Introduction
Pulmonary hypertension (PH) is an insidious and
complex disease involving both the cardiovascular and pulmonary
systems (1). It is a progressive
disease characterized by progressive increasing of pulmonary
vascular resistance, leading to right ventricular hypertrophy,
right heart failure and fatal arrhythmia. According to the PH
guideline of ESC/ERS 2009 and ACCF/AHA, pulmonary arterial
hypertension (PAH) is the most common series of PH (1,2).
Although the imbalance between pulmonary artery vasoconstriction
and vasodilatation is the pathophysiology base of PAH, the
remodeling of pulmonary arteries represents a main pathological
lesion associated with the disease. The abnormal remodeling of the
pulmonary arterial wall includes intimal proliferative, medial
hypertrophy, adventitial thickening and plexiform lesions,
resulting in the obstruction of resistant pulmonary arteries
(3).
The development of PAH involves a complex and
heterogeneous constellation of multiple genetic, molecular, and
humoral abnormalities, presenting a final manifestation of vascular
remodeling in which fibroblasts, smooth muscle and endothelial
cells all have a role (4).
Studies have demonstrated that the proliferation of pulmonary
arterial smooth muscle cells (PASMCs) are a significant feature of
PAH (5). PASMC apoptosis is
suppressed and proliferation is enhanced in experimental PAH
(6). Recently, several groups
have concluded that PAH may be viewed as a disease of excess
proliferation and impaired apoptosis of PASMCs, similarly in some
regards to neoplasia (7). Several
mechanisms have been proposed for the involvement of PASMC abnormal
proliferation in PAH, but the precise mechanisms are not clearly
understood (4,5,8).
The ribosome is a cellular organelle where mRNAs are
translated into proteins in all eukaryotic cells. The ribosome has
about 80 different proteins. Ribosomal proteins (RPs) are important
constituents of the ribosome that have an essential role in the
formation of a fully functional ribosome. It can be predicted that
the defects in RPs can cause ribosome dysfunction (9). Although RPs are essential for cell
growth, the effects of their mutations and their roles in human
diseases have been ignored. With the advancing research during the
past decade, more and more functions of RPs have been discovered.
Studies have shown that RPL11 can form a complex binding with p53
to stabilize p53 protein. RPL11 expression can relieve
HDM2-mediated repression of p53 transactivation and activate
endogenous p53 (10). The defects
of RPs can lead to Diamond-Blackfan anemia (DBA), an inherited red
cell aplasia (11). RPL26 can
bind p53 mRNA to augment its translation and bind Mdm2 to drive its
degradation (12).
Ribosome protein L22 (RPL22), a component of the 60S
eukaryotic ribosomal subunit, is one of the important proteins in
the RP family. RPL22 deficiency may selectively upregulate p53 in
αβ-lineage thymocytes by increasing p53 synthesis. Moreover, RPL22
deficiency has been reported to selectively arrest development of T
cells at the β-selection checkpoint by inducing their death
(13). Our preceding research
showed that iptakalim, a selective KATP channel opener
(14,15), suppressed ET-1-induced HPASMC
proliferation and significantly inhibited RPL22 expression in
HPASMCs (16). It was presumed
that RPL22 may be involved in the proliferation of HPASMCs. In the
present study, siRNA against RPL22 was used to investigate the
effects of RPL22 on HPASMC proliferation and the potential
mechanisms.
Materials and methods
HPASMCs preparation and cultures
Approval for using human specimens in this study was
obtained from the Jiangsu Province Public Hospital Ethical Review
Board (Nanjing, China). Specimens of the human pulmonary arteries
were obtained from the healthy lung segments of the patients
undergoing pulmonary resection at the Department of Cardiothoracic
Surgery, Jiangsu Province Hospital (Nanjing, China). Under sterile
conditions, the intrapulmonary arteries (3rd-4th division) were
dissected from adventitia and opened longitudinally. The
endothelium was separated with a sterile scalpel blade, and the
vessel was cut into 1–3 mm2 pieces. Pieces of the
arteries were incubated in a culture bottle of SMCM with 20% FBS,
100 μg/ml streptomycin, 100 U/ml penicillin (all these were
purchased from ScienCell Ltd.) at 37°C in an atmosphere of 5%
CO2 and 95% air. Cells were confluent after 4 weeks,
then digested with 0.25% trypsin and subcultured. Cells from
passages 4 and 6 were used for the subsequent experiments. The
purity of HPASMCs in the primary cultures was confirmed by positive
staining with a smooth muscle α-actin antibody by
immunocytochemistry as previously described (17).
