Introduction
Estrogen-related receptor α (ERRα, NR3B1) is among
the first orphan nuclear receptors identified by low-stringency
screening of cDNA libraries with a probe encompassing the DNA
binding domain of human estrogen receptor (ER) α (1). ERRα shares only 37% amino acid
identity in its ligand binding domain with ERα (1), which may explain the fact that ERRα
does not bind estrogen. Instead, ERRα activates gene transcription
constitutively in a ligand-independent manner (2). ERRα is involved in various
physiological regulatory processes. It is a regulator of energy
metabolism (3,4), and is essential for adaptive
thermogenesis (5). ERRα is also
related to the growth and progression of several gynecological
cancers (6,7). ERRα is regulated by estrogen in
bone, and it may play a functional role in diseases caused by
estrogen deficiency, such as osteoporosis (8). ERRα is expressed throughout
osteoblastic differentiation and may regulate bone formation both
in vitro (9) and in
vivo (10), and it is also
involved in osteoclast adhesion and transmigration (11). Since osteoblasts arise from
multipotent mesenchymal stem cells (MSCs), several studies have
investigated the role of ERRα in the osteogenic differentiation of
MSCs. Various studies have designated ERRα as an activator of
osteogenic differentiation of MSCs (12,13), whereas other studies have
suggested that ERRα is an inhibitor (14,15). Therefore, the function of ERRα in
osteogenic differentiation of MSCs has not been clearly
understood.
Human periodontal ligament tissue-derived
mesenchymal stem cells (hPDLSCs), first isolated by Seo et
al (16) in 2004, are
multipotent stem cells that have been recently used in stem
cell-mediated therapies and tissue engineering (17,18). Studies have shown that hPDLSCs
have an osteogenic potential both in vitro and in
vivo, but the molecular mechanisms that underlie hPDLSC
differentiation toward an osteoblastic phenotype remain elusive
(19).
Thus, we hypothesized that ERRα may be expressed in
PDLSCs and may be involved in the osteogenic characteristic of
PDLSCs. In the present study, we found that ERRα was expressed in
hPDLSCs. Moreover, the expression level of ERRα was increased
during the late period of osteogenic differentiation of hPDLSC. To
confirm the role of ERRα in osteogenic differentiation of hPDLSCs,
we used lentiviral delivery of miRNA to knock down the expression
of ERRα in hPDLSCs, and found that the osteogenic potential of
hPDLSCs was impaired after ERRα silencing. Our data indicate that
ERRα may play an important role in osteogenic differentiation of
hPDLSCs and may be used to improve the osteogenic potential of
hPDLSCs. Thus, hPDLSCs may be a promising therapeutic target for
the treatment of some bone diseases.
Materials and methods
Cell culture
Periodontal ligament (PDL) tissues were harvested
from healthy premolars extracted for orthodontic reasons. Seven
donors (12–16 years of age; four females and three males) and their
parents provided informed consent. Approval was granted by the
Ethics Committee of the School of Stomatology at the Fourth
Military Medical University, China. PDL tissue attached to the
middle third of the root was removed and cultured in phenol
red-free α-MEM supplemented with 10% charcoal-treated FBS (both
from Gibco-BRL, Rockville, MD, USA), 2 mmol/l glutamine, 100 U/ml
penicillin and 100 μg/ml streptomycin at 37°C in a humidified
atmosphere with 5% CO2. After reaching 100% confluence,
the cells were subcultured. To obtain homogeneous populations of
PDLSCs, the limiting dilution technique was used as described
previously (20). After 2–3 weeks
in culture, single cell-derived clones were harvested and combined.
Multiple colony-derived PDLSCs at passage 3 or 4 were used in
experiments. The human breast adenocarcinoma cell line MCF-7 was
cultured in the same manner and served as a positive control.
Flow cytometric analysis
The isolated putative hPDLSCs were collected and
washed with PBS. To identify the PDLSC phenotype, ~3×106
hPDLSCs were incubated with Alexa Fluor-conjugated monoclonal
antibodies against human STRO-1 (340104), CD29 (303016), CD34
(343518), CD45 (304019) and CD105 (323209) (all from BioLegend, San
Diego, CA, USA) for 2 h on ice. After washing twice and
resuspending in PBS, the cells were analyzed using an Epics XL
(Beckman Coulter, Fullerton, CA, USA).
Adipogenic induction
Adipogenic induction medium consisted of α-MEM
supplemented with 10% FBS, 2 μmol/l insulin (I6279), 0.5 mmol/l
isobutyl-methylxanthine (I5879) and 10 nmol/l dexamethasone (D1756)
(all from Sigma-Aldrich, St. Louis, MO, USA). PDLSCs were incubated
in adipogenic induction medium for 14 days. The medium was replaced
every other day. Intracellular lipid accumulation was detected by
staining with Oil Red O (O0625; Sigma-Aldrich).
