Introduction
Although tuberculosis (TB) is an ancient disease
resulting from infection with Mycobacterium tuberculosis
(M. tuberculosis), it remains a great threat to both
individual and public health throughout the world. It is reported
that approximately one-third of the world’s population has been
latently infected (1). The
prevalence of human immunodeficiency virus has enhanced the spread
of multi-drug resistant and extensively drug resistant tuberculosis
strains, and the morbidity and mortality of TB have been rising
yearly without much curative success using existing anti-TB drugs
(2–4). Therefore, it is a matter of urgency
to discover targets for new anti-TB drugs. Serine acetyltransferase
(CysE) is involved in the biosynthesis of cysteine, which catalyzes
the conversion of acetyl-CoA (AcCoA) and L-serine (L-Ser) to CoA
and O-acetyl-L-serine (OAS) (5,6).
This reaction is the first step in the two-step biosynthesis of
L-cysteine in microorganisms and plants (7,8).
Because of the differing pathways for cysteine anabolism in humans
and microorganisms (9), serine
acetyltransferase exists only in microorganisms. An ideal drug
target should be unique to the pathogen, thus M.
tuberculosis serine acetyltransferase is regarded as a
potential drug target (10,11).
The CysE protein has been purified and characterized
from certain bacteria, such as Escherichia coli (6,12,13), Salmonella typhimurium
(5,14) and Haemophilus influenzae
(15). Bioinformatic analyses
have shown that M. tuberculosis Rv2335 is homologous to
E. coli CysE, S. typhimurium CysE and H.
influenzae CysE. Therefore, M. tuberculosis Rv2335
(GenBank accession no. CAB06152.1) could be a cysE gene that
encodes the CysE protein.
In this study, we cloned and expressed the M.
tuberculosis cysE (Rv2335) gene in E. coli and
characterized the purified M. tuberculosis CysE protein. The
kinetic studies on M. tuberculosis CysE allow for the
screening of its inhibitors in the development of anti-TB
drugs.
Materials and methods
Microorganisms and plasmids
E. coli NovaBlue and E. coli BL21
(DE3) (Novagen) were maintained as the hosts for cloning and
expression, respectively. The cloning plasmid pMD18-T (Takara) with
the ampicillin resistance gene was utilized to clone and sequence
the target gene or DNA fragment. The expression vector pET29b
(Novagen) carrying the kanamycin resistance gene was used for gene
expression in E. coli. M. tuberculosis H37Rv genomic DNA was
supplied by Colorado State University via an NIH contract.
Cloning the cysE (Rv2335) gene from M.
tuberculosis H37Rv genomic DNA
The M. tuberculosis cysE gene was amplified
from M. tuberculosis H37Rv genomic DNA using the following
set of primers: cysE forward, 5′-AACATATGCT GACGGCCATGCGGG-3′
(underlined sequence is the NdeI site) and cysE
reverse primer, 5′-AACTCGAGGATCGAG AAGTCCTCGCCG-3′
(underlined sequence is the XhoI site). The amplified PCR
product was ligated into pMD18-T to generate the plasmid
pMD18-cysE, which was transformed into E. coli
NovaBlue. The positive recombinant plasmid pMD18-cysE was
confirmed by digestion with restriction endonucleases
(EcoRI) and subsequently sequenced. The cysE gene was
subcloned into the NdeI and XhoI sites of pET29b,
yielding the expression vector pET29b-cysE.