The cells were randomly divided into six groups:
control group, treated with culture medium alone; siRNA-RPL22
group, treated with siRNA-RP22; siNT group, treated with nontarget
siRNA; ET-1 group, treated with 10 nM ET-1, a suitable
concentration for ET-1 to trigger HPASMC proliferation as
previously described (17);
siRNA-RPL22+ET-1 group, treated with siRNA-RPL22 plus 10 nM ET-1;
siNT+ET-1 group, treated with nontarget siRNA plus 10 nM ET-1.
Before treatment, cells were cultured in serum-free medium for 24 h
to be synchronized.
Small interference RNA (siRNA)
transfection treatment
The RPL22 siRNA (ON-TARGETplus SMARTpool) and
negative control siRNA (siCONTROL Non-Targeting siRNA pool) were
purchased from Thermo Fisher Scientific, Inc. (Waltham, MA). One
day prior to the experiment, cells were plated at 1×105
cells/well in 6-well plates at 2 ml of medium overnight to achieve
the desired density of 50–60% confluency. On the next day, the
cells were transfected with siRNA (control siRNA and siRNA-RPL22,
final concentration: 100 nM) using the X-tremeGENE siRNA
transfection reagent (Roche, USA), according to the manufacturer’s
instructions: 500 pmol of siRNA and 10 μl X-tremeGENE were diluted
in 750 μl of OptiMEM (Gibco-BRL Life Technologies) in one well.
After pre-incubation for 25 min at 37°C, both solutions were mixed
and incubated for an additional 15 min at room temperature. The
X-tremeGENE/siRNA mixture was subsequently overlaid onto the
HPASMCs and incubated for 6 h. Finally, 2 ml of growth medium (10%
fetal bovine serum) per well replaced the mixture for further
cultivation of the HPASMCs.
Real-time transcriptase-polymerase chain
reaction (real-time PCR)
After treating cells as the above groups,
transcripts were measured by TaqMan real-time quantitative
reverse-transcriptase polymerase chain reaction (RT-PCR). Total-RNA
was isolated from tissue samples using TriPure®
Isolation Reagent (Roche, Indianapolis, IN, USA). The reverse
transcription was performed in 300 ng of total-RNA with TaqMan
reverse transcription reagents kit (Takara, Japan). Real-time PCR
was conducted as follows: 95°C at 10 min for 1 cycle, 95°C for 10
sec and 60°C for 1 min for 40 cycles in the ABI PRISM®
7300 Real-time PCR system in a single capillary tube. Forward and
reverse primers were each designed in a different exon of the
target gene sequence, eliminating the possibility of amplifying
genomic DNA. A positive result was determined by identifying the
threshold cycle value at which reporter dye emission appeared above
background. If the fluorescence signal was not detected within 40
cycles, the result was considered negative. The following gene
primers were used: β-actin forward, 5′-TAAAGACCTCTATGCCAACACAGT-3′
and reverse, 5′-CACGATGGAGGGGCCGGACTCATC-3′; RPL22 forward,
5′-CATGCCACTTAGGCCATGACT-3′ and reverse,
5′-TGGTAGCCCCTTTCAGTTGTCTA-3′; cyclin D1 forward,
5′-GGTCTGCGAGGAACAGAAGTG-3′ and reverse,
5′-TGCAGGCGGCTCTTTTTC-3′.
Western blot analysis
The HPASMCs treated as mentioned above after
incubation were washed twice with ice-cold PBS and lysed in 200 μl
lysis buffer (Beyotime, China). The cell lysates were
microcentrifuged at 14,000 rpm for 5 min at 4°C. After being heated
for 5 min at 95°C, 40 μg of denatured protein for each reaction was
used to load a 12% polyacrylamide SDS-PAGE gel. Equal amounts of
protein were transferred to PVDF membranes (Millipore, MA, USA).
The membranes were blocked with 5% non-fat dry milk in
Tris-buffered saline containing 0.1% Tween-20, and incubated with
respective primary antibodies (RPL22 antibody 1:1,000 dilution,
Abcam, USA; cyclin D1 antibody 1:1,000 dilution, Cell Signaling
Technology, USA). The signals were developed using Super-Signal
West Pico chemiluminescent substrate (Pierce, Rockford, IL, USA)
and visualized by Quantity One 4.6.2 software. GAPDH was used as
the control.