Osteogenic induction
Osteogenic induction medium consisted of α-MEM
supplemented with 10% FBS, 10 mmol/l β-glycerophosphate (G6251), 10
nmol/l dexamethasone, and 50 μg/ml ascorbic acid (A5960) (both from
Sigma-Aldrich). PDLSCs were incubated in osteogenic induction
medium for 21 days. The medium was replaced every other day. For
ALP staining, cells were fixed and stained using a BCIP/NBT
Alkaline Phosphatase Color Development Kit (Beyotime, Haimen,
China). Calcium accumulation was detected by staining with 2%
Alizarin Red S (pH 8.3, A5533; Sigma-Aldrich), and then rinsing
extensively with PBS. After images of the nodules were recorded,
10% (w/v) cetylpyridinium chloride (Sigma-Aldrich) was used to
dissolve the nodules, and the absorbance was examined at 562
nm.
RNA extraction and RT-PCR
Total RNA from PDLSCs was isolated using TRIzol
reagent (Invitrogen, Carlsbad, CA, USA), according to the
manufacturer’s instructions. cDNA was synthesized from mRNA using
M-MLV reverse transcriptase (Invitrogen). Primer sequences for ERRα
and β-actin are shown in Table I.
After predenaturation at 94°C for 5 min, 30 PCR cycles were
performed (94°C for 30 sec; 57°C for 30 sec; and 72°C for 30 sec),
followed by a final extension at 72°C for 10 min. PCR products were
separated on 1.5% agarose gels containing ethidium bromide by
electrophoresis and then visualized by a UV transilluminator.
 | Table IPrimer sequences and product sizes for
RT-PCR and quantitative real-time PCR. |
Table I
Primer sequences and product sizes for
RT-PCR and quantitative real-time PCR.
| Target gene | GeneBank accession
no. | Primer sequences | Size of amplified
product (bp) |
|---|
| ERRα | NM_004451 | F:
GTGGGCGGCAGAAGTACAAG | 234 |
| | R:
GGTCAAAGAGGTCACAGAGGGT | |
| OCN | NM_199173 | F:
AGGGCAGCGAGGTAGTGAA | 151 |
| | R:
TCCTGAAAGCCGATGTGGT | |
| OPN | NM_000582 | F:
CTGATGCTACAGACGAGGACAT | 173 |
| | R:
GCTGTGGGTTTCAGCACTCT | |
| ALP | NM_000478 | F:
AGAACCCCAAAGGCTTCTTC | 74 |
| | R:
CTTGGCTTTTCCTTCATGGT | |
| RUNX2 | NM_004348 | F:
TCTACTATGGCACTTCGTCAGG | 164 |
| | R:
GCTTCCATCAGCGTCAACAC | |
| β-actin | NM_001101 | F:
TCCTTCCTGGGCATGGAGT | 208 |
| | R:
CAGGAGGAGCAATGATCTTGAT | |
Immunocytochemical analysis
PDLSCs and MCF-7 cells (positive control) were
seeded on coverslips at a density of 5×103 cells/ml for
48 h, and then fixed with cold acetone. Immunocytochemical analysis
was performed using the streptavidin-biotin complex method
according to the manufacturer’s protocol (Zhongshan Golden Bridge
Biotechnology Co., Ltd., Beijing, China). DAB was used as the
chromogen. The primary antibody against ERRα was a monoclonal
rabbit anti-human ERRα (ab41868; Abcam, Cambridge, UK) at a 1:100
dilution. For the negative control, the primary antibody was
substituted with a commensurable volume of PBS. The samples were
counterstained with hematoxylin and examined under an Olympus
compound microscope (Olympus, Tokyo, Japan) equipped with a Nikon
digital camera (Nikon, Tokyo, Japan).
Viral vector construction and
transduction
The BLOCK-iT™ RNAi Designer Program (Invitrogen) was
used to design the miRNA sequence targeting the human ERRα gene.
The target sequence was GCTACCCTCTGTGACCTCTTT. The annealed DNA
sequences were cloned into pcDNA6.2-GW/EmGFP-miR (Invitrogen). The
lentiviral vector plasmids were derived from the pLenti6.3/V5-Dest
construct using the BLOCK-iT Lentiviral Pol II miR RNAi Expression
System with EmGFP (Invitrogen), according to the manufacturer’s
instructions. Briefly, pcDNA6.2-GW/EmGFP-ERRα-miR was recombined
into the pLenti6.3/V5-Dest vector. The reaction mixtures were
transformed into DH5α competent cells to select for positive
clones. Sequencing was performed to verify the recombinant
pLenti-ERRα-miR plasmid. Lentiviruses were produced by transient
transfection of 293FT cells using Lipofectamine 2000, lentiviral
vectors, and packaging mix (Invitrogen). Transfection of hPDLSCs
was performed by exposure to viral supernatant at a MOI of 100 in
the presence of Polybrene (8 mg/ml; Sigma-Aldrich) for 48 h. To
produce stably transfected cell lines, the cells were cultured in
selection medium containing 5 μg/ml blasticidin (Invitrogen) for 2
weeks. The cells were cultured in α-MEM supplemented with 10% FBS
and 2.5 μg/ml blasticidin to maintain and expand the cells.
