1 | 10 | 77.46 (a1) | | |
| T2 | 1 | 96.42 (a2) | | |
| T3 | 0.1 | 92.45 (a3) | | |
| C1 | 10 | 44.28 (b1) | | |
| C2 | 5 | 82.14 (b2) | | |
| C3 | 1 | 83.85 (b3) | | |
| T1+C1 | | 30.19 | 24.01 | >0.05 |
| T1+C2 | | 51.39 | 44.54 | >0.05 |
| T1+C3 | | 47.72 | 45.47 | >0.05 |
| T2+C1 | | 40.20 | 29.89 | >0.05 |
| T2+C2 | | 59.96 | 55.44 | >0.05 |
| T2+C3 | | 62.25 | 56.59 | >0.05 |
| T3+C1 | | 37.86 | 40.94 | >0.05 |
| T3+C2 | | 52.26a | 53.15 | <0.05 |
| T3+C3 | | 83.40 | 54.26 | >0.05 |
DNA ladder assay and flow cytometry
DNA fragmentation was seen in the TRAIL and combined
groups, but not in the other 2 groups (Fig. 2C). The apoptosis rates were 0.9,
17.5, 8.1 and 29.3%, respectively, in the control, TRAIL alone,
CPT-11 alone, and combined group. There were significant
differences of apoptosis rates in the combined group vs. the other
groups (P<0.05) (Fig. 2A and
B).
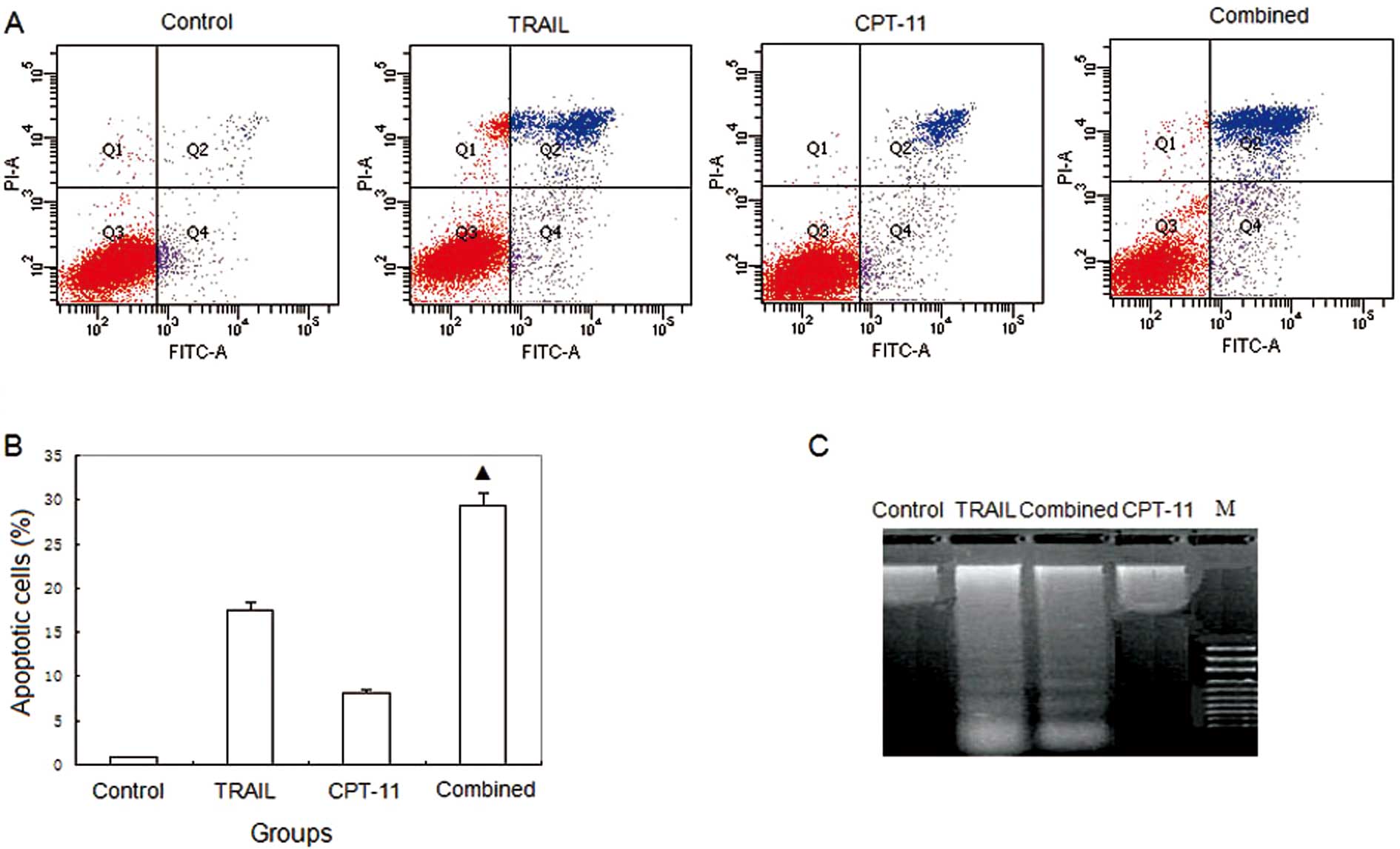 | Figure 2.Flow cytometry and DNA ladder. (A)
Flow cytometry; (B) histogram of flow cytometry; (C) DNA ladder.