Cell counting kit-8
The HPASMCs as described above were planted into
96-well plates in 200 μl of cell culture medium at 3,000 cells/well
supplemented with 2% FBS and incubated for 24 or 48 h. Next, 10 μl
of CCK-8 reagent (Dojindo, Japan) was added to each well 2 h before
the end of incubation. The optical density value (OD) of each
sample was measured at a wavelength of 450 nM on a microplate
reader (Multiskan MK3, Thermo Lab Systems). The results of cell
viability measurement were expressed as the absorbance at
OD450.
Cell cycle phase determination using flow
cytometry
HPASMCs were cultured in culture bottles and brought
to quiescence as described above. After ET-1 (final concentrations
10−8 nM, Sigma) treatment for 24 h, cells were
trypsinized and centrifuged at 1,500 × g for 5 min, and then were
fixed with ice-cold 70% ethanol for 30 min. Finally, the cells were
stained with a mixture of propidium iodide (PI, 50 mg/l) for 30 min
at 4°C protected from light. The minimum of 105 events were
collected and analyzed by flow cytometry in a FACScan
(Becton-Dickinson, Heidelberg, Germany).
PCNA immunofluorescence assay
The HPASMCs were placed onto coverslips, which were
covered in 24-well culture plates with polylysine. After treatment
as described above and incubation for 72 h, the HPASMCs were washed
with PBS, fixed with 4% formaldehyde in PBS for 10 min and blocked
in 1% BSA for 30 min. The cells were incubated with an antibody
against PCNA (1:100 dilution, Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA) overnight at 4°C. Secondary antibodies used were
coupled to goat anti-rabbit IgG Alexa Fluor 488 (1:800 dilution,
A11008; Invitrogen, USA).
Statistical analysis
All data are presented as the mean ± SD of three
independent experiments. Differences between data sets were
assessed by analysis of unpaired Student’s t-test or ANOVA and
post-hoc tests (Student-Newman-Keuls) as indicated. Differences
were considered to be significant at P<0.05.
Results
Expression of RPL22 of HPASMCs with ET-1
condition
To determine RPL22 expression of HPASMC, real-time
PCR and western blot analysis were used to assess RPL22 mRNA and
protein levels of the cells cultured in SMCM alone or combined with
ET-1. Results showed that RPL22 mRNA and protein were increased in
ET-1 medium compared with control groups (P<0.05) (Fig. 1). Consequently, RPL22 expression
increased in ET-1-treated HPASMCs.
Expression of RPL22 after siRNA
interference treatment
To determine the effects of siRNA-RPL22 transfection
on HPASMCs, RPL22 mRNA and protein levels were analyzed by
real-time PCR and western blot analysis after cells were cultured
for 48 h. RPL22 mRNA levels were reduced to 25% in the siRNA-RPL22
group, compared with the siNT group. RPL22 protein levels were
reduced to 38% compared with the siNT group (Fig. 2). Consequently, RPL22 expression
was efficiently inhibited by siRNA-RPL22 transfection.
