Introduction
Protocadherin-10 (PCDH10) belongs to the δ2 subgroup
of the protocadherin subfamily. The human PCDH10 gene, also known
as OL-PCDH or KIAA1400, is located at 4q28.3 on the long arm of
chromosome 4 (1). Cell adhesion
proteins are involved in a wide range of roles, including cell
sorting and recognition, boundary formation, induction and
cell-cell adhesion (1,2). The genes encoding adhesion proteins
are perturbed in a multitude of human cancers and have been shown
to promote survival, migration and metastasis (3,4).
Current literature shows that genetic and epigenetic
deregulation of PCDH10 occurs in a multitude of human cancers.
Failure to express PCDH10 may result in loss of inhibition of cell
migration, thereby contributing to cancer progression (3–8).
In addition, increased angiogenesis has been demonstrated in the
bone marrow (BM) microenvironment in hematological malignancies,
including multiple myeloma (MM), suggesting a potential
pathophysiologic role for angiogenesis in MM (9). MM is a plasma cell malignancy
characterized by a tight relationship between tumor cells and the
BM microenvironment that supports myeloma cell growth and survival
(10). In MM, as in solid tumors,
disease progression is characterized by a pre-angiogenic stage of
slow tumor progression followed by an angiogenic switch and a
subsequent angiogenic stage associated with progressive tumor
growth (11). The chick embryo
chorioallantoic membrane (CAM) is commonly used for the study of
in vivo angiogenesis (12,13). Through the effect of PCDH10 in CAM
angiogenic assays, we found the PCDH10 acts as a novel tumor
suppressor gene involved in angiogenesis.
Although the PCDH10 cDNA was cloned in 2000 and much
is known about how PCDH10 functions, the mechanistic basis
governing the basal expression of PCDH10 has not yet been fully
elucidated (14–16). Here we further identified and
characterized the promoter and elements that regulate PCDH10
expression in human MM patient samples and cancer cells.
Materials and methods
Patients characteristics
We studied 44 patients (23 females, 21 males), with
a median age of 66 years (range 43–78 years)admitted to our
Institution between January, 2010 and February, 2011. The diagnosis
was established according to standard morphological and
immunophenotypic criteria and revised according to the
International Staging System (ISS) classification. All patients
gave informed consent. The study was conducted according to good
clinical and laboratory practice rules and the principles of the
Declaration of Helsinki.
Cell culture and sample collection
The KM3 and RPMI-8226 cell lines, gifts from Jian
Hou (The Second Military Medical University, Shanghai, China) were
routinely maintained in RPMI-1640 media (Gibco, USA), supplemented
with 10% heat-inactivated fetal bovine serum (FBS, Gibco). We
obtained all specimens in accordance with the Research Ethics Board
of the Hematology Laboratory of the First Affiliated Hospital of
Chongqing Medical University (Chongqing, China).
5-Aza-2′-deoxycytidine and trichostatin A
treatment
For the treatment combining 5-aza-2′-deoxycytidine
(Aza; Sigma, USA) and trichostatin A (TSA; Sigma), cells were
treated with Aza (10 μM) for 3 days and subsequently with TSA (100
ng/ml) for 24 h as previously described (17).
Total-RNA isolation and semi-quantitative
reverse transcription (RT-PCR)
Following tumour sample homogenization, RNA was
extracted using the TRIzol reagent (Invitrogen). cDNA was then
synthesized using Go-Taq (Promega, USA) and random hexamer primers.
β-actin served as a control for RNA integrity. PCDH10 expression
was analyzed by PCR. The primers used were PCDH10-F, 5′-ACT GCT ATC
AGG TAT GCC TG-3′ and PCDH10-R, 5′-GTC TGT CAA CTA GAT AGC TG-3′;
β-actin-F, 5′-CTC CAT CCT GGC CTC GCT GT-3′ and β-actin-R, 5′-GCT
GTC ACC TTC ACC GTT CC-3′. RT-PCR was performed for 32 cycles for
PCDH10 and 23 cycles for β-actin.
DNA bisulfite treatment and methylation
analysis
DNA was extracted from MM and healthy adult BM
samples by a standard protocol (Zymo Research, USA). Bisulfite
modification of DNA and the methylation status in the CpG islands
of the PCDH10 promoter were carried out as previously described
(3). PCDH10 primers detecting
methylated (M) or unmethylated (U) alleles of the PCDH10 promoter
were: PCDH10-M1, 5′-TCG TTA AAT AGA TAC GTT ACG C-3′ and PCDH10-M2,
5′-TAA AAA CTA AAA ACT TTC CGC G-3′ for methylated alleles;
PCDH10-U1, 5′-GTT GTT AAA TAG ATA TGT TAT GT-3′ and PCDH10-U2,
5′-CTA AAA ACT AAA AAC TTT CCA CA-3′ for unmethylated alleles.
Methylation-specific PCR (MSP) was performed for 40 cycles using
Ampli Taq-Gold (methylation-specific primer, annealing temperature
600˚C; unmethylation specific primer, annealing temperature 580˚C).