Quantitative real-time PCR analysis
PDLSCs were treated with osteogenic induction
medium, or cultured in standard medium as the control group. The
cells were harvested at day 1, 5, 10, 15 and 20.
Lentivirus-transduced PDLSCs were harvested after infection for 48
h to determine the efficiency of ERRα gene knockdown. After the
production of stably transfected cell lines, osteogenic induction
was performed, and the cells were harvested at day 7, 14 and 21.
Total RNA isolation and first-strand cDNA synthesis were performed
as described above. Real-time PCR was carried out with a
Mastercycler ep Realplex4 (Eppendorf AG, Hamburg, Germany) and
SYBR-Green (Invitrogen). Primer sequences are shown in Table I. Reactions were performed under
the following cycling conditions: 95°C for 10 min, followed by 45
cycles of 95°C for 15 sec and 60°C for 1 min. Expression of the
target genes was calculated using the formula 2−ΔΔCt.
Expression data were normalized to the expression of the β-actin
gene.
Western blot analysis
Western blot analysis was used to confirm gene
silencing as described previously (21). Briefly, cell extracts containing
30 μg total protein were subjected to SDS-PAGE and then transferred
onto PVDF membranes. The membranes were blocked and probed with
primary antibodies that recognized ERRα (ab41868; Abcam) or β-actin
(sc-47778; Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA).
Secondary antibodies were chosen according to the species of origin
of the primary antibodies. After the incubation, the luminescent
signals were detected using an enhanced chemiluminescence kit
(Pierce, Rockford, IL, USA).
Statistical analyses
All experiments were performed at least three times.
Each value is expressed as the mean ± SD. Comparisons between two
groups were performed by the independent samples t-test.
Differences among three or more groups were analyzed by one-way
ANOVA, followed by Dunnett’s test for significance. Data with a
P-value of <0.05 was considered to represent a statistically
significant difference.
Results
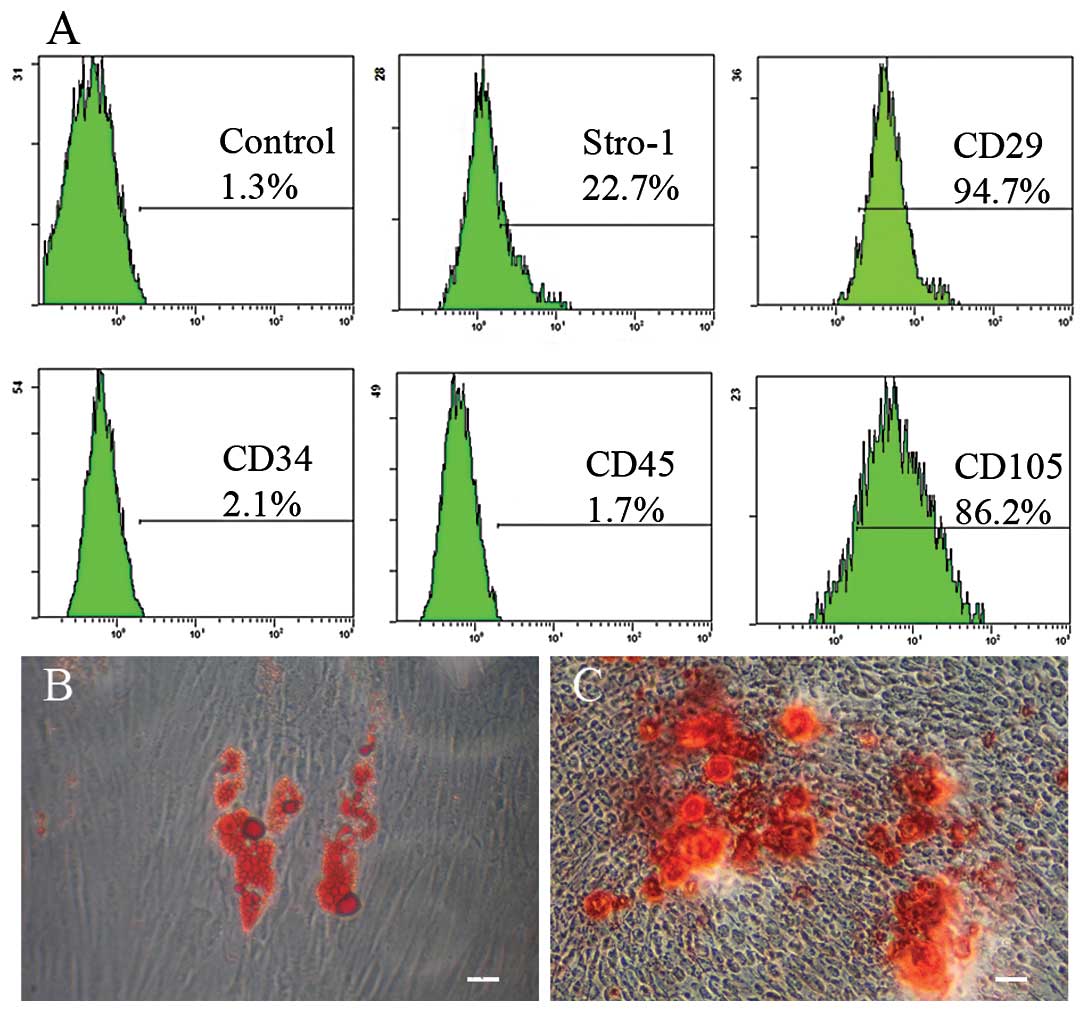
Characterization of hPDLSCs