Role of RPL22 on ET-1 induced HPASMCs
proliferation and cell cycle alteration
To determine the effects of siRNA-RPL22 on ET-1
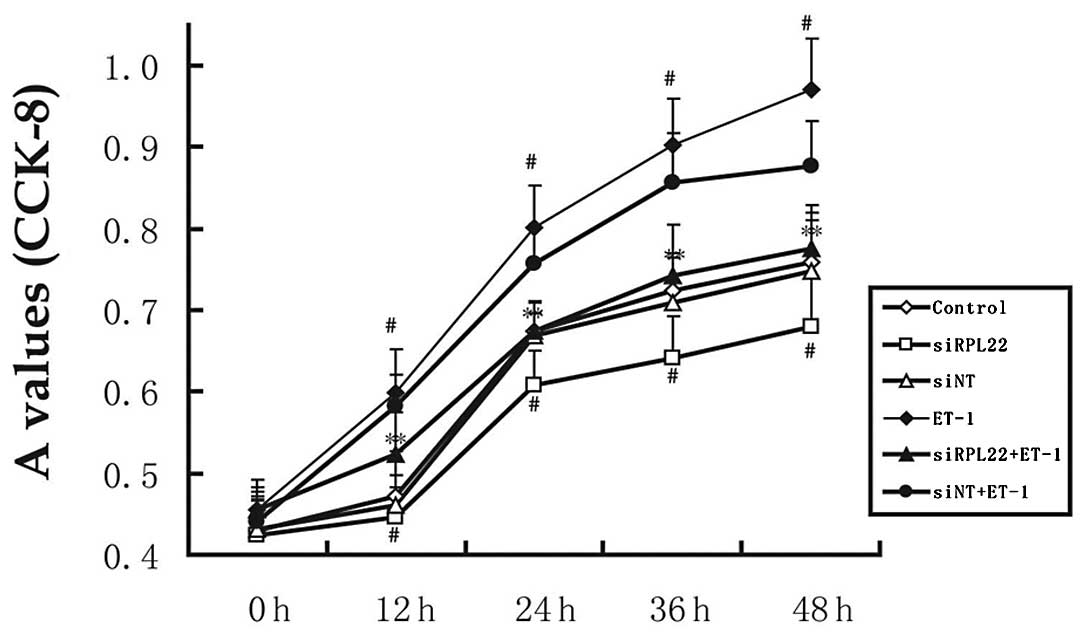
induced proliferation of HPASMCs, the CCK-8 assay was used to
assess HPASMC proliferation (Fig.
3). ET-1 significantly enhanced the proliferation of HPASMCs
(P<0.05 vs. control). However, transient expression of RPL22
inhibited ET-1-induced proliferation of HPASMCs. After incubation
for 24 and 48 h, the effects in the siRPL22 group were
significantly lower compared with the controls (P<0.05).
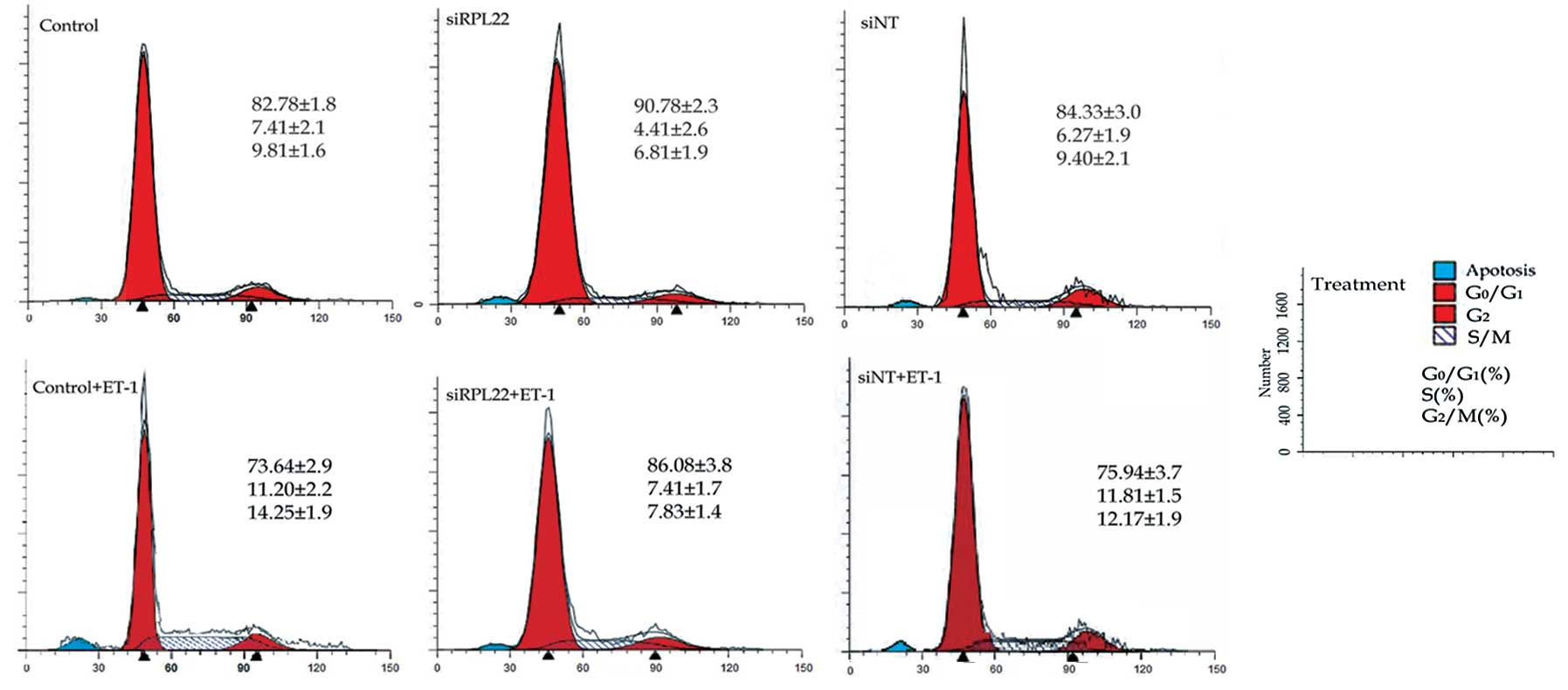
Cell cycle analysis was performed to determine the
cell cycle alteration in the HPASMCs treated as above. When treated
with 10 nM ET-1 alone, HPASMCs were propelled from the static phase
(G0/G1) to the DNA synthesis (S) and mitotic phase (G2/M). The
ratios of the S and G2/M phases were increased by 1.8- and
1.5-fold, respectively compared to the control cells. However, the
ratios of the S and G2/M phases were decreased insignificantly in
the siRPL22 group cells compared to the control group (Fig. 4).
Role of RPL22 on ET-1-induced expression
of PCNA
The expression of PCNA, a cell cycle-associated
protein, was assessed by immunofluorescence (IF) analyses to
further confirm the effects of siRNA-RPL22 on the proliferation of
HPASMCs (Fig. 5). The percentages
of PCNA protein positive cells were significantly reduced in the
ET-1+siRPL22 group compared to the control and siNT groups.
Consequently, siRNA-RPL22 inhibited HPASMC proliferation induced by
ET-1.
Role of RPL22 expression of cyclin D1 in
HPASMCs
Using real-time PCR and western blot analysis, the
mRNA and protein expression of CCND1 were analyzed (Fig. 6). Statistically significant
(P<0.05) increases compared to the control group were noted in
the ET-1 group; statistically significant (P<0.05) changes
compared to the siRPL22+ET-1 group were noted in the siRPL22 and
siNT+ET-1 groups. Consequently, inhibition of RPL22 downregulated
CCND1 expression.
Discussion
The present study provides the first direct evidence
that RPL22 is involved in HPASMC proliferation. The development of
PAH involves a complex constellation of multiple genes and factors,
