Introduction
Colorectal carcinoma (CRC) is one of the leading
causes of cancer-related mortality worldwide. The incidence and
mortality rate of CRC in China have increased rapidly over the past
several decades (1). A
significant number of patients with CRC who undergo curative
surgery develop local recurrence or distant metastasis, leading to
shorter survival (2). A better
understanding of the molecular mechanisms underlying metastasis is
essential to facilitate the prevention and treatment of advanced
CRC.
One of the key molecular steps in the process of
distant metastasis includes epithelial-to-mesenchymal transition
(EMT), which permits invasion and migration in various types of
cancer (3,4), and is associated with a poor
prognosis in CRC (5). During EMT,
epithelial cell-cell adhesion is decreased through the
downregulation of adhesion molecules, such as E-cadherin, and the
cells then acquire a spindle-shaped, highly motile fibroblast-like
phenotype (6,7). Several mechanisms for E-cadherin
transcriptional silencing during EMT have been suggested, including
the direct inhibition by transcriptional repressors, such as Snail,
Twist and zinc finger E-box-binding homeobox (ZEB)1 and ZEB2
(8–10).
MicroRNAs (miRNAs or miRs) are a large family of
highly conserved short (approximately 22 nucleotides in length)
single-stranded non-coding RNAs which regulate the translational
inhibition of target messenger RNAs (mRNAs) by binding to their
3′-untranslated region (UTR) (11). The dysregulation of miRNAs is
common in various carcinomas and plays an important role in cancer
progression by altering normal gene expression (12,13).
Previous studies have suggested that miR-155 is
over-expressed in various solid tumors (14–16). High expression levels of miR-155
have been found to correlate with the poor prognosis of colorectal
cancer and pancreatic tumors (16,17). Moreover, miR-155 has been shown to
enhance tumor invasion and migration, and acts as a mediator of EMT
(18). All these lines of
evidence are consistent with the notion that miR-155 functions as
an oncogenic miRNA in human cancers.
However, the biological roles of miR-155 in CRC
remain poorly understood. Therefore, in this study, the expression
of miR-155 in CRC was examined by real-time PCR and the effects of
miR-155 on cell proliferation, invasion and EMT were also
investigated.
Materials and methods
Patients and tissue samples
Primary CRC tissue and matched normal adjacent
colorectal mucosa were obtained from 76 patients who underwent
colorectal cancer resection without pre-operative treatment at the
First Department of General Surgery, The Affiliated Hospital of
North Sichuan Medical College, Nanchong, China from 2009 to 2011,
after receiving adequate informed consent. All tissue samples were
immediately frozen in liquid nitrogen and stored at −80°C until the
extraction of RNA. The specimens were stained with hematoxylin and
eosin and examined histopathologically. Sections that consisted of
>80% carcinoma cells were used to prepare the total RNA.
Clinicopathological information, including age, gender, pathology,
differentiation and tumor-node-metastasis (TNM) classification, was
available for all patients. The study was approved by the Medical
Ethics Committee of North Sichuan Medical College.
Cell culture
The SW480 human colon cancer cell line was obtained
from the American Type Culture Collection (ATCC; Manassas, VA, USA)
and cultured in RPMI-1640 medium (Gibco, Carlsbad, CA, USA)
supplemented with heat-inactivated 10% fetal bovine serum (FBS)
(Invitrogen, Carlsbad, CA, USA), 100 U/ml penicillin and 100 mg/ml
streptomycin, at 37°C in a humidified atmosphere of 5%
CO2 and 95% air.
Quantitative real-time reverse
transcriptase-PCR (qRT-PCR)
Total RNA was extracted from the tumor tissue and
adjacent normal mucosa by homogenizing the tissue in TRIzol reagent
(Invitrogen), according to the manufacturer’s instructions. For
miRNA qPCR, reverse transcription was performed using the QuantMir
RT kit (System Biosciences, Mountain View, CA, USA). The cDNA then
serves as the template for SYBR real-time PCR using Power
SYBR-Green PCR Master mix (Applied Biosystems, Foster City, CA,
USA). All reactions were run in triplicate on the iCycler iQ
Multicolor Real-Time PCR Detection System (Bio-Rad, Hercules, CA,
USA) using miR-155-specific primers (Applied Biosystems). The
amplification profile was denatured at 95°C for 10 min, followed by
40 cycles of denaturation at 95°C for 15 sec, annealing at 60°C for
30 sec, and extension at 72°C for 1 min. The comparative cycle
threshold (CT) method was applied to quantify the miRNA
expression levels. The relative amount of miR-155 to small nuclear
U6 RNA was calculated using the equation
2−ΔCT where ΔCT = (CT
miR−155 − CT U6 RNA). The fold change of gene
expression was calculated using the 2−ΔΔCT method. U6
small nuclear RNA was used as the internal standard.