Control, HT-29 treated with saline; TRAIL, HT-29 treated with 0.1
μg/ml TRAIL; CPT-11, HT-29 treated with 5 μg/ml CPT-11; combined,
HT-29 treated with 0.1 μg/ml TRAIL and 5 μg/ml CPT-11; M, marker.
The apoptosis rates were 0.9, 17.5, 8.1 and 29.3%, respectively in
the control, TRAIL, CPT-11, and combined group.
▲P<0.05 compared with the control, TRAIL and CPT-11
groups. DNA fragmentation was seen in the TRAIL and combined
groups, but not in the other 2 groups. |
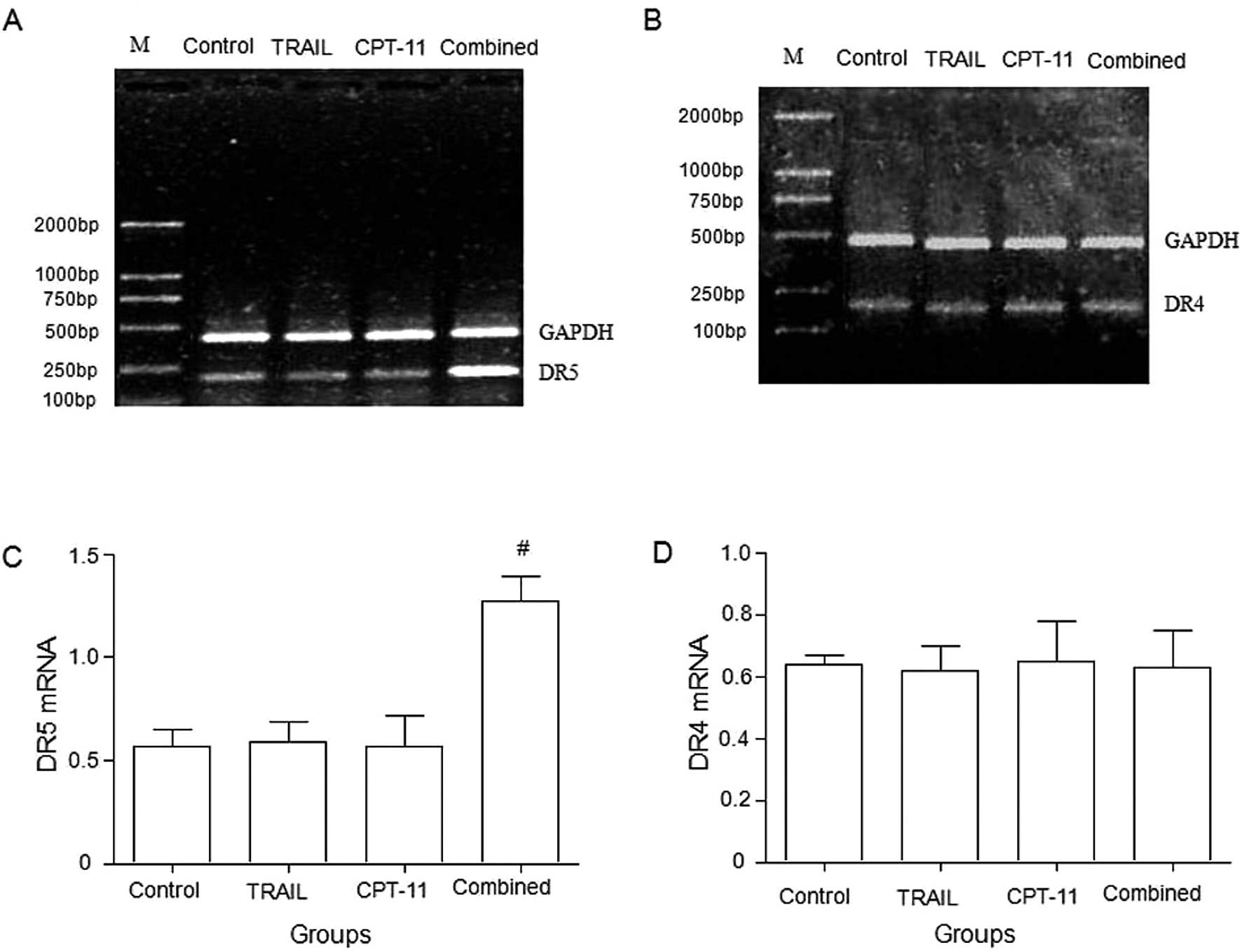
DR5 and DR4 mRNA expression
Compared with the control or TRAIL alone or CPT-11
alone groups, the combination of TRAIL and CPT-11 significantly
upregulated the expression of DR5 mRNA in HT-29 cells (Fig. 3A and C), whereas DR4 mRNA in HT-29
cells showed no significant difference among the groups (Fig. 3B and D).