which interact with each other and subsequently activate
intracellular signaling pathways that eventually result in the
remodeling of pulmonary artery (4,5,8).
Many factors, such as ET-1 (18),
serotonin (19), potassium
channel (20), caveolins
(21), VEGF (22), PDGF (23), bone morphogenetic protein (BMP)
(24) involved in PAH progression
are thought to play roles on HPASMC proliferation. However, the
precise mechanisms of PAH are still not completely unclear.
Pulmonary artery vasoconstriction and vascular remodeling are
pathological bases of PAH. Pulmonary arterial smooth muscle
hypertrophy, the main cause of pulmonary artery vascular
remodeling, is characterized by enhanced HPASMC proliferation
(4,8).
Our previous study showed that iptakalim (14,15), inhibited ET-1 synthesis in
cultured endothelial cells and suppressed ET-1-induced
proliferation of HPASMCs in vitro (17,25). In a proteomic study, it was found
that iptakalim significantly inhibited protein expression of RPL22
in HPASMCs (16). Therefore, we
presumed that RPL22 may be involved in the proliferation of
HPASMCs. In this study, we firstly demonstrated that HPASMC
proliferation was suppressed by siRNA-RPL22 interference. To
investigate the effects of RPL22 on HPASMC proliferation, ET-1 was
used as an activation factor.
ET-1 is a vasoactive peptide upregulated in PAH
patients and exerts diverse actions on HPASMCs by interacting with
the G protein-coupled receptors (26,27). It is synthesized and released
particularly by endothelial cells and PASMCs in the pulmonary
circle (28). Previous studies
have confirmed that ET-1 acts as the most potent vasoconstrictor
and directly modulates HPASMCs growth by acting as an autocrine and
paracrine secretion mitogen (29). Its effects include cell
proliferation and contraction and it mediates cellular effects via
two receptors, ETA and ETB. ETA receptor is
the predominant subtype of HPASMCs. It acts as a strong
growth-promoting factor affecting HPASMC proliferation. ET-1 has
also been reported to have anti-apoptotic effects on HPASMCs
(30). To assess the effects on
HPASMCs, we chose ET-1 as the activation factor. In the present
study, ET-1 significantly enhanced proliferation of HPASMCs.