MSP primers were first checked for not amplifyling any unbisulfited
DNA and the specificity of MSP was further confirmed by direct
sequencing of some PCR products. PCR reactions were resolved on a
2% agarose gel.
Construction of expression plasmids
pcDNA3.1(+)TP53 was constructed by subcloning the
full-length wild-type TP53 from plasmid pC53-SN (a gift of Bert
Vogelstein) into pcDNA3.1(+). pcDNA3.1(+)PCDH10 was constructed by
cloning the PCR product generated from the full-length clone of
KIAA1400 (gift from Kazusa DNA Research Institute, Japan) with
AccuPrime Pfx DNA Polymerase (Invitrogen). All the plasmid
sequences and orientations were confirmed by sequencing.
Colony formation assays
For the colony formation assay using monolayer
culture, cells (2×105/well) were plated on a 6-well
plate and transfected with expression plasmids or the empty vector
(2 μg each), using Lipofectamine 2000 (Invitrogen). Cells were
collected and plated in a 5-cm dish 48 h post-transfection, and
selected after 21 days with G418 (0.4 mg/ml). Surviving colonies
(50 cells/colony) were counted under a fluorescence microscope.
For the colony formation assay using semi-solid
medium, cells were transfected as above. At 48 h post-transfection,
cells were suspended in RPMI-1640 containing 1% methyl cellulose,
35% FBS and 0.8 mg/ml G418 in a 5-cm dish. The dish was placed in a
sealed chamber and incubated at 37˚C in a 5% CO2
incubator for 21 days. The number of colonies/cm3 was
counted under an inverted microscope. Total-RNA from the
transfected cells was extracted, treated with DNAse I and analyzed
by RT-PCR and western blotting to confirm the ectopic expression of
PCDH10. All the experiments were performed in triplicate wells
three times.
Cell cycle analysis
PCDH10-8226 or Vector-8226 cells were cultured in
RPMI-1640 medium and 10% FBS with G418 (0.4 mg/ml). These cells
were harvested and fixed in ice-cold 70% ethanol for 1 h. The cell
cycle profiles were assayed by the Elite ESP flow cytometer and
data were analyzed with the CellQuest software (BD Biosciences,
USA).
Protein extraction and western blot
analysis
For western blot analysis, total cellular extracts
were obtained by lysis of cells in a lysis buffer and a protease
inhibitor cocktail. Protein concentrations of the cell lysates were
determined by the Bradford method (Bio-Rad, Hercules, CA). An equal
volume of 2X sodium dodecyl sulfate (SDS) loading buffer was added,
and the samples were boiled for 5 min. Protein samples (70 μg/lane)
were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE)
and transferred to nitrocellulose filters (Amersham Biosciences,
Piscataway, NJ). The filters were blocked with Tris-HCl buffer
saline containing Tween-20 buffer (pH 7.6, 10 mM Tris-HCl buffer,
0.15 M NaCl and 0.05% Tween-20) and 5% skim milk at room
temperature for 1 h and then incubated with the primary antibody at
4˚C overnight [1:800 dilution of mouse monoclonal antibody against
human PCDH10, obtained from Abnova; 1:1,000 mouse monoclonal
antibody against glyceraldehyde-3-phosphate dehydrogenase (GAPDH),
Epitomics, Inc., USA], followed by the addition of a horseradish
peroxidase-conjugated antibody (Cell Signaling Technology,
1:2,000). The bands were visualized using the enhanced
chemiluminescence substrate (Cell Signaling Technology).
Chick chorioallantoic membrane (CAM)
assays
White fertilized eggs (56–64 g) with a surface free
of pathogens and without damage were incubated for 8 days. The eggs
were placed in a 37˚C incubator upward and allowed to hatch for 24
h, ensuring 40–60% relative humidity. The eggs were rotated each
morning and evening.
On Day 9, the chick embryos were randomly divided
into 4 groups of 3 embryos each. The chick embryo chorioallantoic
membrane surface spaces (avoiding the vascular chorioallantoic
membrane) were respectively exposed to 100 μl RPMI-1640 medium
(control group), 100 μl of transfected RPMI-8226 cell culture
supernatant (positive control group), 100 μl of transfected
PCDH10-treated RPMI-8226 cell culture supernatant (transfection
group) or 100 μl of RPMI-8226 cell culture supernatant after
transfection with empty plasmid (empty plasmid group), at 38˚C for
48 h. The sample fluid was extracted from 1×106 cells
cultured for 72 h, followed by freezing and dehydration, and cells
were dissolved in 2 ml RPMI-1640 and filtered. All operations were
strictly aseptic.
After 12 days exposure of the embryos to test
material, the chorioallantoic membrane was removed and fixed to
allow the observation of the vascular zone. The vascular networks
in the CAMs were blindly scored for the presence or absence of an
avascular zone larger than 5 mm.
Statistical analysis
All statistical calculations were performed using
SAS version 3.1 for Windows (SAS, Institute, USA). The results are
expressed as values of mean ± SEM. Differences between the
subgroups were tested with the 2-tailed t-test, P-values <0.05
were considered to indicate significant differences.