Expression, purification and
identification of CysE protein
The plasmid pET29b-cysE was transformed into
E. coli BL21 (DE3). BL21 (DE3)/pET29b-cysE culture
was induced with 1 mM IPTG at 37°C for 3 h. The cells were
harvested and suspended in lysis buffer (20 mM Tris-HCl pH 8.0, 100
mM NaCl, 25 mM MgCl2, 5% (v/v) glycerol, 1 mM EDTA, 1 mM
β-mercaptoethanol and 1 mM PMSF). The cells were homogenized by
sonication and the cell lysate was centrifuged at 20,000 × g for 20
min. The supernatant was then loaded onto a 1-ml Ni-NTA agarose
column (Qiagen). The column was then washed with 20 ml of wash
buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 20% glycerol, 60 mM
imidazole and 1 mM PMSF), and the CysE protein with a His-tag at
its C-terminus was eluted with 10 ml of elution buffer (20 mM
Tris-HCl pH 8.0, 500 mM NaCl, 20% glycerol, 300 mM imidazole and 1
mM PMSF) and examined by SDS-PAGE and western blotting. The
purified CysE protein was further confirmed by matrix-assisted
laser desorption/ionization-time of flight mass spectrometry
(MALDI-TOF-MS) (BIG, China).
Enzyme assays
The serine acetyltransferase activity of the CysE
protein was determined by monitoring the increase in the absorbance
of Ellman’s reagent (DTNB) due to its reaction with CoA (16,17). Briefly, a 50-μl reaction mixture
(50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 0.4 mM AcCoA, 2 mM
L-Ser and 0.037 μg purified CysE protein) in a 96-well microtiter
plate was incubated at 37°C for 20 min. A blank control without
L-Ser and AcCoA, and a positive control containing standard CoA
(0.2 mM) only were included. The reaction was terminated with 50 μl
of stop solution (50 mM Tris-HCl pH 7.5, 6 M guanidine
hydrochloride). Fifty microliters of Ellman’s reagent (50 mM
Tris-HCl pH 7.5, 0.2 mM DTNB and 1 mM EDTA) was added to the
reaction mixture. The mixture was incubated at room temperature for
10 min. The absorbance values were obtained using a microplate
reader (Multiskan Ascent; Thermo Scientific) at a wavelength of 405
nm (18). One unit of specific
enzyme activity was defined as 1 μmol of CoA-SH produced by 1 mg
protein/min under specific conditions.
Characterization of M. tuberculosis
CysE
The maximum velocity (Vmax) and Michaelis
constant (Km) of M. tuberculosis CysE were
measured by a colorimetric assay coupled with DTNB. Based on the
concentration curves and time-course curves of CysE, the range of
CysE initial velocities was measured. The concentration curves of
CysE were plotted by measuring the reaction velocities at varying
CysE concentrations and reaction times. The reactions were
performed in 50 mM Tris-HCl buffer (pH 7.5) containing AcCoA, L-Ser
and different concentrations of purified CysE (0.74, 1.48, 2.22,
2.96 and 3.70 μg/ml) at 37°C for 5, 15 and 25 min. The time-course
curves were plotted by measuring the amount of CoA at different
reaction times (5, 10, 15, 20 and 25 min) and different
concentrations of CysE (0.74, 2.22 and 3.70 μg/ml) at 37°C. To
further characterize the CysE, the effect of pH, temperature, and
Mg2+ concentration on CysE were evaluated by measuring
CysE activity in different pH buffers (3–11),
at various temperatures (16–80°C) and concentrations of
Mg2+ (0–20 mM), respectively.
In dual-substrate reactions, the steady-state
kinetic parameters Km and Vmax were
calculated by double reciprocal plots prepared by varying the
concentration of one substrate while the second substrate was in
excess under optimal conditions.