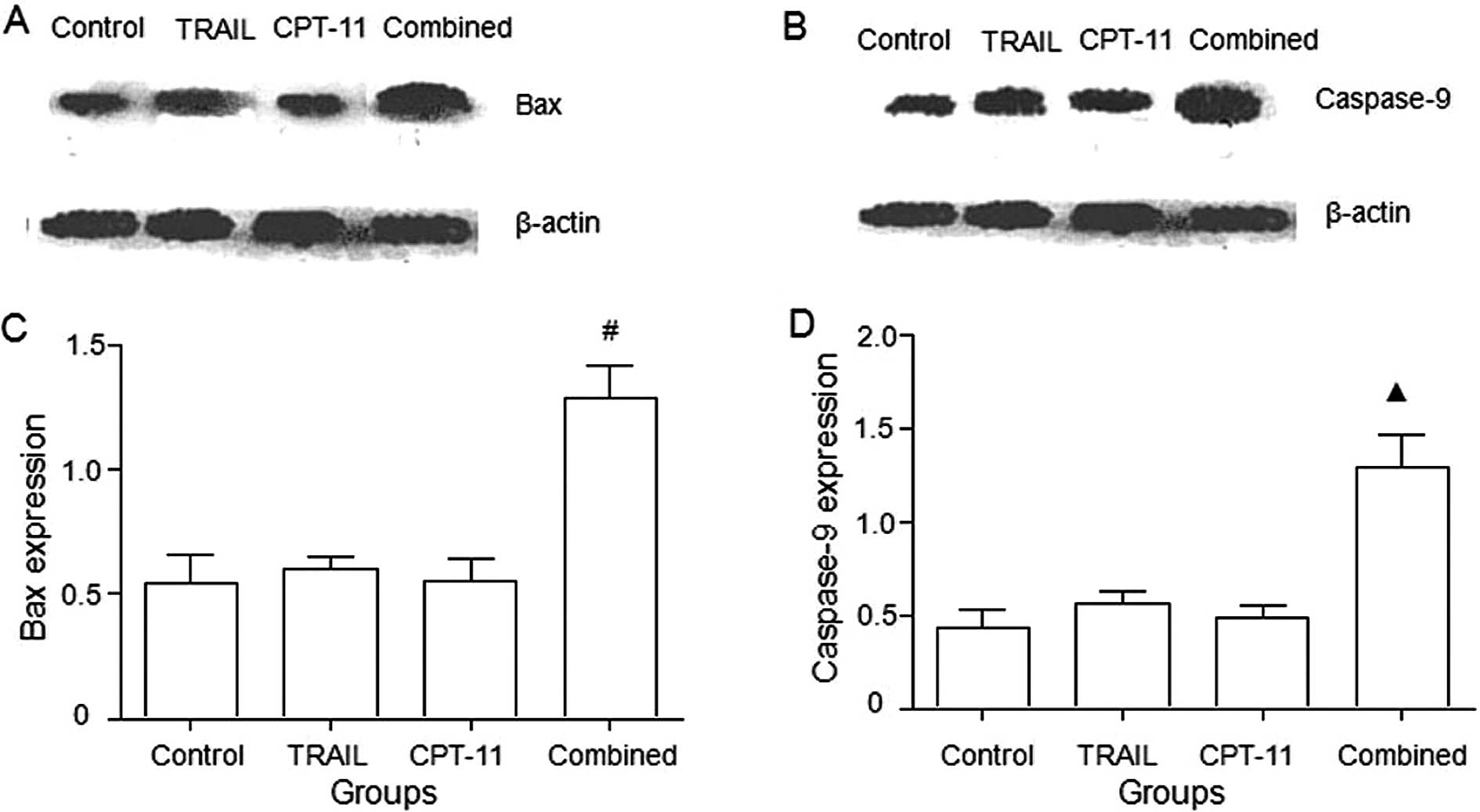
Bax and caspase-9 protein expression
As shown in Fig.
4, the combination treatment of TRAIL and CPT-11 significantly
upregulated Bax and caspase-9 proteins (P<0.05).