The ribosome is a cellular organelle, where mRNAs
carrying DNA message assemble with ribosomal subunits and associate
with tRNA, responsible for protein synthesis. In all eukaryotic
cells, the ribosome consists of a small 40S subunit and a large 60S
subunit (31). These subunits are
composed of 4 RNAs and ∼80 distinct proteins (32). Ribosomal proteins are the basic
structure of the ribosome. They play an essential role in the
formation of a fully functional ribosome. RPL22 is a component of
the 60S large subunit and is encoded by the RPL22 gene. It has been
confirmed to be located on chromosome 1 in eukaryotes (9). In the present study, we designed an
siRNA against the RPL22 gene to inhibit RPL22 expression.
By real-time PCR and western blot analysis, results
showed that siRNA-RPL22 transfection significantly inhibited RPL22
expression in HPASMCs. To confirm the effects of RPL22 deficiency
on HPASMC proliferation, we examined HPASMC proliferation by
different ways including PCNA immunocytochemistry, the CCK-8 test
and flow cytometry analysis. Proliferating cell nuclear antigen
(PCNA) is a protein that acts as a processivity factor for DNA
polymerase. In this study, by PCNA immunocytochemical staining, we
found that compared with the control group, HPASMC proliferation
was significantly increased in the ET-1 group and was inhibited in
the RPL22-deficient groups with or without ET-1. The results of the
CCK-8 assay and of the cell flow cytometry analysis also showed
that HPASMC proliferation was significantly suppressed by
inhibiting RPL22 expression. These results confirmed that
inhibiting RPL22 expression can inhibit HPASMC proliferation and
inhibit the proliferation effect of ET-1 on HPASMCs.
Furthermore, it was demonstrated that RPL22
deficiency can reduce CCND1 expression. CCND1 is one of the key
proteins in the CCND protein family which control the progression
of cells through the cell cycle by activating cyclin dependent
kinases (CDKs) and plays an important role in the transition from
the G1 to the S phase of the cell cycle during cell division. CCND1
is required in human cell proliferation and plays a key role at the
G1 phase (33). In this study, we
found that when HPASMCs were transfected with siRNARPL22, the CCND1
expression was significantly suppressed and the proliferation
ability of HPASMCs was significantly impaired. This result show
that RPL22 downregulation can inhibit CCND1 synthesis.
Ribosome gene disruption is not lethal as shown for
other ribosomal proteins, and further supported by the fact that
the effects of deficiency of some ribosomal protein can cause
critical diseases (34). Our
study showed that RPL22 deficiency can influence the synthesis of
an proliferation factor of HPASMCs. RPL22 deficiency can inhibit
CCND1 synthesis. It can be concluded that inhibiting CCND1
synthesis is one of the effects of RPL22 on HPASMC
proliferation.
In summary, our present study confirms that RPL22
expression can be inhibited by siRNA-RPL22 transfection. The
results demonstrate that siRNA-RPL22 can inhibit ET-1-induced
HPASMC proliferation. These findings provide evidence to support
that RPL22 is involved in HPASMC proliferation. This novel finding
needs to be further explored.
Acknowledgements
We thank Dr G. Hu (Department of
Pharmacology, Nanjing Medical University, Nanjing, China) for
excellent technical assistance. This study was supported by the
State Natural Science Funds Commission and by the Funds of the
National Natural Science Foundation of China (Grant no.
30971319).