Results
PCDH10 downregulation and promoter
hypermethylation in MM cell line
To examine if PCDH10 is downregulated in MM, we
first examined its expression in KM3 cells, 3 normal adult BM
samples and 9 MM samples by semi-quantitative RT-PCR. PCDH10 was
silenced in MM cells and 9/9 (100%) MM samples while it was readily
detected in normal adult BM samples (0/3) (Fig. 1A). The PCDH10 CpG island (CGI) was
methylated in KM3 and RPMI-8226 cells (Fig. 1B), while no methylation was
detected in normal adult BM (Fig.
3B), samples showing that silencing of PCDH10 expression in MM
was correlated with its methylation status.
Pharmacological demethylation reactivates
the silenced PCDH10
To evaluate the effect of promoter CGI methylation
on the expression of PCDH10, KM3 and RPMI-8226 cells were treated
with a DNA methytransferase inhibitor, Aza for 3 days together with
a histone deacetylase inhibitor TSA for 1 day. PCDH10 mRNA
expression was dramatically induced after the treatment. This
reactivation was associated with an increase of unmethylated
alleles and a decrease of methylated alleles of the PCDH10
promoter, as assessed by MSP (Fig.
2). These results show a direct link between CGI methylation
and PCDH10 silencing.
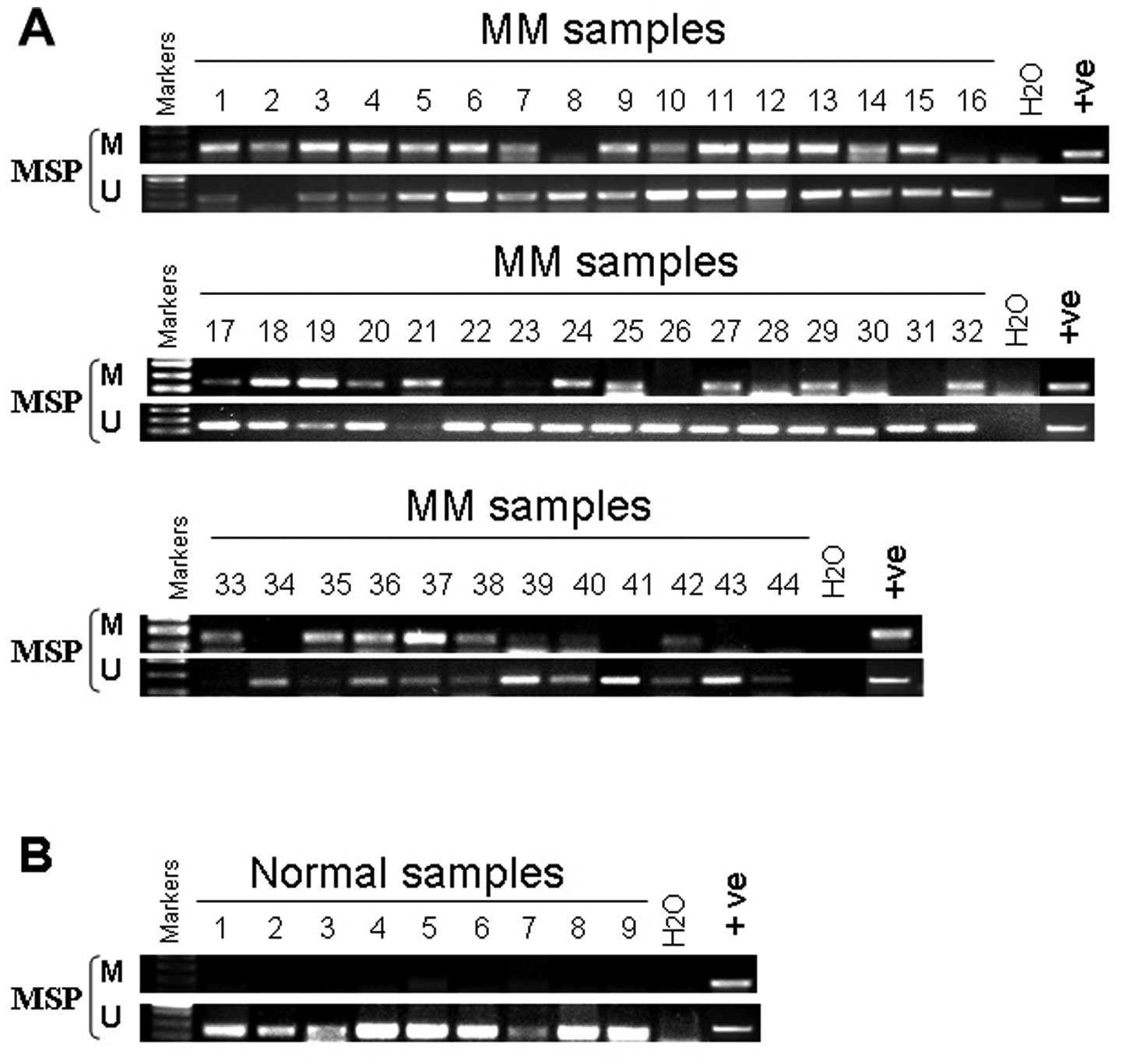
Hypermethylation and disease
evolution
We examined the dynamics of PCDH10 hypermethylation
during disease evolution in MM. We examined sequential samples from
44 patients at diagnosis and after a median of 12 months (range
1–30 months), (Fig. 3A) either
during routine follow-up (n=6), or in the presence of disease
progression (n=8), with blast counts increasing from 6.9±1.7 to
31.7±7.5% (mean ± SEM, P<0.05). We found that the methylation
status of PCDH10 did not change (Table I).