Tumor growth curves and inhibitive rates
of tumor growth
The treatment began on Day 10 after the injection of
tumor cells. The tumor volumes were recorded every 5 days and the
tumor growth curves were depicted. The tumor growth curves and
inhibitive rates are shown in Figs.
5 and 6, respectively. The
results show that the 2 combined treatment groups could inhibit the
tumor growth significantly compared with the other groups
(P<0.05).
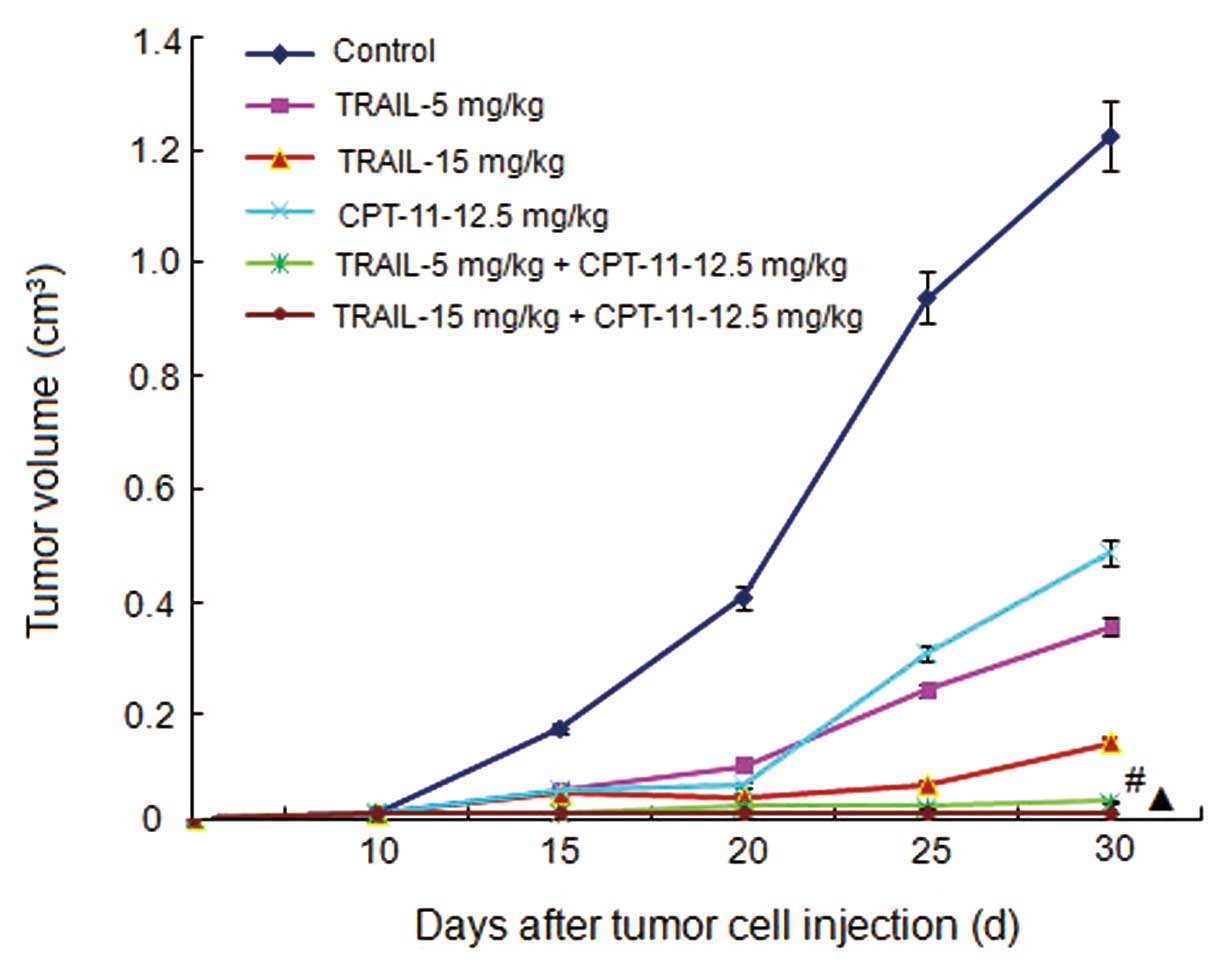 | Figure 5.Tumor growth curves. Control, mice
treated with normal saline; TRAIL-5 mg/kg, mice treated with TRAIL
with a concentration of 5 mg/mouse weight (kg); TRAIL-15 mg/kg,
mice treated with TRAIL with a concentration of 15 mg/mouse weight
(kg); CPT-11-12.5 mg/kg, mice treated with CPT-11 with a
concentration of 12.5 mg/mouse weight (kg); TRAIL-5 mg/kg +
CPT-11-12.5 mg/kg, mice treated with TRAIL with a concentration of
5 mg/mouse weight (kg) and CPT-11 with a concentration of 12.5
mg/mouse weight (kg); TRAIL-15 mg/kg + CPT-11-12.5 mg/kg, mice
treated with TRAIL with a concentration of 15 mg/mouse weight (kg)
and CPT-11 with a concentration of 12.5 mg/mouse weight (kg).