Results
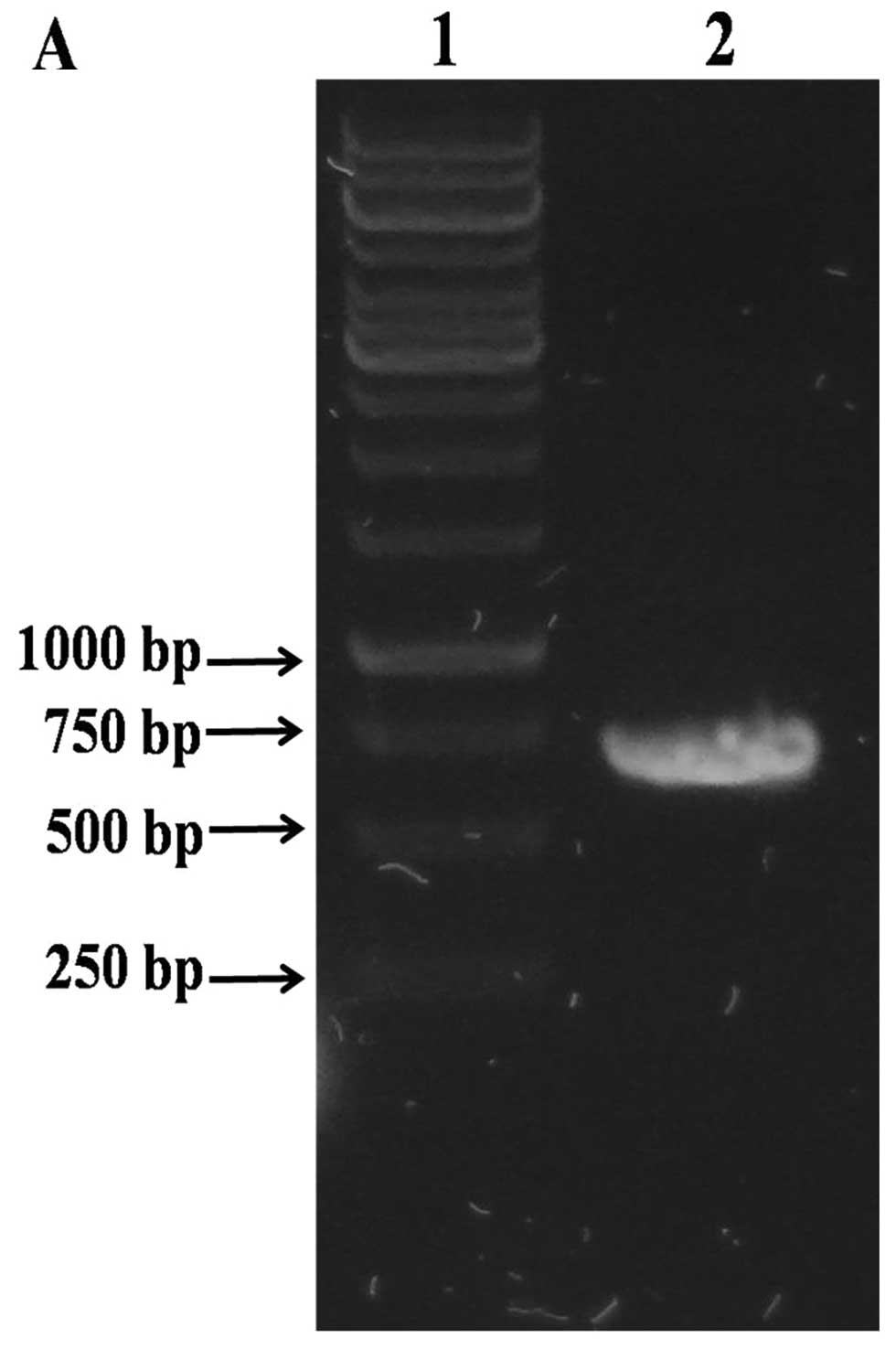
Cloning of the M. tuberculosis cysE
gene
The PCR product for the cysE gene was
obtained from the genomic DNA of M. tuberculosis H37Rv
(Fig. 1A). The size of the PCR
product (cysE gene plus NdeI and XhoI
recognition sites) was 700 bp.
Expression, purification and
identification of the CysE protein
The soluble M. tuberculosis CysE protein was
expressed in E. coli BL21 (DE3) by induction with 1 mM IPTG.
The purified CysE protein was detected by SDS-PAGE (Fig. 1B) and western blotting (Fig. 1C). The band of the CysE protein
appeared at 30 kDa, which was higher than the theoretical molecular
mass (24.6 kDa) of the CysE protein. The purified CysE protein was
further confirmed by MALDI-TOF-MS analysis (data not shown).