References
|
1.
|
N GalièMM HoeperM HumbertGuidelines for
the diagnosis and treatment of pulmonary hypertensionEur Heart
J30249325372009
|
|
2.
|
VV McLaughlinSL ArcherDB BadeschACCF/AHA
2009 expert consensus document on pulmonary
hypertensionCirculation11922502294200910.1161/CIRCULATIONAHA.109.192230
|
|
3.
|
S StewartD RasslAdvances in the
understanding and classification of pulmonary
hypertensionHistopathology54104116200910.1111/j.1365-2559.2008.03180.x19187180
|
|
4.
|
M HumbertNW MorrellSL ArcherKR StenmarkMR
MacLeanIM LangBW ChristmanEK WeirO EickelbergNF VoelkelM
RabinovitchCellular and molecular pathobiology of pulmonary
arterial hypertensionJ Am Coll Cardiol43Suppl
12S13S24200410.1016/j.jacc.2004.02.02915194174
|
|
5.
|
M MandegarYC FungW HuangCV RemillardLJ
RubinJX YuanCellular and molecular mechanisms of pulmonary vascular
remodeling: role in the development of pulmonary
hypertensionMicrovasc
Res6875103200410.1016/j.mvr.2004.06.00115313118
|
|
6.
|
E GurbanovX ShiliangThe key role of
apoptosis in the pathogenesis and treatment of pulmonary
hypertensionEur J Cardiothorac
Surg30499507200610.1016/j.ejcts.2006.05.02616870458
|
|
7.
|
SL ArcherEK WeirMR WilkinsBasic science of
pulmonary arterial hypertension for clinicians: new concepts and
experimental
therapiesCirculation12120452066201010.1161/CIRCULATIONAHA.108.84770720458021
|
|
8.
|
NW MorrellS AdnotSL ArcherCellular and
molecular basis of pulmonary arterial hypertensionJ Am Coll
Cardiol54Suppl 1S20S31200910.1016/j.jacc.2009.04.01819555855
|
|
9.
|
T UechiT TanakaN KenmochiA complete map of
the human ribosomal protein genes: assignment of 80 genes to the
cytogenetic map and implications for human
disordersGenomics72223230200110.1006/geno.2000.647011401437
|
|
10.
|
Y ZhangGW WolfK BhatA JinT AllioWA
BurkhartY XiongRibosomal protein L11 negatively regulates
oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress
checkpoint pathwayMol Cell
Biol2389028912200310.1128/MCB.23.23.8902-8912.200314612427
|
|
11.
|
HT GazdaMR SheenA VlachosV ChoesmelMF
O’DonohueH SchneiderN DarrasC HasmanCA SieffPE NewburgerRibosomal
protein L5 and L11 mutations are associated with cleft palate and
abnormal thumbs in Diamond-Blackfan anemia patientsAm J Hum
Genet83769780200810.1016/j.ajhg.2008.11.00419061985
|
|
12.
|
Y Ofir-RosenfeldK BoggsD MichaelMB KastanM
OrenMdm2 regulates p53 mRNA translation through inhibitory
interactions with ribosomal protein L26Mol
Cell32180189200810.1016/j.molcel.2008.08.03118951086
|
|
13.
|
SJ AndersonJP LauritsenMG HartmanAM
FousheeJM LefebvreSA ShintonB GerhardtRR HardyT OraveczDL
WiestAblation of ribosomal protein L22 selectively impairs αβ T
cell development by activation of a p53-dependent
checkpointImmunity26759772200717555992
|
|
14.
|
Z PanJ HuangW CuiC LongY ZhangH
WangTargeting hypertension with a new adenosine
triphosphate-sensitive potassium channel opener iptakalimJ
Cardiovasc
Pharmacol56215228201010.1097/FJC.0b013e3181e23e2b20410832
|
|
15.
|
H WangY TangYL ZhangHypoxic pulmonary
hypertension (HPH) and iptakalim, a novel ATP-sensitive potassium
channel opener targeting smaller arteries in hypertensionCardiovasc
Drug Rev23293316200510.1111/j.1527-3466.2005.tb00174.x16614730
|
|
16.
|
MX YangZX LiuS ZhangY JingSJ ZhangWP XieL
MaCL ZhuH WangProteomic analysis of the effect of iptakalim on
human pulmonary arterial smooth muscle cell proliferationActa
Pharmacol Sin30175183200910.1038/aps.2008.3019169269
|
|
17.
|
YM ZhuSJ ZhangWP XieQL LiYJ ZhouH
WangIptakalim inhibited endothelin-1-induced proliferation of human
pulmonary arterial smooth muscle cells through the activation of
KATP channelVascul
Pharmacol489299200810.1016/j.vph.2008.01.00118276195
|
|
18.
|
S SakaoK TatsumiNF VoelkelEndothelial
cells and pulmonary arterial hypertension: apoptosis,
proliferation, interaction and transdifferentiationRespir
Res1095103200910.1186/1465-9921-10-95
|
|
19.