 | Table ICorrelation between PCDH10 gene
methylation and the clinical characteristics of MM patients. |
Table I
Correlation between PCDH10 gene
methylation and the clinical characteristics of MM patients.
| Status of PCDH10
methylation | | |
|---|
|
| | |
|---|
| Patient
parameters | Methylated
(n=34) | Unmethylated
(n=10) | Total (n=44) | P-value |
|---|
| Age at
diagnosis |
| Mean ± SD | 62.3±9.4 | 68.7±6.9 | | |
| Range | 54–77 | 43–78 | 43–78 | 0.074 |
| Gender, n (%) |
| Female | 19 (43.2) | 4 (9.1) | 23 (52.3) | |
| Male | 15 (34.1) | 6 (13.6) | 21 (47.7) | |
| ISS stage, n
(%) | | | | 0.153 |
| I | 12 (27.3) | 3 (6.8) | 15 (34.1) | |
| II | 15 (34.1) | 3 (6.8) | 18 (40.9) | |
| III | 7 (15.9) | 4 (9.1) | 11 (25) | |
| Platelets
(×109) | 162±77 | 231±86 | | 0.885 |
| Hemoglobin
(g/l) | 95.7±23.0 | 93.2±25.2 | | 0.800 |
| Serum calcium
(mg/dl) | 2.23±0.39 | 2.24±0.20 | | 0.599 |
| Serum creatinine
(SCr.) (mg/dl) | 128.0±124.9 | 117.0±116.3 | | 0.801 |
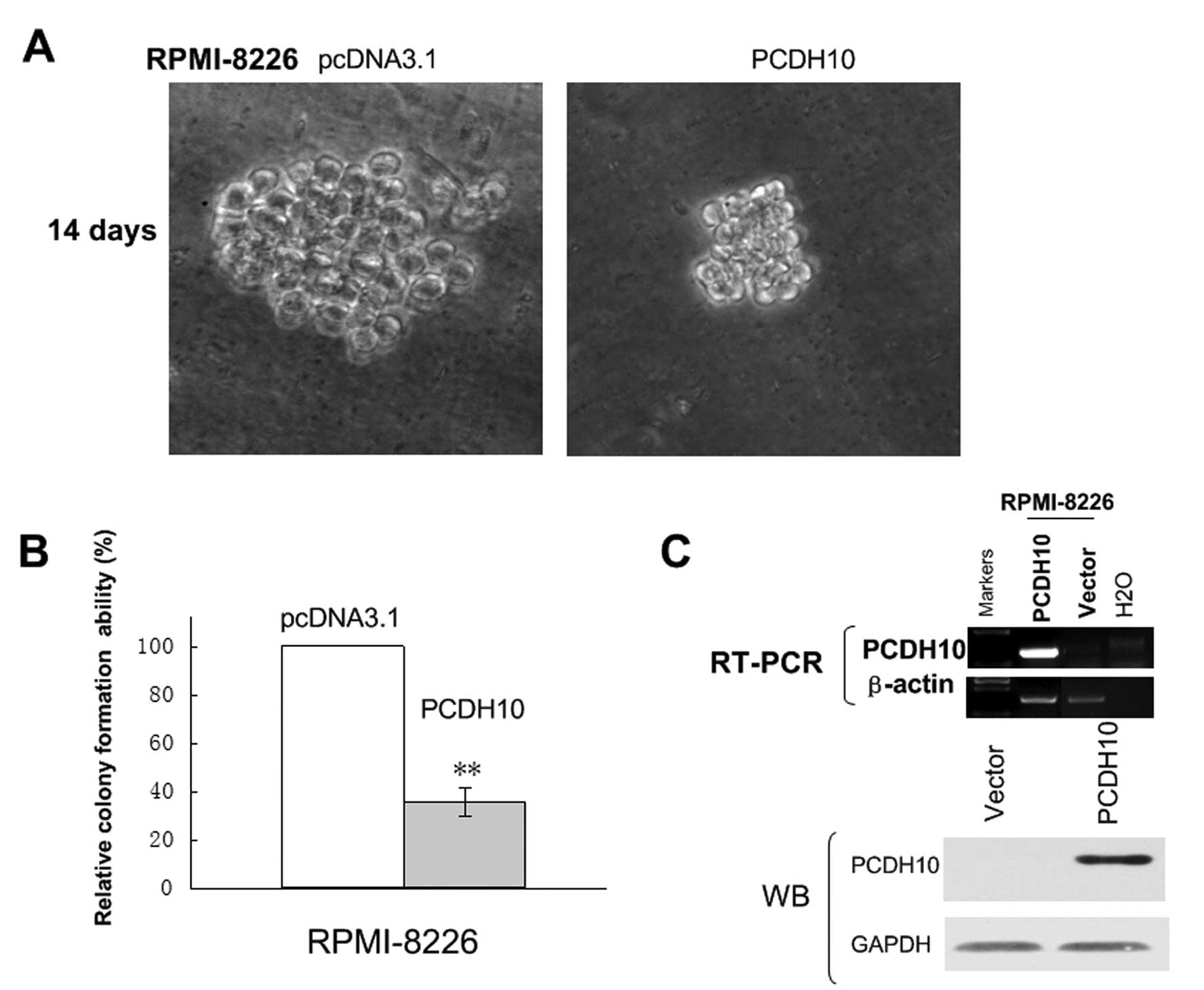
Ectopic expression of PCDH10 inhibits
tumor cell clonogenicity and induces G1 cell cycle arrest
To evaluate the role of PCDH10 as a tumor suppressor
gene (TSG) in MM, we thus sought to establish whether ectopic