Serine acetyltransferase activity of M.
tuberculosis CysE protein
The serine acetyltransferase activity of M.
tuberculosis CysE protein was detected. The specific activity
of the serine acetyltransferase was 10.66±0.44 μmol/min/mg
(Table I).
 | Table ISpecific activity and kinetic
parameters of M. tuberculosis CysE. |
Table I
Specific activity and kinetic
parameters of M. tuberculosis CysE.
| Specific activity
(μmol·min−1·mg−1) | Vmax
(mM·min−1) | KAcCoA
(mM) | Kser
(mM) | Kcat
(sec−1) |
|---|
| M.
tuberculosis CysE | 10.66±0.44 | 0.0073±0.0005 | 0.0513±0.0050 | 0.0264±0.0006 | 81.36±5.22 |
Characterization of M. tuberculosis
CysE
The reaction velocity was proportional to the
concentration of M. tuberculosis CysE when the reaction time
was 5 min (Fig. 2A). At 15 or 25
min reaction times, the reaction velocity gradually slowed and
became non-linear with the CysE concentration. Therefore, the
initial velocity of CysE was within 5 min.
Within a maximum concentration limit of 0.74 μg/ml,
the concentration of CoA was proportional to the reaction time
(Fig. 2B). As the CysE
concentration reached 2.22 or 3.70 μg/ml, the rate of CoA formation
gradually decreased with reaction time. The optimal concentration
for characterizing CysE was 0.74 μg/ml.
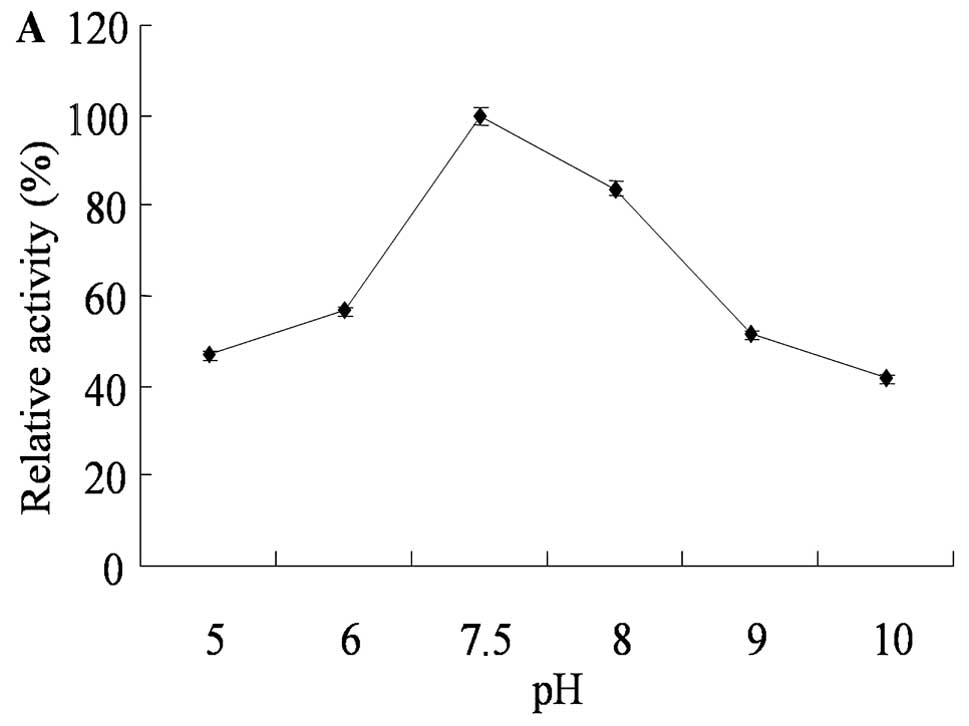
The CysE activity was determined at varying pHs with
appropriate buffer systems (3–11)
after the initial velocity and optimal CysE protein concentration
were set (Fig. 3A). The optimal
pH for CysE was 7.5. The optimal temperature for CysE was
investigated from 16 to 80°C (Fig.