|
MR MacleanY DempsieThe serotonin
hypothesis of pulmonary hypertension revisitedAdv Exp Med
Biol661309322201010.1007/978-1-60761-500-2_2020204739
|
|
20.
|
ED BurgCV RemillardJX YuanPotassium
channels in the regulation of pulmonary artery smooth muscle cell
proliferation and apoptosis: pharmacotherapeutic implicationsBr J
Pharmacol153Suppl 1S99S111200810.1038/sj.bjp.070763518084317
|
|
21.
|
YY ZhaoAB MalikA novel insight into the
mechanism of pulmonary hypertension involving caveolin-1 deficiency
and endothelial nitric oxide synthase activationTrends Cardiovasc
Med19238242200910.1016/j.tcm.2010.02.00320382348
|
|
22.
|
L Taraseviciene-StewartY KasaharaL AlgerP
HirthG McMahonJ WaltenbergerNF VoelkelRM TuderInhibition of the
VEGF receptor 2 combined with chronic hypoxia causes cell
deathdependent pulmonary endothelial cell proliferation and severe
pulmonary hypertensionFASEB
J15427438200110.1096/fj.00-0343com11156958
|
|
23.
|
RT SchermulyE DonyHA GhofraniS
PullamsettiR SavaiM RothA SydykovYJ LaiN WeissmannW SeegerF
GrimmingerReversal of experimental pulmonary hypertension by PDGF
inhibitionJ Clin Invest11528112821200510.1172/JCI2483816200212
|
|
24.
|
G LagnaPH NguyenW NiA HataBMP-dependent
activation of caspase-9 and caspase-8 mediates apoptosis in
pulmonary artery smooth muscle cellsAm J Physiol Lung Cell Mol
Physiol291L1059L1067200610.1152/ajplung.00180.200617030903
|
|
25.
|
WP XieH WangJH DingH WangG
HuAnti-proliferating effect of iptakalim, a novel KATP
channel opener, in cultured rabbit pulmonary arterial smooth muscle
cellsEur J Pharmacol5118187200510.1016/j.ejphar.2005.01.039
|
|
26.
|
B RohitMT RubinMH PaulEndothelial
dysfunction in pulmonary
hypertensionCirculation109159165200410.1161/01.CIR.0000102381.57477.5014734504
|
|
27.
|
N DavieSJ HaleenPD UptonJM PolakMH
YacoubNW MorrellJ WhartonETA and ETB receptors modulate
the proliferation of human pulmonary artery smooth muscle cellsAm J
Respir Crit Care Med1653984052002
|
|
28.
|
E TronicDJ WebbEndothelium-derived
endothelin-1Pflug Arch Eur J
Phy459951958201010.1007/s00424-009-0763-y
|
|
29.
|
I KomuroH KuriharaT SugiyamaM YoshizumiF
TakakuY YazakiEndothelin stimulates c-fos and c-myc expression and
proliferation of vascular smooth muscle cellsFEBS
Lett238249252198810.1016/0014-5793(88)80489-73139457
|
|
30.
|
RP JankovC KantoresR BelcastroM YiAK
TanswellEndothelin-1 inhibits apoptosis of pulmonary arterial
smooth muscle in the neonatal ratPediatr
Res60245251200610.1203/01.pdr.0000233056.37254.0b16857764
|
|
31.
|
S RobledoRA IdolDL CrimminsJH LadensonPJ
MasonM BesslerThe role of human ribosomal proteins in the
maturation of rRNA and ribosome
productionRNA1419181929200810.1261/rna.113200818697920
|
|
32.
|
D Hernandez-VerdunAssembly and disassembly
of the nucleolus during the cell
cycleNucleus2189194201110.4161/nucl.2.3.1624621818412
|
|
33.
|
DW StaceyCyclin D1 serves as a cell cycle
regulatory switch in actively proliferating cellsCurr Opin Cell
Biol15158163200310.1016/S0955-0674(03)00008-512648671
|
|
34.
|
KA McGowanJZ LiCY ParkV BeaudryHK TaborAJ
SabnisW ZhangH FuchsMH de AngelisRM MyersRibosomal mutations cause
p53-mediated dark skin and pleiotropic effectsNat
Genet40963970200810.1038/ng.18818641651
|