expression of PCDH10 could inhibit tumor cell clonogenicity. The
expression vector encoding full-length PCDH10 or vector alone were
transfected into RPMI-8226 cells, in which PCDH10 was fully
silenced by methylation. After G418 seletion for 3 weeks, stable
overexpression of PCDH10 as shown by RT-PCR and western blotting,
was successfully obtained (Fig.
4C).
A colony formation assay (Fig. 4A) was used to evaluate the
suppressor function of PCDH10 in vector of PCDH10-transfected
cells. Ectopic expression of PCDH10 dramatically reduced the colony
formation efficiencies of RPMI-8226 cells (Fig. 4B) down to 40–50% of vector
controls (P<0.05), indicating that PCDH10 functions as a TSG in
KM3 and RPMI-8226 cells.
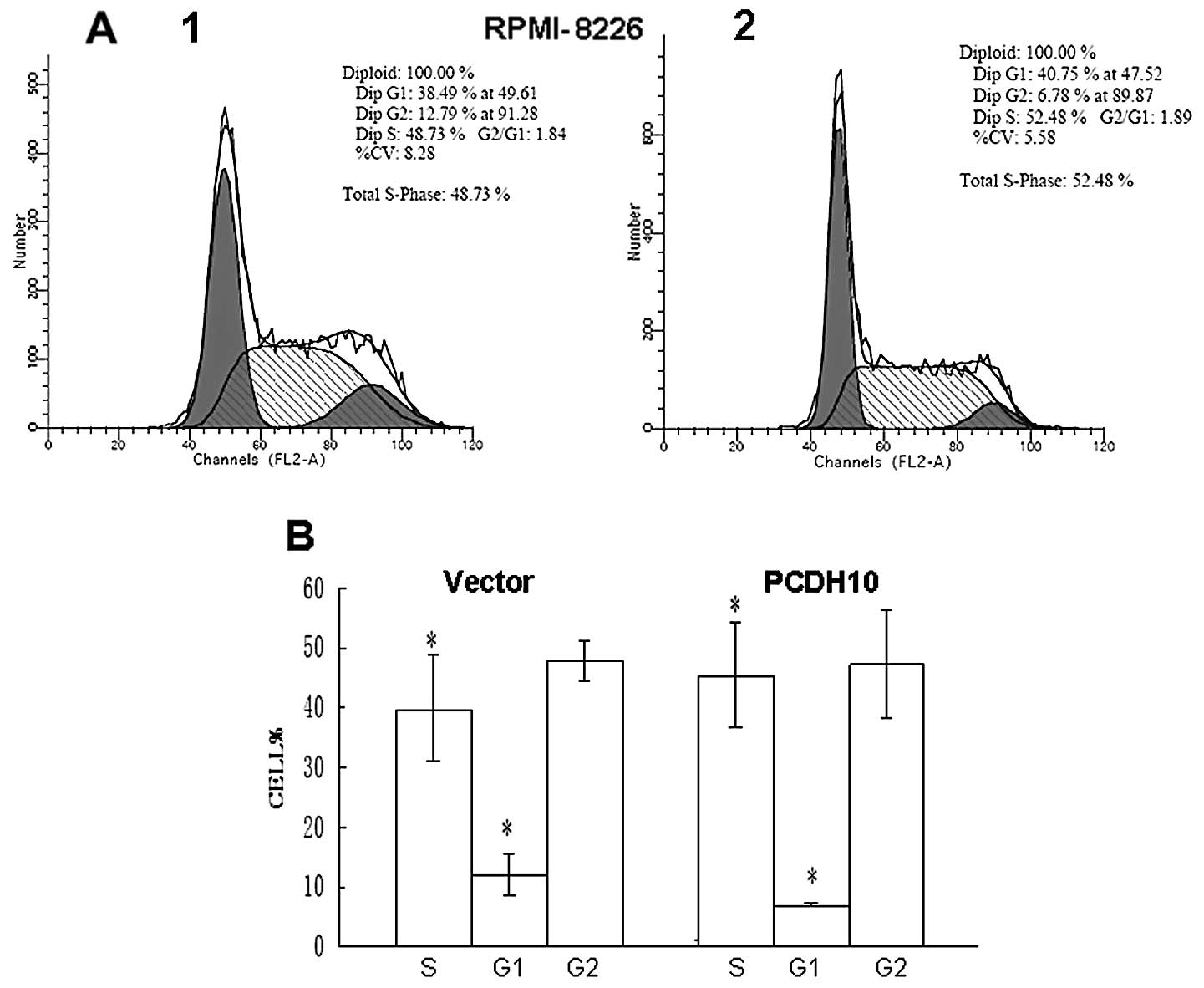
To further explore the mechanism by which PCDH10
suppresses colony formation, we investigated the effect of PCDH10
on cell cycle distribution by flow cytometry. The percentage of
cells in the G1 phase was increased in PCDH10-transfected cells
compared with vector RPMI-8226 cells (P<0.05), indicating that
the effect of PCDH10 was likely to be independent of the cell cycle
(Fig. 5). Thus, collectively the
results demonstrate that PCDH10 has growth inhibitory activity and
is a functional TSG in MM.