Stem-like cells were successfully isolated from the
human PDL tissue. To characterize the phenotypic stem cell markers
of single colony-derived PDLSCs, the expression levels of STRO-1,
CD29, CD34, CD45 and CD105 were analyzed by flow cytometry. We
observed that the hPDLSC population showed a high percentage of
cells expressing markers of MSCs: STRO-1, CD29 and CD105 (Fig. 1A). In contrast, the cells were
negative for the hematopoietic lineage marker CD34 and the
leukocyte common antigen CD45 (Fig.
1A). To evaluate the multipotency of PDLSCs, we performed Oil
Red O staining after culturing in adipogenic induction medium for
14 days and Alizarin Red S staining after culturing in osteogenic
induction medium for 21 days. The results showed that hPDLSCs had
strong adipogenic and osteogenic differentiation capacities
(Fig. 1B and C).
Expression of ERRα in hPDLSCs
Immunocytochemistry and RT-PCR analyses were
employed to examine the expression of ERRα in cultured hPDLSCs. As
shown in Fig. 2A, a clear band
representing ERRα was detected in the hPDLSCs at the same molecular
weight as that in positive control cells (MCF-7 cells).
Immunocytochemical staining confirmed the positive expression of
ERRα protein in the hPDLSCs (Fig.
2B) and MCF-7 cells (Fig.
2C). Positive signals in the nuclei were stronger than those in
the cytoplasm of the hPDLSCs (Fig.
2B). In contrast, no positive signal was found in the negative
control (Fig. 2D).
Temporal expression of ERRα in hPDLSCs
treated with osteogenic induction medium
To investigate the role of ERRα in osteogenic
differentiation of hPDLSCs, we measured the expression of ERRα mRNA
during osteogenic induction (day 1, 7, 14 and 21) of hPDLSCs by
quantitative real-time PCR (Fig.
3). There were no significant differences between the
osteogenic induction group and the control group at day 1 and 7. As
the cells entered the mineralization stage after culturing for 14
days, ERRα mRNA expression in the osteogenic induction group was
significantly increased (P<0.05), and at day 21, it reached a
peak level compared with that in the control group
(P<0.001).