Transfection of miRNA
miR-155 precursor (pre-miR-155) and pre-miR miRNA
precursor negative control (pre-miR-n.c.) were purchased from
Applied Biosystems. For transfection, 2×105 cells were
seeded into each well of a 6-well plate. When the cells were 70–80%
confluent the following day, they were transfected with pre-miR-155
and pre-miR-n.c. using Lipofectamine 2000 reagent (Invitrogen)
following the manufacturer’s instructions.
MTT assay
Following transfection of the SW480 cells with
pre-miR-155 or pre-miR-n.c. for various periods of time (20 and 44
h), the SW480 cells were re-seeded at a density of 2×104
cells/well in 96-well plates containing 0.2 ml RPMI-1640 (with 10%
FBS). Subsequently, 20 μl of
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
were added followed by incubation for a further 4 h at 37°C. A
total of 150 μl of dimethyl sulfoxide was then added to each
well and the absorbance was measured at 570 nm on an enzyme
immunoassay analyzer (Bio-Rad).
Migration and invasion assays
To measure cell migration and invasion,
5×104 SW480 cells transfected with either pre-miR-155 or
pre-miR-n.c. were seeded into Transwell chambers (8.0 μm
pore size; Corning Inc., Corning, NY, USA) uncoated or coated with
Matrigel (BD Biosciences, Bedford, MA, USA).Medium containing 10%
FBS in the lower chamber served as the chemoattractant. After the
cells were incubated for 48 h at 37°C in a humidified incubator
with 5% CO2, cells that did not migrate through the
pores were mechanically removed by a cotton swab. The migrated
cells attached to the bottom of the membrane insert were fixed in
methanol at room temperature for 5 min and stained with
hematoxylin. The number of migrated or invaded cells on the lower
surface of the membrane was then counted under a microscope at a
magnification of ×400.
Western blot analysis
To isolate the proteins, cells collected from 6-well
plates were washed twice in PBS and lysed in RIPA lysis buffer
(ProMab Biotechnologies, Inc., Richmond, CA, USA). The lysates were
kept on ice for 30 min, and then centrifuged at 13,000 × g for 30
min. The surpernatant was collected and then 20 μg of each
of the proteins was separated by SDS-PAGE on 10% gels and
transferred onto nitrocellulose membranes. After being blocked in
5% skimmed milk, the membranes were incubated with the respective
antibodies: goat anti-E-cadherin (Santa Cruz Biotechnology, Inc.,
Santa Cruz, CA, USA), goat anti-ZEB1 (Santa Cruz Biotechnology,
Inc.), mouse anti-GAPDH (Santa Cruz, Biotechnology, Inc.) and
rabbit anti-claudin-1 antibody (Zymed/Invitrogen). After incubation
with the appropriate secondary antibody, the bands were visualized
using ECL-Plus reagents (GE Healthcare Bio-Science Corp.,
Piscataway, NJ, USA). The density of the E-cadherin, ZEB1,
claudin-1 and GAPDH bands was measured using Image J software, and
values were normalized to the densitometric values of GAPDH in each
sample. The fold change in protein amount was then calculated for
the experimental sets compared to the control.
Statistical analysis
Continuous variables are expressed as the means ±
SD. The gene expression levels in the CRC samples were compared
with those in normal adjacent mucosa using the Wilcoxon test.
Measurement data were analyzed using the Student’s t-test, while
categorical data were examined using the χ2 test.
P-values <0.05 were considered to indicate statistically
significant differences. All statistical analyses were performed
using SPSS 16.0 software (SPSS; Chicago, IL, USA).