3B), with the highest activity observed as the temperature
reached 37°C. The catalytic activity of CysE was not significantly
changed by varying the Mg2+ concentration (Fig. 3C), indicating that Mg2+
had no effect on the CysE activity.
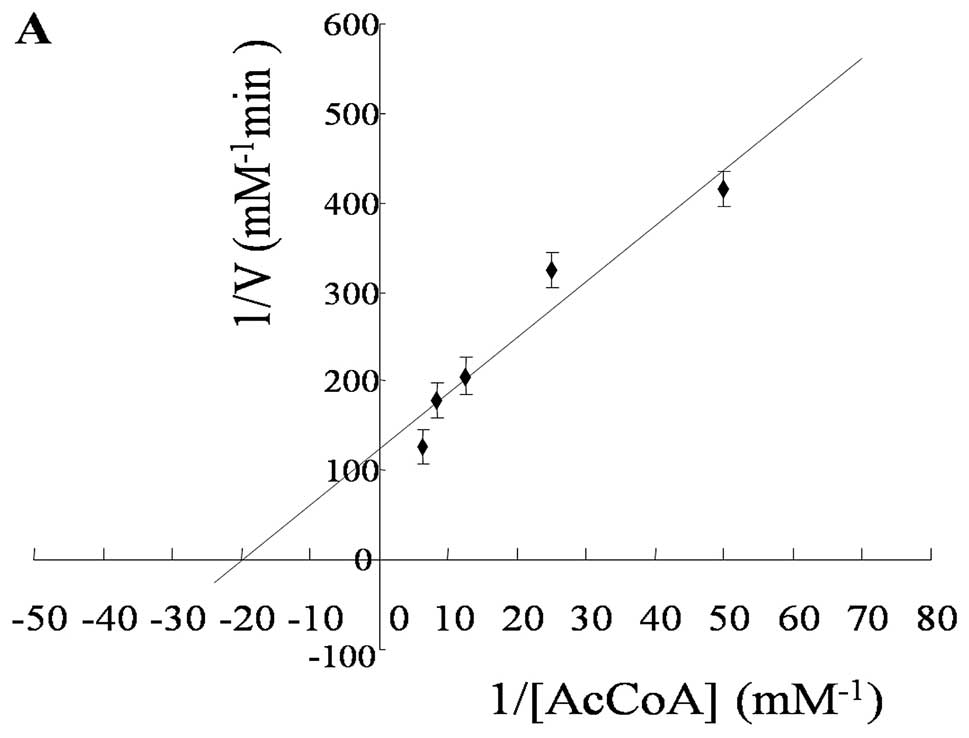
The steady-state kinetic constants were determined
under the optimal conditions and the initial velocity by a double
reciprocal plot (Fig. 4). The
Vmax value of CysE was 0.0073±0.0005 mM/min. The
Km of CysE against AcCoA was 0.0513±0.0050 mM, while the
Km value of L-Ser was 0.0264±0.0006 mM (Table I).
Discussion
Serine acetyltransferase is an enzyme involved in
cysteine biosynthesis, and it plays an important role in the growth
of M. tuberculosis (10).
In addition, this enzyme only exists in microorganisms and plants
(9), making serine
acetyltransferase a potential anti-TB drug target.
M. tuberculosis Rv2335 is predicted to be a
cysE gene encoding serine acetyltransferase. Bioinformatic
analyses have shown that the M. tuberculosis Rv2335 protein
is 45% identical to E. coli CysE, S. typhimurium CysE
and H. influenzae CysE using the Basic Local Alignment
Search Tool (BLAST). Serine acetyltransferase is a member of the
hexapeptide acetyltransferase family (19). This protein family has a conserved
active left-handed-β-helix (LβH) domain, which is composed of a
six-peptide ([LIV]-[GAED]-X2[STAV]-X) tandem repeat (15,20,21). The M. tuberculosis Rv2335
protein contained the tandem repeat and showed LβH structure when
modeled using the NCBI Conserved Domain Search (data not
shown).