Knockdown of the expression of ERRα
results in decreased osteogenic differentiation of hPDLSCs
To further analyze the biological function of ERRα,
we used lentiviral vectors driving the expression of miRNA against
the ERRα gene. DNA sequencing results revealed that the inserted
fragments were correct, and no mutations were found in the
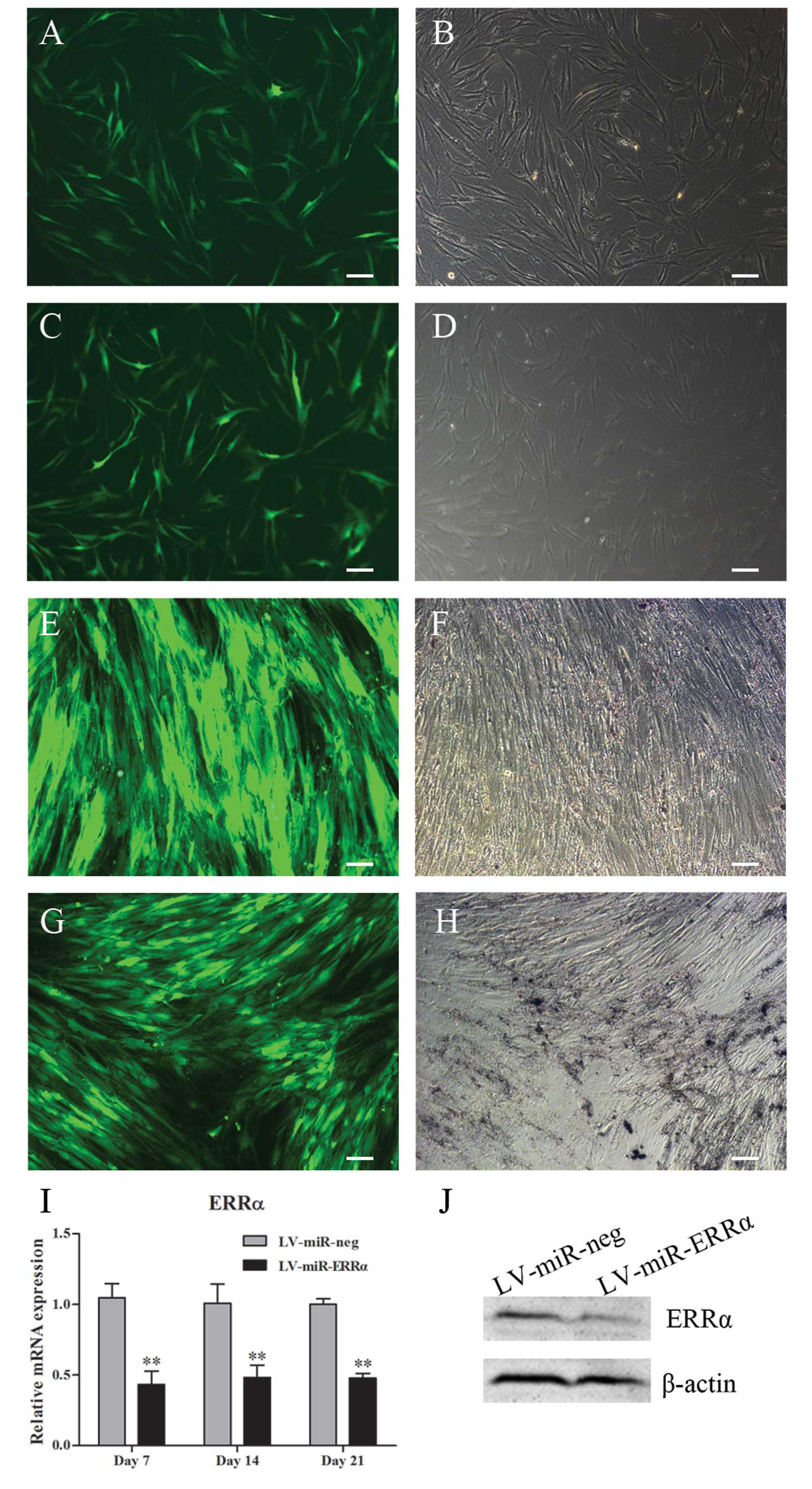
recombinant plasmids. Successful transfection of recombinant
plasmids into hPDLSCs was confirmed by detecting EmGFP expression
under a fluorescence microscope (Fig.
4A-D). After stably transfected cell lines were produced by
culturing in selection medium containing blasticidin for 2 weeks,
flow cytometric analysis showed that the transfection rate of
LV-miR-neg and LV-miR-ERRα was >90%. Fig. 4F-H shows the EmGFP expression in
hPDLSCs stably transfected with LV-miR-neg or LV-miR-ERRα. Gene
silencing was confirmed by real-time PCR after culturing in
osteogenic induction medium for 7, 14 and 21 days, and by western
blot analysis at day 7 (Fig. 4I and
J). The results showed that transfection with LV-miR-ERRα
decreased ERRα mRNA and protein expression by ~50%.
Next, we determined the osteogenic capacity of
hPDLSCs transfected with LV-miR-ERRα, and used the cells
transfected with LV-miR-neg as the control. ALP and Alizarin Red S
staining were performed to detect the mineralization of hPDLSCs
(Fig. 5A). The density of ALP
staining at day 14 was lower in the ERRα-knockdown group than that
in the control group (P<0.01) and Alizarin Red S staining at day
21 also showed a significant decrease in the mineralization of
hPDLSCs transfected with LV-miR-ERRα (P<0.01).
We next monitored the mRNA expression of some
mineralization-related genes in hPDLSCs cultured in osteogenic
induction medium at day 7, 14 and 21. We observed significant
decreases in ALP, RUNX2, OCN and OPN mRNA levels in ERRα-knockdown
cells (P<0.05) (Fig. 5B).
Taken together, our data indicate that ERRα promotes osteogenic
differentiation of hPDLSCs.
Discussion
ERRα is capable of regulating the transcription of
genes involved in multiple cellular and physiological processes
(3,8,22).