#P<0.05 compared with the control, TRAIL, and CPT-11
groups. ▲P<0.05 compared with the control, TRAIL, and
CPT-11 groups. |
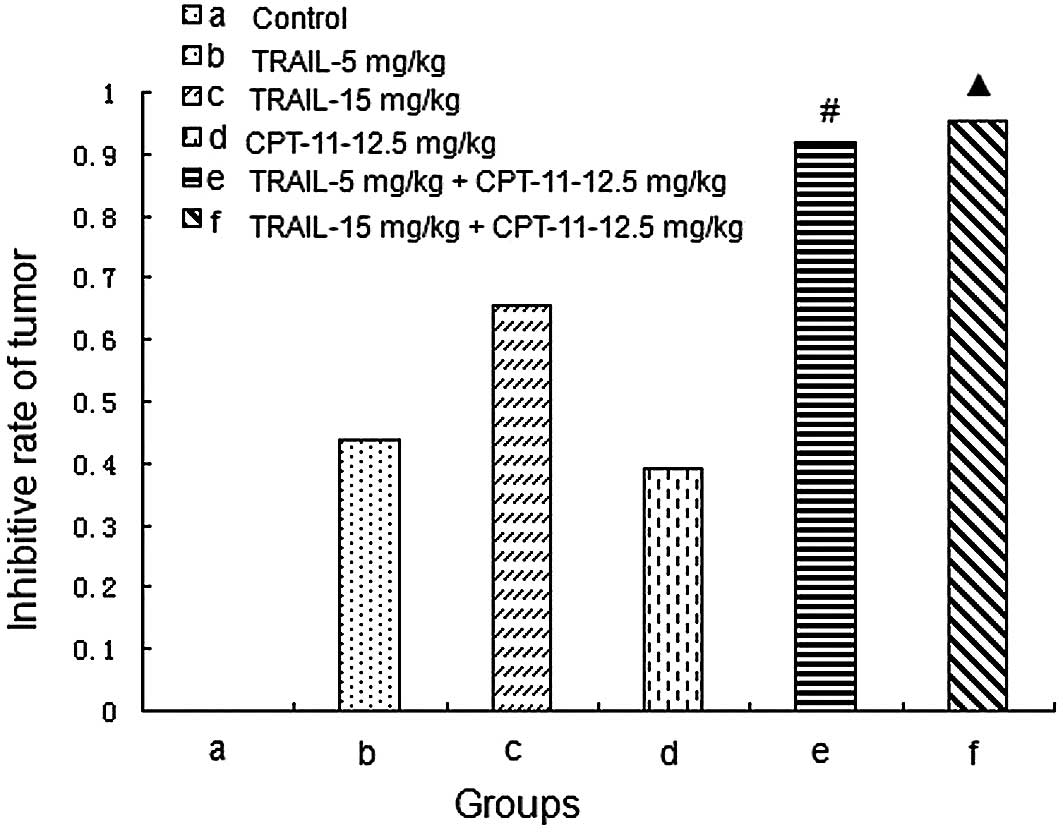 | Figure 6.Inhibitive rates of tumor growth. (a)
Control, mice treated with normal saline; (b) TRAIL-5 mg/kg, mice
treated with TRAIL with a concentration of 5 mg/mouse weight (kg);
(c) TRAIL-15 mg/kg, mice treated with TRAIL with a concentration of
15 mg/mouse weight (kg); (d) CPT-11-12.5 mg/kg, mice treated with
CPT-11 with a concentration of 12.5 mg/mouse weight (kg); (e)
TRAIL-5 mg/kg + CPT-11-12.5 mg/kg, mice treated with TRAIL with a
concentration of 5 mg/mouse weight (kg) and CPT-11 with a
concentration of 12.5 mg/mouse weight (kg); (f) TRAIL-15 mg/kg +
CPT-11-12.5 mg/kg, mice treated with TRAIL with a concentration of
15 mg/mouse weight (kg) and CPT-11 with a concentration of 12.5
mg/mouse weight (kg). #P<0.05 compared with the
control, TRAIL, and CPT-11 groups. ▲P<0.05 compared
with the control, TRAIL, and CPT-11 groups. |
Discussion
TRAIL induced apoptotic cell death in a variety of
tumorigenic or transformed cell lines but not in normal cells
(14). The selective killing of
tumor cells by TRAIL has made TRAIL receptors attractive targets
for cancer treatment. In preclinical models, recombinant soluble
TRAIL demonstrated notable anticancer activity (15,16). However, an increasing number of
studies have demonstrated TRAIL resistance in primary human tumor
cells especially in those of solid tumor entities (17). Thus, novel treatment strategies
are urgently required to overcome this resistance in tumor cells.