Results
Clinicopathological significance of
miR-155 in CRC patients
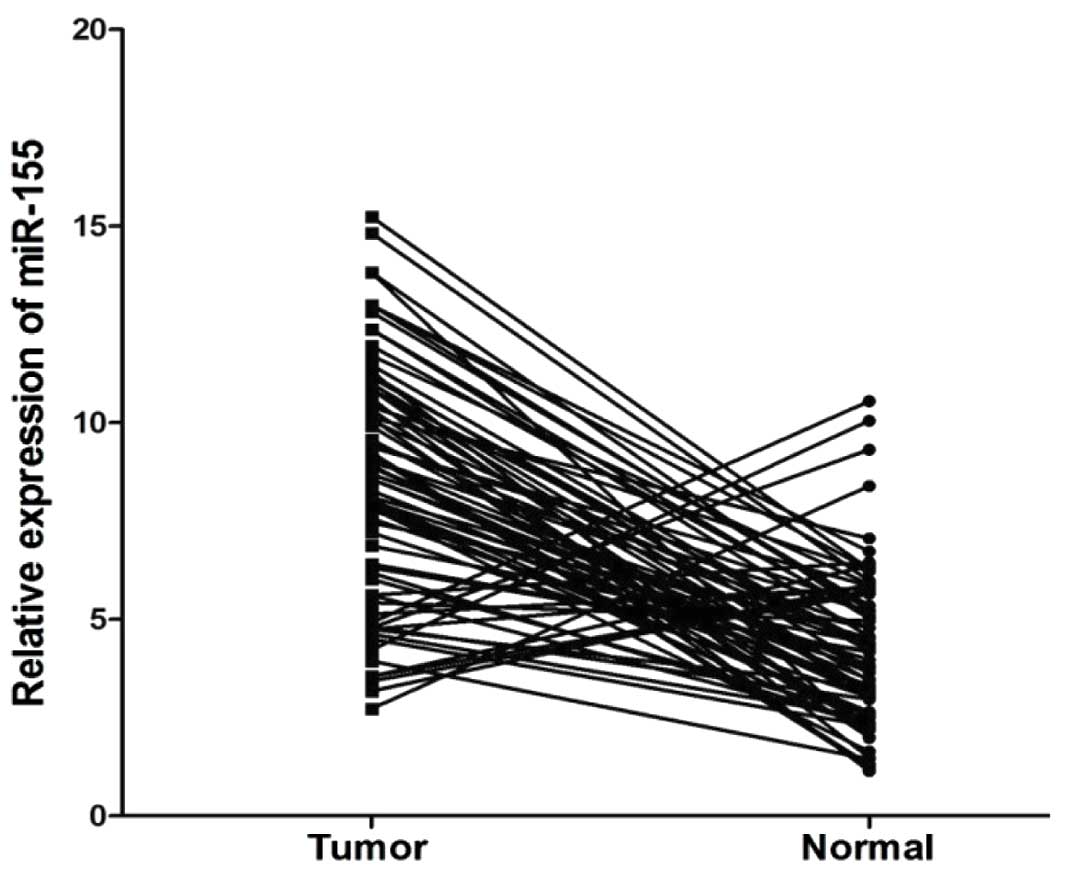
Using qRT-PCR, we found that the miR-155 expression
levels were significantly upregulated in the cancer tissues from
patients with CRC compared to those from the normal adjacent mucosa
(P<0.01, Fig. 1). The
expression levels of miR-155 were categorized as low or high in
relation to the median value. The high expression levels of miR-155
in CRC with respect to several standard clinicopathological
characteristics are presented in Table I. Our results demonstrated that
the high expression of miR-155 significantly correlated with an
advanced TNM stage, lymph node and distant metastasis (P<0.05,
Table I). However, there was no
significant correlation between miR-155 expression and other
clinicopathological characteristics, such as age, gender, tumor
size, histological type, depth of invasion, tumor location, or
lymph node invasion (P>0.05, Table
I).
 | Table IAssociation of miR-155 expression with
the clinicopathological characteristics of colorectal cancer
patients. |
Table I
Association of miR-155 expression with
the clinicopathological characteristics of colorectal cancer
patients.