Overexpression of PCDH10 has tumor
inhibitory effect on the colony formation of tumor cells
The frequent silencing of PCDH10 in MM cell lines,
compared with its broad expression in normal tissues, suggests that
PCDH10 might have tumor suppressor function. We investigated
whether restoration of PCDH10 could suppress the clonogenicity of
MM cell lines RPMI-8226 which are methylated and silenced for
PCDH10. Two weeks after transfection and subsequent selection of
drug resistant colonies (18,19), the numbers of colonies produced by
PCDH10-transfected cells was significantly less than that by empty
vector-transfected cells, suggesting that PCDH10 does suppress the
colony formation of tumor cells (Fig.
5).
Chick chorioallantoic membrane (CAM)
assays
Addition of a PCDH10 gene transfection cell culture
supernatant in chick embryo CAM and comparison of the vascular map
between this transfection group and the negative control group,
indicated that the vascular branches decreased. On the other hand,
in the positive control group and the blank control group, vascular
branch growth increased, and abnormal shapes were observed
resulting in abnormal vascular network structure.
Statistical analysis show that the chick embryo
chorioallantoic membrane vascular branching points, in the blank
group and positive control group compared with the control and
transfection groups increased about 40–60% (Fig. 6).
The total length of the vascular branches in the
transfection group compared with the control group decreased by 43%
(RPMI-8226) and 66% (KM3), lower than that in the blank group and
positive control group. The increases in the positive control group
compared with the blank group were approximately 21 and 20%
(Fig. 6). The branch vessel total
length of the positive control group and blank group significantly
increased compared to that of the transfection group. The results
show that, PCDH10 can inhibit the growth of chick embryo
chorioallantoic membrane angiogenesis.
Discussion
In this study, we found that PCDH10, located at an
important tumor suppressor locus 4q28.3, was totally silenced in
KM3 and RPMI-8226 cell lines, but widely expressed in normal adult
bone samples. PCDH10 silencing in cancer cell lines was well
correlated with its promoter CpG methylation, which could be
restored by pharmacological demethylation, suggesting that promoter
methylation of PCDH10 plays an important role in its inactivation
in MM (6–8). Moreover, in patients with MM, there
were no associations between hypermethylation and clinical
characteristics, including IPSS score, ISS classification and
cytogenetics. Whether this suggests that in early onset MM, the
methylation of PCDH10 may have changed, or it indicates a change
predicting the pathogenesis of MM, remains to be elucidated. The
present study has proven that PCDH10 is an active member of the MM
gene methylation profiling.
We located the core promoter of PCDH10 to a 462-bp
segment of the 5′-flanking region characterized by a high GC
content. It has also been reported that Sp1/Sp3 and CBF/NF-Y
transcription factors play a crucial role in the basal expression
of the human PCDH10 gene (4).
Gene-specific hypermethylation was observed at the myeloma stage
(20). Methylation was evident at
the development of myeloma, particularly of genes involved in
cell-cell signaling and cell adhesion, which may contribute to
independence from the BM microenvironment. More specifically,
methylation subgroups defined by translocations and hyperdiploidy
were found, with t(4;14) myeloma having the greatest impact on DNA
methylation (21,22). Thus as one of the significant
cadherins, PCDH10 may have the potential to be a epigenetic
regulator.