To identify the function of M. tuberculosis
CysE, the M. tuberculosis cysE (Rv2335) gene was amplified
with high fidelity DNA polymerase, and the soluble CysE protein was
expressed in E. coli. SDS-PAGE and western blotting showed
that the molecular weight of the expressed CysE protein (~30 kDa)
was higher than predicted. This finding could be due to the
auxiliary fusion of six histidines to the recombinant M.
tuberculosis CysE protein generated from the pET29b vector. The
six consecutive histidines impart a strong positive charge that may
retard the mobility of the CysE protein in SDS-PAGE.
As indicated in Table
I, M. tuberculosis CysE demonstrated serine
acetyltransferase activity of 10.66 μmol/min/mg. The specific
activity of E. coli serine acetyltransferase has been
reported as 71.6 μmol/min/mg (22). The specific activity of M.
tuberculosis CysE is lower than that of E. coli CysE,
possibly because of the different methods of purification. M.
tuberculosis CysE exhibited its highest acetyltransferase
activity at pH 7.5 and 37°C. The optimal pH is consistent with
those reported for other bacteria, but the optimal temperature is
different from those reported for other bacteria such as S.
typhimurium (25°C) (14),
E. coli (25°C) (12) and
H. influenzae (25°C) (15). The Km for L-serine
(Kser) of M. tuberculosis CysE (0.026 mM) is
lower than the Kser of S. typhimurium CysE (0.7
mM) and E. coli CysE (1.17 mM) (12,23). The Km for AcCoA
(KAcCoA) of M. tuberculosis CysE (0.051 mM) is
also lower than that of S. typhimurium CysE (0.1 mM) and
E. coli CysE (0.2 mM) (12,23). In the present study, the
KAcCoA of M. tuberculosis CysE was 0.051 mM,
while the Kser was 0.026 mM. This finding suggests that
CysE had higher affinity for L-Ser than AcCoA, and CysE was bound
more easily to L-Ser than to AcCoA in M. tuberculosis.
Cysteine is reported to inhibit the activity of serine
acetyltransferase in its biosynthetic pathway by a feedback
mechanism (7,12,15). Furthermore, cysteine was found to
bind E. coli CysE at the serine substrate site rather than
at the acetyl-CoA substrate site from the structural study on
acetyltransferase (20). This
finding indicates that it is preferable to screen and design
compounds against the L-serine site to inhibit the activity of
CysE.
In summary, serine acetyltransferase CysE was
encoded by the cysE (Rv2335) gene in M. tuberculosis.
We investigated the kinetic parameters and optimal catalytic
conditions of CysE using simple and rapid enzyme assays. The CysE
assay and kinetic properties of CysE will facilitate the
high-throughput screening of inhibitors against CysE. However,
there are currently no reports of the crystal structure and active
sites of M. tuberculosis CysE. The expressed soluble CysE
protein will be available to further elucidate its crystal
structure and active sites.
Acknowledgements
This study was supported by a grant from the
National Natural Science Foundation of China (31070066) and the
National Basic Research Program of China (2012CB518803).
Abbreviations:
|
DTNB
|
5,5′-dithio-bis-(2-nitrobenzoic
acid)
|
|
EDTA
|
ethylendiaminetetraacetic acid
|
|
IPTG
|
isopropyl
β-D-thiogalactopyranoside
|
|
PMSF
|
phenymethylsulfonyl fluoride
|
|
NIH
|
National Institutes of Health
|
References
|
1
|
Donald PR and van Helden PD: The global
burden of tuberculosis - combating drug resistance in difficult
times. N Engl J Med. 360:2393–2395. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Migliori GB, Matteelli A, Cirillo D and
Pai M: Diagnosis of multidrug-resistant tuberculosis and
extensively drug-resistant tuberculosis: current standards and
challenges. Can J Infect Dis Med Microbiol. 19:169–172.
2008.PubMed/NCBI
|
|
3
|
Harrington M: From HIV to tuberculosis and
back again: a tale of activism in 2 pandemics. Clin Infect Dis.