| Characteristics | miR-155 expression
| |
|---|
| Low (n=38) | High (n=38) | P-value |
|---|
| Age (years) | 64.1±13.9 | 62.3±11.3 | 0.511 |
| Gender | | | 0.222 |
| Male | 28 | 23 | |
| Female | 10 | 15 | |
| Tumor size | | | 0.159 |
| ≤5 cm | 26 | 20 | |
| >5 cm | 12 | 18 | |
| Histological
type | | | 0.243 |
| Well, moderate | 25 | 20 | |
| Poor, mucinous | 13 | 18 | |
| Depth of
invasion | | | 0.063 |
| T1, T2 | 20 | 12 | |
| T3, T4 | 18 | 26 | |
| TNM stage | | | 0.020a |
| I, II | 21 | 11 | |
| III, IV | 17 | 27 | |
| Location | | | 0.622 |
| Colon | 11 | 13 | |
| Rectum | 27 | 25 | |
| Lymph node
metastasis | | | 0.011a |
| Absent | 23 | 12 | |
| Present | 15 | 26 | |
| Lymph node
invasion | | | 0.169 |
| Absent | 22 | 16 | |
| Present | 16 | 22 | |
| Distant
metastasis | | | 0.033a |
| Absent | 35 | 28 | |
| Present | 3 | 10 | |
miR-155 modulates migration and invasion
but not proliferation in CRC cells
To investigate the mechanism by which miR-155
promotes lymph node and distant metastasis, we examined the role of
miR-155 in CRC cell migration and invasion. Total RNA was isolated
from the cells 48 h after transfection; the expression of miR-155
was significantly increased in the cells transfected with
pre-miR-155, compared with that in the cells transfected with
pre-miR-n.c. (P<0.01, Fig.
2A). We observed that cell migration was significantly
increased following transfection with pre-miR-155 compared with the
negative control (P<0.01, Fig.
2C). We then examined the effect of miR-155 on cell invasion
across an extracellular matrix and showed that in the SW480 cells,
the overexpression of miR-155 markedly enhanced the invasive