Substantial evidence has shown that tumor growth and
metastasis are angiogenesis-dependent (12,13,23). In MM, the increased BM
angiogenesis is due to the aberrant expression of angiogenic
factors by myeloma cells, the subsequent increase in pro-angiogenic
activity of normal plasma cells as a result of myeloma cell
angiogenic activity, and the increased number of plasma cells
overall. Molecular mechanisms underlying the angiogenic switch in
MM have recently been identified. The BM microenvironment is
hypoxic and the hypoxia induced transcription factor HIF-1α is
critically involved in the production of angiogenic factors by
myeloma cells. In addition, myeloma cells, stem and progenitor
cells directly produce or induce several pro-angiogenic molecules
in the microenvironment, including VEGF, bFGF, Ang-1, OPN, HGF,
HOXB7, IL-8, and PGE2 (9,24,25).
This complex pathogenesis of myeloma-induced
angiogenesis suggests that several pro-angiogenic molecules and
related genes in the myeloma microenvironment are potential
therapeutic targets (9). Our
results show that PCDH10, an important member of the cadherin
family, may directly induce several pro-angiogenic molecules in the
microenvironment, and plays a positive role in inhibiting tumor
angiogenesis. This indicates that silencing of a tumor suppressor
may be implicated in its role in angiogenesis, and the recovery of
some tumor suppressor gene expression may inhibit angiogenesis so
as to exert the tumor suppressor function (26,27). It is also possible that
epigenetics and angiogenesis have a potential connection, worthy of
further research and discovery.
Vascular disrupting agents (VDAs) target the
cytoskeleton and tubulin network of endothelial cells, thereby
causing vascular disruption and subsequent tumor cell death
(12,13). Moreover, the novel VDA,
plinabulin, inhibits tumor growth in human plasmacytoma mouse
xenograft models, at well-tolerated doses. These studies provide
the rationale for the development of plinabulin as a novel therapy
to improve patient outcome in MM (23,28).
Treatment modalities targeting genes perturbed by
epigenetic mechanisms, such as decitabine, have entered clinical
trials for cancers, such as chronic myeloid leukemia, and other
myelodysplastic syndromes (29–32). As genome-wide approaches begin to
unravel the epigenomic landscape of MM, the utility of targeting
genes and pathways deregulated by epigenetics will be uncovered
(20,32). In the present study, we have shown
that PCDH10 represents a tumour suppressor gene in MM which may
benefit from such an approach. Moreover, PCDH10 involved in
angiogenesis, with its epigenetic silencing correlated with MM.
Thus, further study of the molecular mechanism of PCDH10 is
necessary, and will be the focus of subsequent research in our
laboratory.
Acknowledgements
We thank Dr Qian Tao (State Key Laboratory in
Oncology in South China/Cancer Epigenetics Laboratory; Sir Y.K.
Pao, Center for Cancer; Department of Clinical Oncology; Hong Kong
Cancer Institute and Li Ka Shing Institute of Health Sciences;
Chinese University of Hong Kong, Hong Kong) for guidance, Dr Jian
Hou (the Second Military Medical University, Shanghai, P.R. China)
for the MM cells. We are also thankful to Dr Guo-sheng Ren and Dr
Ting-xiu Xiang (Molecular Oncology and Epigenetics Laboratory, the
First Affiliated Hospital of Chongqing Medical University,
Chongqing, P.R. China) for technical assistance and
encouragement.
References
|
1
|
T WolvertonM LalandeIdentification and
characterization of three members of a novel subclass of
protocadherinsGenomics766672200110.1006/geno.2001.659211549318
|
|
2
|
SY KimS YasudaH TanakaK YamagataH
KimNon-clustered protocadherinCell Adh
Migr597105201110.4161/cam.5.2.1437421173574
|
|
3
|
J YingH LiTJ SengFunctional epigenetics
identifies a protocadherin PCDH10 as a candidate tumor suppressor
for nasopharyngeal, esophageal and multiple other carcinomas with
frequent
methylationOncogene2510701080200610.1038/sj.onc.1209154
|
|
4
|
Z LiJ XieW LiIdentification and
characterization of human PCDH10 gene
promoterGene4754956201110.1016/j.gene.2011.01.00121237250
|
|
5
|
KC BertrandSC MackPA NorthcottPCDH10 is a
candidate tumour suppressor gene in medulloblastomaChilds Nerv
Syst2712431249201110.1007/s00381-011-1486-x21597995
|
|
6
|
PA JonesSB BaylinThe fundamental role of
epigenetic events in cancerNat Rev Genet3415428200212042769
|
|
7
|
PA JonesPW LairdCancer epigenetics comes
of ageNat Genet21163167199910.1038/59479988266
|
|
8
|
SB BaylinJG HermanDNA hypermethylation in
tumorigenesis: epigenetics joins geneticsTrends