Studies have established the key roles of ERRα in regulating
mitochondrial biogenesis (3),
fatty acid oxidation (23) and
oxidative phosphorylation (3) and
have correlated ERRα with various types of cancer and metabolic
disorders (22). ERRα has also
been proposed as an important regulatory factor of bone metabolism
(8) and is regarded as a
potential therapeutic target for treating osteoporosis (24).
Previous studies have shown that ERRα is selectively
expressed in a variety of cell types during development and in
adult tissues, and ERRα expression increases according to metabolic
demands (8,23). ERRα is highly expressed during the
formation of ossification zones during mouse development in
vivo (25), as well as in
primary rat calvarial cells in vitro (9). Nevertheless, its expression in
periodontal tissues has not been studied. PDLSCs are isolated from
the PDL that connects two types of hard tissues, tooth cementum and
alveolar bone, and thus express an array of
cementoblastic/osteoblastic markers. PDLSCs are capable of forming
cementum/PDL-like tissue, and participate in the repair of alveolar
bone (16). In addition, healthy
cells can be easily obtained from adolescents who require teeth
extraction for orthodontic reasons. Therefore, PDLSCs are ideal
seed cells for periodontal tissue engineering therapies. Since PDL
is also a highly metabolically active tissue with peculiar
mechanical/functional demands (26), we investigated ERRα expression in
PDLSCs in vitro.
Osteogenic differentiation of hPDLSCs is a complex
process regulated by multiple signals at different levels. Since
ERRα was expressed in hPDLSCs, we assumed that ERRα was also
involved in osteogenic differentiation of hPDLSCs. We monitored the
expression of ERRα as the cells were induced to differentiate in
osteogenic induction medium, and found a gradual increase in the
expression levels of ERRα. We next used RNA interference to inhibit
ERRα expression in hPDLSCs. Downregulation of ERRα significantly
inhibited the mineralization capacity of hPDLSCs. We further
detected the expression of mineralization-related genes during the
osteogenic differentiation of hPDLSCs. ALP and OCN are regarded as
indicators of early and late osteogenesis, respectively. RUNX2 is a
key transcription factor essential for the commitment of
multipotent mesenchymal cells to the osteoblast lineage, and serves
as an early transcriptional regulator of osteogenic differentiation
(27). OPN is a non-collagenous
bone matrix protein, a marker of the late stages of osteoblastic
differentiation, and its promoter can be transactivated by ERRα
(28). These
mineralization-related genes were downregulated by
lentiviral-mediated ERRα knockdown, suggesting that ERRα initiates
early-stage osteogenic differentiation and maintains late-stage
osteogenic differentiation of hPDLSCs in vitro.
In a recent study, Rajalin et al (12) also proposed a positive role for
ERRα in osteoblastic differentiation of MSCs using ERRα-knockout
(KO) mice. Auld et al (13) showed similar results regarding the
role of ERRα in mineralization of human MSCs and they found that
native ERRα represses Wnt signaling, which has been shown to
suppress osteogenesis in the human MSC system (29). Our results also showed that
lentiviral-mediated ERRα knockdown reduced the osteogenic
differentiation of PDLSCs that are tissue-specific MSCs. However,
the mechanism by which ERRα enhances the osteogenic potential of
MSCs is still unclear. Previous studies have shown that
overexpression of ERRα leads to induction of the p21 cell cycle
inhibitor, and inhibition of proliferation in breast cell lines
(30,31). Based on these studies, we inferred
that, upon silencing in MSCs, ERRα may enhance cell expansion and
lead to delayed differentiation. However, a report by Delhon et
al (14) showed a negative
effect of ERRα on bone formation both in vivo and in
vitro. Teyssier et al (15) also found that ERRα negatively
regulates osteogenic differentiation in vitro and
demonstrated a gender-dependent effect of ERRα in ERRα-KO mice.
These contradictions may result from differences in the tissues and
species, variability of the primary cell cultures, differences in
the osteogenic culture conditions, the genetic backgrounds of the
ERRα-KO mice, or potential gender-dependent effects of ERRα
(12).