potential compared with the control (P<0.01, Fig. 2D). However, the overexpression of
miR-155 in the SW480 cells had no significant effect on cell
proliferation (P>0.05, Fig.
2B). These results demonstrate that miR-155 promotes CRC cell
migration and invasion but has no effect on cell proliferation.
Upregulation of miR-155 alters cell
morphology and inhibits E-cadherin and enhances ZEB1 expression in
CRC cells
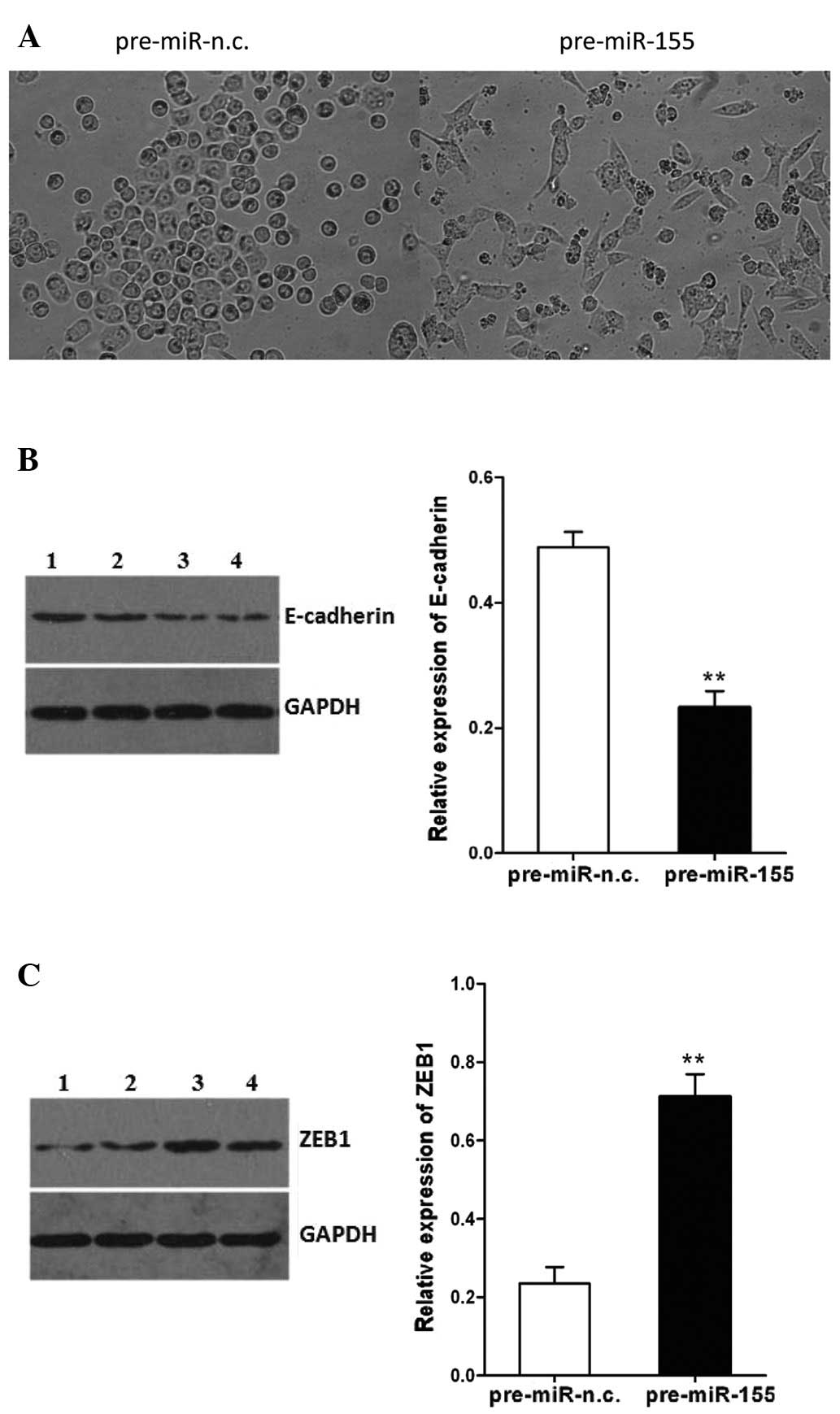
The SW480 cells transfected with pre-miR-155
underwent an EMT-like transformation, evidenced by the loss of
cell-cell adhesion and an alteration of morphology, from a round
compact shape to a spindle shape, which may facilitate cell
migration (Fig. 3A). In addition,
the expression of E-cadherin in the SW480 cells following
transfection with pre-miR-155 was decreased, whereas the expression
of ZEB1 was increased (P<0.01, Fig. 3B and C). These effects of miR-155
on CRC cells may involve the induction of EMT. These findings
demonstrate the potential role of miR-155 in promoting cell
migration and invasion.
Regulation of claudin-1 expression by
miR-155 in CRC cells
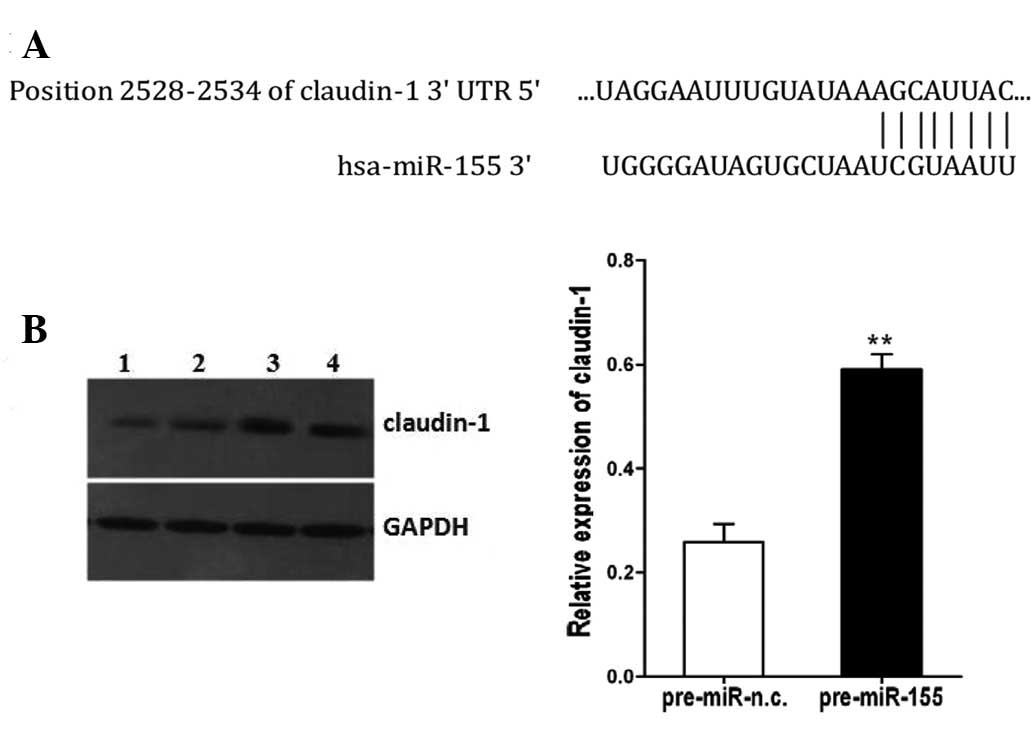
Through bioinformatics analyses with TargetScan
version 6.2, we found that E-cadherin and ZEB1 are not the targets
of miR-155. However, the 3′UTR of claudin-1 mRNA contains a highly
conserved binding site of miR-155 (Fig. 4A), and claudin-1 is predicted to
be a potential target for miR-155. To validate this hypothesis, we
compared claudin-1 expression in the SW480 cells transfected with
pre-miR-155 or pre-miR-n.c. Indeed, the protein level of claudin-1
was increased in the SW480 cells transfected with pre-miR-155
compared with the control-transfected cells (P<0.01, Fig. 4B).