Several reports have described the ability of subtoxic
concentrations of chemotherapeutic drugs to sensitize tumor cells
that are resistant to TRAIL (18). The antitumor properties of TRAIL
can be significantly enhanced when used in combination with
chemotherapy, as has been demonstrated in many studies using
different tumor cell lines and mouse models (15).
In the present study, we focused on the synergistic
effects of low-dose CPT-11 and TRAIL on the HT-29 TRAIL resistant
colon cancer cell line. The SRB assay demonstrated that HT-29 cells
were relatively resistant to TRAIL and sensitive to CPT-11. The
antitumor effects of TRAIL could be significantly enhanced by a
low-dose of CPT-11 on HT-29 cells. The DNA ladder assay and flow
cytometry confirmed that low-dose CPT-11 could significantly
enhance the apoptosis induction effects of TRAIL on HT-29
cells.
Previous studies showed that chemotherapeutic agents
could increase the expressions of DR4 and DR5 in tumor cells
(19). The upregulation of DR4
and DR5 enhanced the responsiveness of cells to TRAIL (20,21) and cell death was enhanced when
these agents were combined with TRAIL (19). Our data showed that the
combination of TRAIL and CPT-11 could upregulate the expression of
DR5 mRNA in HT-29 cells, whereas DR4 mRNA in HT-29 cells showed no
significant difference among the groups.
TRAIL-induced apoptosis appears to require
expression of one or both of its death domain containing receptors
DR4 or DR5 (14). In addition,
DR4 and DR5 actively promote apoptosis upon TRAIL binding (22). Although DR4, DR5, DcR1 and OPG
show similar affinities for TRAIL at 4°C, their rank-ordered
affinities are substantially different at 37°C with DR5 having the
highest affinity (14). The
findings of Wen et al (23) indicate that etoposide, Ara-C, or
doxorubicin that are sensitive to TRAIL-induced apoptosis in human
acute leukemia cells are associated with upregulation of DR5 but
not DR4. In addition, some results also showed that
chemotherapeutic agents could augment TRAIL-induced apoptosis by
upregulating DR5 (24,25). Collectively, our data indicate
that the synergistic apoptotic effect of CPT-11 and TRAIL is
mediated by a transcriptional induction of DR5.
Apoptosis is now understood to involve 2 major
signaling pathways, one initiated by DNA damage and one initiated
by death receptors (DRs) (26).
The synergistic cytotoxic effect of genotoxic drugs and TRAIL was
proposed to be p53 dependent (18,20). The genes involved in p53-mediated
apoptosis include Bax (27,28) and caspase-9 (28,29). Bax is essential for death
receptor-mediated apoptosis in cancer cells. LeBlanc et al
(30) found that HCT116 human
colon carcinoma cells are completely dependent upon Bax for
apoptosis induction by death-receptor ligands. Caspases play a
critical role in the execution of apoptosis (31). Caspase-9, the major initiator
caspase identified to date (32),
is an essential downstream component of p53-induced apoptosis. In
addition, caspase-9 is involved in death induced by cytotoxic
agents (33). Some results showed
that overexpression of caspase-9 alone induced apoptosis in U-87MG
glioma cells (28). Our results
showed that the combination treatment of TRAIL and CPT-11 could
significantly upregulate Bax and caspase-9 proteins. These results
indicate that anticancer drugs increase the ability of TRAIL to
trigger a caspase-dependent cell apoptosis, in accordance with the
results of Lacour et al (18).
The in vivo experiments also showed that the
antitumor effects of TRAIL could be enhanced significantly by a
low-dose of CPT-11. The 2 combined groups yielded very strong
effects and tumor growth was thoroughly inhibited. In this study,
the dose of CPT-11 used was 12.5 mg/kg which was lower than other
studies (25 or 50 mg/kg) (24,34). However, the inhibitive rates of
tumor in the combined groups were still notable. TRAIL and CPT-11
showed significant synergistic effects and could be a promising
treatment for TRAIL resistant HT-29 colon carcinoma.
In conclusion, the antitumor effect of TRAIL can be
significantly enhanced by low-dose CPT-11 on TRAIL-resistant HT-29
cells both in vitro and in vivo. The synergistic
apoptotic effect of CPT-11 and TRAIL was proposed to be mediated by
upregulating DR5 mRNA expression and increasing expression of Bax
and caspase-9 proteins. The data suggest that the combination of
TRAIL with low-dose CPT-11 may be a promising therapeutic approach
for HT-29 colon carcinoma.