ERRα has crosstalk with ERs and estrogen and
modulates ER-mediated signaling pathways (8). Studies have shown that ERRα is a
potential therapeutic target of postmenopausal osteoporosis
(24). Since postmenopausal
osteoporosis is a well-known systemic inflammatory environment, and
periodontitis is also a chronic inflammatory microenvironment, we
theorized that ERRα may be involved in periodontal disease
(32). Previous studies by our
group demonstrated that hPDLSCs derived from patients with chronic
periodontitis (P-PDLSCs) display an impaired osteogenic potential
compared with that of hPDLSCs derived from healthy donors
(H-PDLSCs) (33). We also
observed that ERRα expression is decreased in P-PDLSCs, and when
H-PDLSCs are treated with TNF-α, the main proinflammatory factor of
periodontitis, the expression of ERRα is significantly decreased
along with the attenuation of osteogenic differentiation
(unpublished data). Similarly, Bonnelye et al (34) found that ERRα mRNA expression is
downregulated in the subchondral bone of mice with induced joint
inflammation, which is paralleled by downregulation of markers of
bone formation. Therefore, ERRα may be involved in impaired
osteogenic differentiation of hPDLSCs in periodontal disease, and
we believe that ERRα may also be a promising therapeutic target for
treating inflammatory bone diseases such as local periodontitis and
systemic osteoporosis. However, the pathway through which
inflammatory diseases affect ERRα expression and the relationship
between ERRα, ERs and estrogen are far from clear and need further
investigation.
In conclusion, we detected the expression of ERRα in
hPDLSCs in vitro and observed positive effects of ERRα on
the osteogenic differentiation of hPDLSCs. This result suggests
that ERRα regulates osteogenic differentiation of PDLs and may be
involved in the pathogenesis of estrogen-related periodontal
disease. Further studies are required to investigate the specific
functions of ERRα in periodontal tissues including periodontal
ligaments, gingiva and alveolar bone under different physiological
and pathophysiological conditions, and the crosstalk between ERRs
and ERs to elucidate the mechanisms through which ERRα acts both in
bone loss due to estrogen deficiency and in periodontal issues.
Acknowledgements
This study was supported by the Nature Science
Foundation of China (grant 30872913). We thank Dr Jiaxing Zhou for
generously providing the MCF-7 cell line. We also acknowledge
Professor Hui Xu for critical reading of the manuscript.
References
|
1
|
Giguère V, Yang N, Segui P and Evans RM:
Identification of a new class of steroid hormone receptors. Nature.
331:91–94. 1988.
|
|
2
|
Vanacker JM, Bonnelye E, Chopin-Delannoy
S, Delmarre C, Cavaillès V and Laudet V: Transcriptional activities
of the orphan nuclear receptor ERRα (estrogen receptor-related
receptor-α). Mol Endocrinol. 13:764–773. 1999.
|
|
3
|
Huss JM, Torra IP, Staels B, Giguere V and
Kelly DP: Estrogen-related receptor α directs peroxisome
proliferator-activated receptor α signaling in the transcriptional
control of energy metabolism in cardiac and skeletal muscle. Mol
Cell Biol. 24:9079–9091. 2004.
|
|
4
|
Luo J, Sladek R, Carrier J, Bader JA,
Richard D and Giguère V: Reduced fat mass in mice lacking orphan
nuclear receptor estrogen-related receptor α. Mol Cell Biol.
23:7947–7956. 2003.PubMed/NCBI
|
|
5
|
Villena JA, Hock MB, Chang WY, Barcas JE,
Giguère V and Kralli A: Orphan nuclear receptor estrogen-related
receptor α is essential for adaptive thermogenesis. Proc Natl Acad
Sci USA. 104:1418–1423. 2007.
|
|
6
|
Fujimoto J and Sato E: Clinical
implication of estrogen-related receptor (ERR) expression in
uterine endometrial cancers. J Steroid Biochem Mol Biol. 116:71–75.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu D, Kiriyama Y, Lee KY and Giguère V:
Transcriptional regulation of the estrogen-inducible pS2 breast
cancer marker gene by the ERR family of orphan nuclear receptors.
Cancer Res. 61:6755–6761. 2001.PubMed/NCBI
|
|
8
|
Bonnelye E and Aubin JE: Estrogen
receptor-related receptor α: a mediator of estrogen response in
bone. J Clin Endocrinol Metab. 90:3115–3121. 2005.
|
|
9
|
Bonnelye E, Merdad L, Kung V and Aubin JE:
The orphan nuclear estrogen receptor-related receptor α (ERRα) is
expressed throughout osteoblast differentiation and regulates bone
formation in vitro. J Cell Biol. 153:971–984. 2001.
|
|
10
|
Bonnelye E and Aubin JE: Differential
expression of estrogen receptor-related receptor α and estrogen
receptors α and beta in osteoblasts in vivo and in vitro. J Bone
Miner Res. 17:1392–1400. 2002.
|
|
11
|
Bonnelye E, Saltel F, Chabadel A, Zirngibl
RA, Aubin JE and Jurdic P: Involvement of the orphan nuclear
estrogen receptor-related receptor α in osteoclast adhesion and
transmigration. J Mol Endocrinol. 45:365–377. 2010.
|
|
12
|
Rajalin AM, Pollock H and Aarnisalo P:
ERRα regulates osteoblastic and adipogenic differentiation of mouse
bone marrow mesenchymal stem cells. Biochem Biophys Res Commun.