Discussion
miR-155 is considered an important miRNA which is
located in the BIC gene and is highly expressed in a variety of
solid tumors, including breast, lung, colon, pancreatic and thyroid
cancers (14–16). In the present study, we also
confirmed that the expression of miR-155 was significantly
increased in CRC tissues compared with normal adjacent mucosa,
which suggests that the overexpression of miR-155 correlates with
the carcinogenesis of CRC. Moreover, we found that miR-155
expression was associated with an advanced clinical stage and lymph
node and distant metastases, strongly suggesting that miR-155 is
involved in the development, progression and metastasis of
colorectal cancer. However, Shibuya et al reported that the
expression level of miR-155 only correlated with lymph node
metastasis in colorectal cancer (17). These inconsistent results may be
due to the use of different detection methods, specimen collection
and patient clinicopathological data evaluation.
Cell motility and invasion are required for the
dissemination of tumor cells from their primary location to lymph
or blood vessels during metastasis (19). Our data demonstrate that miR-155
promotes cell migration and invasion, but not cell proliferation,
which is consistent with our tissue correlation study in which the
level of miR-155 was found to be associated with lymph node and
distant metastasis, but not the T stage and tumor size. In
addition, the results of this study revealed that tumors with a
high expression of miR-155 had a spindle-like morphology, and that
the upregulation of miR-155 decreased the expression of E-cadherin
and increased the expression of ZEB1, suggesting that miR-155 is
involved in the regulation of EMT. These findings demonstrate that
miR-155 exerts its effects on CRC progression by promoting the
metastatic ability of CRC cells.
The claudin family of proteins is the principal
constituent of the tight junction (20). Claudins are dynamic proteins and
help regulate a variety of cellular functions, such as
proliferation, migration and EMT, in addition to their tight
junction function (21–23). It has been reported that claudin-1
expression is highly upregulated and mislocalized in colon cancer
and correlates with tumor progression and metastasis (24). Kinugasa et al also reported
that claudin-1 levels were upregulated in hepatic metastatic
lesions of colorectal cancer (25). In the present study, transfection
with miR-155 significantly upregulated claudin-1 expression in CRC
cells. Using TargetScan analysis, miR-155 was found to target the
claudin-1 gene directly. Thus, the overexpression of miR-155
promotes the migration and invasive ability of CRC cells, at least
in part through the regulation of claudin-1 expression. A previous
study reported that claudin-1 upregulated the repressor ZEB-1,
inhibiting E-cadherin expression in colon cancer cells (26). These data suggest that the ectopic
expression of miR-155 upregulates claudin-1 expression, thus
increasing ZEB-1 expression levels and inhibiting E-cadherin, thus
facilitating cell migration and invasion. Thus, the data presented
in this study suggest that miR-155 plays a role in the metastatic
progression of CRC through the regulation of claudin-1
expression.
In conclusion, the overexpression of miR-155 plays
an important role in the migration and invasion of CRC; its effects
on CRC cells are possibly associated with the regulation of the
claudin-1/ZEB-1/E-cadherin axis. The results from this study
provide important clues as to the mechanisms involved in metastasis
in CRC. However, further studies are required to fully elucidate
the mechanisms involved in metastatic progression in CRC.
Acknowledgements
This study was supported by a grant
from the Scientific Research Program of the North Sichuan Medical
College of China (CBY11-A-ZP-05).
References
|
1
|
Jemal A, Siegel R, Ward E, et al: Cancer
statistics. CA Cancer J Clin. 58:71–96. 2008.
|
|
2
|
Kobayashi H, Mochizuki H, Sugihara K, et
al: Characteristics of recurrence and surveillance tools after
curative resection for colorectal cancer: a multicenter study.