Genet16168174200010.1016/S0168-9525(99)01971-X10729832
|
|
9
|
N GiulianiP StortiM BolzoniBD PalmaS
BonominiAngiogenesis and multiple myelomaCancer MicroenvironJuly
72011(Epub ahead of print)
|
|
10
|
KC AndersonRD CarrascoPathogenesis of
myelomaAnnu Rev
Pathol6249274201110.1146/annurev-pathol-011110-130249
|
|
11
|
K AsosinghH De RaeveE MenuAngiogenic
switch during 5T2MM murine myeloma tumorigenesis: role of CD45
heterogeneityBlood10331313137200410.1182/blood-2003-08-294615070695
|
|
12
|
JS KimHK YuJH AhnHuman apolipoprotein(a)
kringle V inhibits angiogenesis in vitro and in vivo by interfering
with the activation of focal adhesion kinasesBiochem Biophys Res
Commun313534540200410.1016/j.bbrc.2003.11.14814697222
|
|
13
|
E CrivellatoB NicoA VaccaV DjonovM PrestaD
RibattiRecombinant human erythropoietin induces intussusceptive
microvascular growth in
vivoLeukemia18331336200410.1038/sj.leu.240324614671634
|
|
14
|
M EstellerRA RisquesM ToyotaPromoter
hypermethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is
associated with the presence of G:C to A:T transition mutations in
p53 in human colorectal tumorigenesisCancer
Res6146894692200111406538
|
|
15
|
M CravoP FidalgoAD PereiraDNA methylation
as an intermediate biomarker in colorectal cancer: modulation by
folic acid supplementationEur J Cancer
Prev3473479199410.1097/00008469-199411000-000047858479
|
|
16
|
PW LairdR JaenischDNA methylation and
cancerHum Mol Genet3Spec No: 1487–14951994
|
|
17
|
HK KuoJD GriffithKN Kreuzer5-Azacytidine
induced methyltransferase-DNA adducts block DNA replication in
vivoCancer
Res6782488254200710.1158/0008-5472.CAN-07-103817804739
|
|
18
|
I SonodaI ImotoJ InoueFrequent silencing
of low density lipoprotein receptor-related protein 1B (LRP1B)
expression by genetic and epigenetic mechanisms in esophageal
squamous cell carcinomaCancer
Res6437413747200410.1158/0008-5472.CAN-04-0172
|
|
19
|
I ImotoY YukiI SonodaIdentification of
ZASC1 encoding a Kruppel-like zinc finger protein as a novel target
for 3q26 amplification in esophageal squamous cell carcinomasCancer
Res6356915696200314522885
|
|
20
|
PA JonesD TakaiThe role of DNA methylation
in mammalian
epigeneticsScience29310681070200110.1126/science.106385211498573
|
|
21
|
BA WalkerCP WardellL ChiecchioAberrant
global methylation patterns affect the molecular pathogenesis and
prognosis of multiple
myelomaBlood117553562201110.1182/blood-2010-04-27953920944071
|
|
22
|
E Martinez-GarciaR PopovicDJ MinThe MMSET
histone methyl transferase switches global histone methylation and
alters gene expression in t(4;14) multiple myeloma
cellsBlood117211220201110.1182/blood-2010-07-29834920974671
|
|
23
|
AV SinghM BandiN RajeA novel vascular
disrupting agent plinabulin triggers JNK-mediated apoptosis and
inhibits angiogenesis in multiple myeloma
cellsBlood11756925700201110.1182/blood-2010-12-323857
|
|
24
|
A VaccaD RibattiAngiogenesis and
vasculogenesis in multiple myeloma: role of inflammatory
cellsRecent Results Cancer
Res1838795201110.1007/978-3-540-85772-3_421509681
|
|
25
|
Y ZhangY DaakaPGE2 promotes angiogenesis
through EP4 and PKA Cγ pathwayBlood11853555364201121926356
|
|
26
|
M PotenteH GerhardtP CarmelietBasic and
therapeutic aspects of
angiogenesisCell146873887201110.1016/j.cell.2011.08.03921925313
|
|
27
|
FJ CarmonaM EstellerIDIBELL Cancer
conference on metastasis and angiogenesisCancer
Res7160976101201110.1158/0008-5472.CAN-11-218021933886
|
|
28
|
KS CohenAngiogenesis and multiple myeloma:
is there a role for angiogenic biomarkers in the context of
autologous stem cell transplant?Leuk
Lymphoma5211731175201110.3109/10428194.2011.59100921699381
|
|
29
|
Y OkiJ JelinekL ShenHM KantarjianJP
IssaInduction of hypomethylation and molecular response after
decitabine therapy in patients with chronic myelomonocytic
leukemiaBlood11123822384200810.1182/blood-2007-07-10396018055864
|
|
30
|
K BalassianoS LimaM JenabAberrant DNA
methylation of cancer-associated genes in gastric cancer in the
European Prospective Investigation into Cancer and Nutrition
(EPIC-EURGAST)Cancer
Lett3118595201110.1016/j.canlet.2011.06.03821831520
|
|
31
|
DJ StewartJP IssaR KurzrockDecitabine
effect on tumor global DNA methylation and other parameters in a
phase I trial in refractory solid tumors and lymphomasClin Cancer
Res1538813888200910.1158/1078-0432.CCR-08-219619470736
|
|
32
|
E FabianiG LeoneM GiacheliaAnalysis of
genome-wide methylation and gene expression induced by
5-aza-2′-deoxycytidine identifies BCL2L10 as a frequent methylation
target in acute myeloid leukemiaLeuk Lymphoma51227522842010
|