396:477–482. 2010.
|
|
13
|
Auld KL, Berasi SP, Liu Y, et al:
Estrogen-related receptor α regulates osteoblast differentiation
via Wnt/β-catenin signaling. J Mol Endocrinol. 48:177–191.
2012.
|
|
14
|
Delhon I, Gutzwiller S, Morvan F, et al:
Absence of estrogen receptor-related-α increases osteoblastic
differentiation and cancellous bone mineral density. Endocrinology.
150:4463–4472. 2009.
|
|
15
|
Teyssier C, Gallet M, Rabier B, et al:
Absence of ERRα in female mice confers resistance to bone loss
induced by age or estrogen-deficiency. PLoS One. 4:e79422009.
|
|
16
|
Seo BM, Miura M, Gronthos S, et al:
Investigation of multipotent postnatal stem cells from human
periodontal ligament. Lancet. 364:149–155. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Trubiani O, Orsini G, Zini N, et al:
Regenerative potential of human periodontal ligament derived stem
cells on three-dimensional biomaterials: a morphological report. J
Biomed Mater Res A. 87:986–993. 2008. View Article : Google Scholar
|
|
18
|
Liu Y, Zheng Y, Ding G, et al: Periodontal
ligament stem cell-mediated treatment for periodontitis in
miniature swine. Stem Cells. 26:1065–1073. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chadipiralla K, Yochim JM, Bahuleyan B, et
al: Osteogenic differentiation of stem cells derived from human
periodontal ligaments and pulp of human exfoliated deciduous teeth.
Cell Tissue Res. 340:323–333. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pan F, Zhang R, Wang G and Ding Y:
Oestrogen receptors are involved in the osteogenic differentiation
of periodontal ligament stem cells. Biosci Rep. 31:117–124. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liang L, Yu JF, Wang Y, Wang G and Ding Y:
Effect of estrogen receptor beta on the osteoblastic
differentiation function of human periodontal ligament cells. Arch
Oral Biol. 53:553–557. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ariazi EA and Jordan VC: Estrogen-related
receptors as emerging targets in cancer and metabolic disorders.
Curr Top Med Chem. 6:203–215. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sladek R, Bader JA and Giguère V: The
orphan nuclear receptor estrogen-related receptor α is a
transcriptional regulator of the human medium-chain acyl coenzyme A
dehydrogenase gene. Mol Cell Biol. 17:5400–5409. 1997.
|
|
24
|
Gallet M and Vanacker JM: ERR receptors as
potential targets in osteoporosis. Trends Endocrinol Metab.
21:637–641. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bonnelye E, Vanacker JM, Dittmar T, et al:
The ERR-1 orphan receptor is a transcriptional activator expressed
during bone development. Mol Endocrinol. 11:905–916. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hassell TM, Rateitschak KH, Wolf HF and
Rateitschak-Pluss EM: Color Atlas of Dental Medicine:
Periodontology. Wolf HF and Rateitschak KH: Thieme; Stuttgart:
2005
|
|
27
|
Komori T: Regulation of bone development
and extracellular matrix protein genes by RUNX2. Cell Tissue Res.
339:189–195. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vanacker JM, Delmarre C, Guo X and Laudet
V: Activation of the osteopontin promoter by the orphan nuclear
receptor estrogen receptor related α. Cell Growth Differ.
9:1007–1014. 1998.
|
|
29
|
Liu G, Vijayakumar S, Grumolato L, et al:
Canonical Wnts function as potent regulators of osteogenesis by
human mesenchymal stem cells. J Cell Biol. 185:67–75. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bianco S, Lanvin O, Tribollet V, Macari C,
North S and Vanacker JM: Modulating estrogen receptor-related
receptor-α activity inhibits cell proliferation. J Biol Chem.
284:23286–23292. 2009.
|
|
31
|
Castet A, Herledan A, Bonnet S, Jalaguier
S, Vanacker JM and Cavaillès V: Receptor-interacting protein 140
differentially regulates estrogen receptor-related receptor
transactivation depending on target genes. Mol Endocrinol.
20:1035–1047. 2006. View Article : Google Scholar
|
|
32
|
Geurs NC: Osteoporosis and periodontal
disease. Periodontol 2000. 44:29–43. 2007. View Article : Google Scholar
|
|
33
|
Liu Y, Liu W, Hu C, et al: MiR-17
modulates osteogenic differentiation through a coherent
feed-forward loop in mesenchymal stem cells isolated from
periodontal ligaments of patients with periodontitis. Stem Cells.
29:1804–1816. 2011. View
Article : Google Scholar
|
|
34
|
Bonnelye E, Laurin N, Jurdic P, Hart DA
and Aubin JE: Estrogen receptor-related receptor-α (ERR-α) is
dysregulated in inflammatory arthritis. Rheumatology. 47:1785–1791.
2008.
|