Surgery. 141:67–75. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: at the crossroads of development
and tumour metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jou J and Diehl AM: Epithelial-mesenchymal
transitions and hepatocarcinogenesis. J Clin Invest. 120:1031–1034.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Spaderna S, Schmalhofer O, Hlubek F, et
al: A transient, EMT-linked loss of basement membranes indicates
metastasis and poor survival in colorectal cancer.
Gastroenterology. 131:830–840. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thiery JP: Epithelial-mesenchymal
transitions in development and pathologies. Curr Opin Cell Biol.
15:740–746. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Eades G, Yao Y, Yang M, et al: miR-200a
regulates SIRT1 expression and epithelial to mesenchymal transition
(EMT)-like transformation in mammary epithelial cells. J Biol Chem.
286:25992–26002. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kurashige J, Kamohara H, Watanabe M, et
al: MicroRNA-200b regulates cell proliferation, invasion, and
migration by directly targeting ZEB2 in gastric carcinoma. Ann Surg
Oncol. 19(Suppl 3): S656–S664. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang J, Mani SA, Donaher JL, et al: Twist,
a master regulator of morphogenesis, plays an essential role in
tumor metastasis. Cell. 117:927–939. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Eger A, Aigner K, Sonderegger S, et al:
DeltaEF1 is a transcriptional repressor of E-cadherin and regulates
epithelial plasticity in breast cancer cells. Oncogene.
24:2375–2385. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang T, Liu M, Wang C, et al:
Down-regulation of miR-206 promotes proliferation and invasion of
laryngeal cancer by regulating VEGF expression. Anticancer Res.
31:3859–3863. 2011.PubMed/NCBI
|
|
13
|
Wu J, Wu G, Lv L, et al: MicroRNA-34a
inhibits migration and invasion of colon cancer cells via targeting
to Fra-1. Carcinogenesis. 33:519–528. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gironella M, Seux M, Xie MJ, et al: Tumor
protein 53-induced nuclear protein 1 expression is repressed by
miR-155 and its restoration inhibits pancreatic tumor development.
Proc Natl Acad Sci USA. 104:16170–16175. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shibuya H, Iinuma H, Shimada R, et al:
Clinicopathological and prognostic value of microRNA-21 and
microRNA-155 in colorectal cancer. Oncology. 79:313–320. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kong W, Yang H, He L, et al: MicroRNA-155
is regulated by the transforming growth factor beta/Smad pathway
and contributes to epithelial cell plasticity by targeting RhoA.
Mol Cell Biol. 28:6773–6784. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bacac M and Stamenkovic I: Metastatic
cancer cell. Annu Rev Pathol. 3:221–247. 2008. View Article : Google Scholar
|
|
20
|
Tsukita S and Furuse M: Claudin-based
barrier in simple and stratified cellular sheets. Curr Opin Cell
Biol. 14:531–536. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ikari A, Takiguchi A, Atomi K, et al:
Decrease in claudin-2 expression enhances cell migration in renal
epithelial Madin-Darby canine kidney cells. J Cell Physiol.
226:1471–1478. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yoon CH, Kim MJ, Park MJ, et al: Claudin-1
acts through c-Abl-protein kinase CI (PKCI) signaling and has a
causal role in the acquisition of invasive capacity in human liver
cells. J Biol Chem. 285:226–233. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee JW, Hsiao WT, Chen HY, et al:
Up-regulated claudin-1 expression confers resistance to cell death
of nasopharyngeal carcinoma cells. Int J Cancer. 126:1353–1366.
2010.PubMed/NCBI
|
|
24
|
Dhawan P, Singh AB, Deane NG, et al:
Claudin-1 regulates cellular transformation and metastatic or in
colon cancer. J Clin Invest. 115:1765–1776. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kinugasa T, Akagi Y, Ochi T, et al:
Increased claudin-1 protein expression in hepatic metastatic
lesions of colorectal cancer. Anticancer Res. 32:2309–2314.
2012.PubMed/NCBI
|
|
26
|
Singh AB, Sharma A, Smith JJ, et al:
Claudin-1 up-regulates the repressor ZEB-1 to inhibit E-cadherin
expression in colon cancer cells. Gastroenterology. 141:2140–2153.
2011. View Article : Google Scholar : PubMed/NCBI
|