50(Suppl 3): S260–S266. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cole ST and Riccardi G: New tuberculosis
drugs on the horizon. Curr Opin Microbiol. 14:570–576. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kredich NM, Becker MA and Tomkins GM:
Purification and characterization of cysteine synthetase, a
bifunctional protein complex, from Salmonella typhimurium. J
Biol Chem. 244:2428–2439. 1969.PubMed/NCBI
|
|
6
|
Kredich NM and Tomkins GM: The enzymic
synthesis of L-cysteine in Escherichia coli and
Salmonella typhimurium. J Biol Chem. 241:4955–4965.
1966.PubMed/NCBI
|
|
7
|
Kredich NM: Biosynthesis of cysteine.
Escherichia coliand Salmonella typhimurium: Cellular
and Molecular Biology. 1. Neidhardt FC, Curtiss R, Ingraham JL, Lin
ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M and
Umberger E: 2nd edition. American Society for Microbiology;
Washington D.C: pp. 514–527. 1996
|
|
8
|
Hell R: Molecular physiology of plant
sulfur metabolism. Planta. 202:138–148. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Meisenberg G and Simmons W: Princples of
Medical Biochemistry. Mosby Elsevier; Philadelphia: 2006
|
|
10
|
Schnell R and Schneider G: Structural
enzymology of sulphur metabolism in Mycobacterium
tuberculosis. Biochem Biophys Res Commun. 396:33–38. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Raman K, Yeturu K and Chandra N: targetTB:
a target identification pipeline for Mycobacterium
tuberculosis through an interactome, reactome and genome-scale
structural analysis. BMC Syst Biol. 2:1092008.PubMed/NCBI
|
|
12
|
Hindson VJ: Serine acetyltransferase of
Escherichia coli: substrate specificity and feedback control
by cysteine. Biochem J. 375:745–752. 2003.PubMed/NCBI
|
|
13
|
Mino K, Yamanoue T, Sakiyama T, Eisaki N,
Matsuyama A and Nakanishi K: Effects of bienzyme complex formation
of cysteine synthetase from Escherichia coli on some
properties and kinetics. Biosci Biotechnol Biochem. 64:1628–1640.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Leu LS and Cook PF: Kinetic mechanism of
serine transacetylase from Salmonella typhimurium.
Biochemistry. 33:2667–2671. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Johnson CM, Huang B, Roderick SL and Cook
PF: Kinetic mechanism of the serine acetyltransferase from
Haemophilus influenzae. Arch Biochem Biophys. 429:115–122.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ellman GL: A colorimetric method for
determining low concentrations of mercaptans. Arch Biochem Biophys.
74:443–450. 1958. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Riddles PW, Blakeley RL and Zerner B:
Reassessment of Ellman’s reagent. Methods Enzymol. 91:49–60.
1983.
|
|
18
|
Zhou Y, Xin Y, Sha S and Ma Y: Kinetic
properties of Mycobacterium tuberculosis bifunctional GlmU.
Arch Microbiol. 193:751–757. 2011.
|
|
19
|
Downie JA: The nodL gene from Rhizobium
leguminosarum is homologous to the acetyl transferases encoded
by lacA and cysE. Mol Microbiol. 3:1649–1651. 1989.PubMed/NCBI
|
|
20
|
Pye VE, Tingey AP, Robson RL and Moody PC:
The structure and mechanism of serine acetyltransferase from
Escherichia coli. J Biol Chem. 279:40729–40736. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Beaman TW, Sugantino M and Roderick SL:
Structure of the hexapeptide xenobiotic acetyltransferase from
Pseudomonas aeruginosa. Biochemistry. 37:6689–6696. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wigley DB, Derrick JP and Shaw WV: The
serine acetyltransferase from Escherichia coli
Over-expression, purification and preliminary crystallographic
analysis. FEBS Lett. 277:267–271. 1990. View Article : Google Scholar
|
|
23
|
Baecker PA and Wedding RT: Purification of
serine acetyltransferase, a component of a multienzyme complex, by
immunoadsorption and selective dissociation of the complex. Anal
Biochem. 102:16–21. 1980. View Article : Google Scholar : PubMed/NCBI
|