Acknowledgements
This study was supported by grants
from the National Natural Scientific Foundation of China (no.
81070313).
References
|
1.
|
SJ OosterlingGJ van der BijM BogelsS ten
RaaJA PostGA MeijerRH BeelenM van EgmondAnti-beta1 integrin
antibody reduces surgery-induced adhesion of colon carcinoma cells
to traumatized peritoneal surfacesAnn
Surg2478594200810.1097/SLA.0b013e3181588583
|
|
2.
|
I HennebelleC TerretE ChatelutR BugatP
CanalS GuichardCharacterization of CPT-11 converting
carboxylesterase activity in colon tumor and normal tissues:
comparison with p-nitro-phenylacetate converting carboxylesterase
activityAnticancer
Drugs11465470200010.1097/00001813-200007000-00007
|
|
3.
|
AM SaeternM BrandlWH BakkelundB
SveinbjornssonCytotoxic effect of different camptothecin
formulations on human colon carcinoma in vitroAnticancer
Drugs15899906200410.1097/00001813-200410000-0001115457131
|
|
4.
|
AD SanliogluE DiriceO ElpekAF KorcumM
OzdoganI SuleymanlarMK BalciTS GriffithS SanliogluHigh TRAIL death
receptor 4 and decoy receptor 2 expression correlates with
significant cell death in pancreatic ductal adeno-carcinoma
patientsPancreas38154160200910.1097/MPA.0b013e31818db9e318981952
|
|
5.
|
A AshkenaziP HollandSG
EckhardtLigand-based targeting of apoptosis in cancer: the
potential of recombinant human apoptosis ligand 2/Tumor necrosis
factor-related apoptosis-inducing ligand (rhApo2L/TRAIL)J Clin
Oncol2636213630200810.1200/JCO.2007.15.7198
|
|
6.
|
J YooSS ParkYJ LeePretreatment of
docetaxel enhances TRAIL-mediated apoptosis in prostate cancer
cellsJ Cell Biochem10416361646200810.1002/jcb.2172918404675
|
|
7.
|
PG OliverAF LoBuglioKR ZinnH KimL NanT
ZhouW WangDJ BuchsbaumTreatment of human colon cancer xenografts
with TRA-8 anti-death receptor 5 antibody alone or in combination
with CPT-11Clin Cancer
Res1421802189200810.1158/1078-0432.CCR-07-139218381960
|
|
8.
|
R RaviAJ JainRD SchulickV PhamTS ProuserH
AllenEG MayerH YuDM PardollA AshkenaziA BediElimination of hepatic
metastases of colon cancer cells via p53-independent cross-talk
between irinotecan and Apo2 ligand/TRAILCancer
Res6491059114200410.1158/0008-5472.CAN-04-248815604280
|
|
9.
|
K SugamuraJF GibbsA Belicha-VillanuevaC
AndrewsEA RepaskyBL HylanderSynergism of CPT-11 and Apo2L/ TRAIL
against two differentially sensitive human colon tumor
xenograftsOncology74188197200810.1159/00015136618714167
|
|
10.
|
W ZhouJ HuH TangD WangX HuangC HeH
ZhuSmall interfering RNA targeting mcl-1 enhances proteasome
inhibitor-induced apoptosis in various solid malignant tumorsBMC
Cancer11485201110.1186/1471-2407-11-48522078414
|
|
11.
|
DD CuiY HuangSH MaoSC ChenM QiuLL JiC
YiSynergistic antitumor effect of TRAIL and adriamycin on the human
breast cancer cell line MCF-7Braz J Med Biol
Res42854862200919738990
|
|
12.
|
YA YehM HerenyiovaG WeberQuercetin:
synergistic action with carboxyamidotriazole in human breast
carcinoma cellsLife
Sci5712851292199510.1016/0024-3205(95)02085-W7674820
|
|
13.
|
B LiuXC PengXL ZhengJ WangYW QinMiR-126
restoration down-regulate VEGF and inhibit the growth of lung
cancer cell lines in vitro and in vivoLung
Cancer66169175200910.1016/j.lungcan.2009.01.01019223090
|
|
14.
|
A TrunehS SharmaC SilvermanS KhandekarMP
ReddyKC DeenMM McLaughlinSM SrinivasulaGP LiviLA
MarshallTemperature-sensitive differential affinity of TRAIL for
its receptors. DR5 is the highest affinity receptorJ Biol
Chem2752331923325200010.1074/jbc.M91043819910770955
|
|
15.
|
FA KruytTRAIL and cancer therapyCancer
Lett2631425200810.1016/j.canlet.2008.02.00318329793
|
|
16.
|
RW JohnstoneAJ FrewMJ SmythThe TRAIL
apoptotic pathway in cancer onset, progression and therapyNat Rev
Cancer8782798200810.1038/nrc246518813321
|
|
17.
|
R KoschnyH WalczakTM GantenThe promise of
TRAIL-potential and risks of a novel anticancer therapyJ Mol
Med85923935200710.1007/s00109-007-0194-117437073
|
|
18.
|
S LacourA HammannA WotawaL CorcosE
SolaryMT Dimanche-BoitrelAnticancer agents sensitize tumor cells to
tumor necrosis factor-related apoptosis-inducing ligand-mediated
caspase-8 activation and apoptosisCancer Res61164516512001
|
|
19.
|
ES HensonJB JohnstonSB GibsonThe role of
TRAIL death receptors in the treatment of hematological
malignanciesLeuk
Lymphoma492735200810.1080/1042819070171365518203008
|
|
20.
|
RK SrivastavaTRAIL/Apo-2L: mechanisms and
clinical applications in
cancerNeoplasia3535546200110.1038/sj.neo.790020311774036
|
|
21.
|
TR SinghS ShankarX ChenM AsimRK
SrivastavaSynergistic interactions of chemotherapeutic drugs and
tumor necrosis factor-related apoptosis-inducing ligand/Apo-2
ligand on apoptosis and on regression of breast carcinoma in
vivoCancer Res63539054002003
|
|
22.
|
ER McDonald IIIPC ChuiPF MartelliDT
DickerWS El-DeiryDeath domain mutagenesis of KILLER/DR5 reveals
residues critical for apoptotic signalingJ Biol
Chem2761493914945200110.1074/jbc.M10039920011279061
|
|
23.
|
J WenN RamadeviD NguyenC PerkinsE
WorthingtonK BhallaAntileukemic drugs increase death receptor 5
levels and enhance Apo-2L-induced apoptosis of human acute leukemia
cellsBlood9639003906200011090076
|
|
24.
|
T NakaK SugamuraBL HylanderMB WidmerYM
RustumEA RepaskyEffects of tumor necrosis factor-related
apoptosis-inducing ligand alone and in combination with
chemotherapeutic agents on patients’ colon tumors grown in SCID
miceCancer Res62580058062002
|
|
25.
|
M NaganeG PanJJ WeddleVM DixitWK CaveneeHJ
HuangIncreased death receptor 5 expression by chemotherapeutic
agents in human gliomas causes synergistic cytotoxicity with tumor
necrosis factor-related apoptosis-inducing ligand in vitro and in
vivoCancer Res608478532000
|
|
26.
|
W LiuE BodleJY ChenM GaoGD RosenVC
BroaddusTumor necrosis factor-related apoptosis-inducing ligand and
chemotherapy cooperate to induce apoptosis in mesothelioma cell
linesAm J Respir Cell Mol
Biol25111118200110.1165/ajrcmb.25.1.447211472983
|
|
27.
|
T MiyashitaJC ReedTumor suppressor p53 is
a direct transcriptional activator of the human bax
geneCell80293299199510.1016/0092-8674(95)90412-37834749
|
|
28.
|
N ShinouraS SakuraiA AsaiT KirinoH
HamadaCaspase-9 transduction overrides the resistance mechanism
against p53-mediated apoptosis in U-87MG glioma
cellsNeurosurgery491771877200111440440
|
|
29.
|
MS SoengasRM AlarconH YoshidaAJ GiacciaR
HakemTW MakSW LoweApaf-1 and caspase-9 in p53-dependent apoptosis
and tumor
inhibitionScience284156159199910.1126/science.284.5411.15610102818
|
|
30.
|
H LeBlancD LawrenceE VarfolomeevK TotpalJ
MorlanP SchowS FongR SchwallD SinicropiA AshkenaziTumor-cell
resistance to death receptor-induced apoptosis through mutational
inactivation of the proapoptotic Bcl-2 homolog BaxNat
Med8274281200210.1038/nm0302-27411875499
|
|
31.
|
G NunezMA BenedictY HuN InoharaCaspases:
the proteases of the apoptotic
pathwayOncogene1732373245199810.1038/sj.onc.12025819916986
|
|
32.
|
SH KaufmannWC EarnshawInduction of
apoptosis by cancer chemotherapyExp Cell
Res2564249200010.1006/excr.2000.483810739650
|
|
33.
|
R NimmanapalliCL PerkinsM OrlandoE
O’BryanD NguyenKN BhallaPretreatment with paclitaxel enhances apo-2
ligand/tumor necrosis factor-related apoptosis-inducing
ligand-induced apoptosis of prostate cancer cells by inducing death
receptors 4 and 5 protein levelsCancer Res617597632001
|
|
34.
|
B GliniakT LeTumor necrosis factor-related
apoptosis-inducing ligand’s antitumor activity in vivo is enhanced
by the chemotherapeutic agent CPT-11Cancer Res59615361581999
|