Introduction
Hyperthermia (HT) is a well-established treatment
modality that is used alone or in combination with other therapies,
such as radiotherapy and chemotherapy, for the treatment of a
variety of tumors (1). It has
been indicated that only cancer cells treated at temperatures
>42.5°C, the inflection point of HT, will undergo cell death as
the temperature increases, whereas temperatures in the mild
hyperthermia (MHT) range <42.5°C induce little or no
cytotoxicity (2). However, HT
monotherapy, maintaining a cancer temperature >42.5°C for direct
cytotoxicity, has been associated with higher toxicity and less
efficacy against cancer. Therefore, MHT, in combination with
radiotherapy and/or chemotherapy, has been used for the treatment
of various types of cancer, and its antitumor effects have been
verified by several clinical trials (3–7).
In addition, recent studies have demonstrated that an antitumor
immune system including heat shock protein (HSP) chaperone tumor
antigens may be effective as a cancer immunotherapy based on MHT
(8,9).
The heat shock response, a universal cellular
response to elevated temperatures, is a very important cellular
event for cell adaptation. It has been well recognized that heat
elicits a wide variety of stress responses, such as the induction
of HSPs, DNA and RNA damage, protein aggregation, cell growth
arrest and cell death (10–12). HSPs, which are are induced by heat
in particular, behave as strong cytoprotective molecules preventing
cell death (12) and participate
in thermoresistance (13,14). Although heat affects several
biological processes, the overall responses to heat in mammalian
cells are not yet completely understood. To this end, a number of
studies have found that the most effective approach for
investigating heat responses is to use global-scale microarrays and
bioinformatics tools. Several genes and biological functions in a
wide variety of cell types affected by heat stress have thus been
reported (15–27).
In the present study, in order to further elucidate
the molecular mechanisms underlying cellular responses to MHT in
cancer cells, we compared the gene expression profiles by using
high-density oligonucleotide microarrays and computational gene
expression analysis tools in two human cancer cell lines, HeLa
cervical squamous cell carcinoma (SCC) and HSC-3 oral SCC cells,
exposed to MHT at 41°C. Moreover, common expression patterns
responsive to MHT in two other human cell lines, MCF-7 breast
adenocarcinoma and HO-1-N-1 oral SCC, in addition to the HeLa and
HSC-3 cell lines were determined by real-time quantitative
polymerase chain reaction (qPCR).
Materials and methods
Cell culture
The human oral SCC cell lines, HSC-3 (derived from
tongue carcinoma) and HO-1-N-1 (derived from buccal mucosa) were
obtained from the Human Science Research Resources Bank of the
Japan Health Sciences Foundation (Tokyo, Japan). The human cervical
SCC cell line, HeLa, and the breast adenocarcinoma cell line,
MCF-7, were provided by the RIKEN BioResource Center through the
National BioResource Project of the Ministry of Education, Culture,
Sports, Science and Technology (MEXT) (Tsukuba, Japan). The HeLa
and MCF-7 cells were cultured in Dulbecco’s modified Eagle’s medium
(DMEM; Life Technologies Co., Grand Island, NY, USA) supplemented
with 10% fetal bovine serum (FBS). The HSC-3 and HO-1-N-1 cells
were cultured in Eagle’s minimum Essential medium (Wako Pure
Chemical Industries, Ltd., Osaka, Japan) and DMEM/Ham’s F-12 medium
(Wako Pure Chemical Industries, Ltd.), respectively, each
supplemented with 10% FBS. The cells were maintained at 37°C in a
humidified atmosphere with 5% CO2 and 95% air.
Exposure to MHT
The cells were exposed to MHT by immersing plastic
culture vessels containing the attached cells in a water bath at
41°C for 30 min. Following exposure to heat, the cells were
incubated for 0–24 h at 37°C.
Analyses of cell viability and cell
cycle
For cell viability, the trypan blue dye exclusion
test was performed. The cells excluding the dye were counted using
a hematocytometer. For the cell cycle, cells were fixed with 70%
ice-cold ethanol and then treated with RNase A and propidium iodide
(PI). The samples were run on an Epics XL flow cytometer (Beckman
Coulter, Fullerton, CA, USA) as previously described (28).
RNA isolation
Total RNA was extracted from the cells using an
RNeasy Total RNA Extraction kit (Qiagen, Valencia, CA, USA) along
with On-Column DNase I treatment. The RNA concentration was
measured using a Nanodrop spectrophotometer (Thermo Fisher
Scientific Inc., Waltham, MA, USA). RNA integrity was determined
using a Bioanalyzer 2100 (Agilent Technologies, Inc., Santa Clara,
CA, USA). RNA samples with RNA integrity number (RIN) values
>9.5 were considered acceptable.
Global-scale gene expression
analysis
Global-scale gene expression analysis was performed
using a GeneChip® microarray system with a Human Genome
U133-plus 2.0 array, which was spotted with 54,675 probe sets
(Affymetrix Inc., Santa Clara, CA, USA) according to the
manufacturer’s instructions. In brief, 500 ng of total RNA were
used to synthesize cRNA with a GeneChip® 3′ IVT Express
kit (Affymetrix Inc.). Following fragmentation, biotin-labeled cRNA
was hybridized to the array at 45°C for 16 h. The arrays were
washed, stained with streptavidin-phycoerythrin, and scanned using
a probe array scanner. The obtained hybridization intensity data
were analyzed using GeneSpring® GX (Agilent Technologies
Inc.) to extract the significant genes. To examine gene ontology,
including biological processes, cellular components, molecular
functions and gene networks, the obtained data were analyzed using
Ingenuity® Pathway Analysis tools (Ingenuity Systems
Inc., Mountain View, CA, USA), a web-delivered application that
enables the identification, visualization and investigation of
molecular interaction networks in gene expression data (29,30).
Real-time qPCR assay
The reverse transcriptase reaction was carried out
with total RNA using a random 6-mer and an oligo(dT) primer.
Real-time qPCR was performed on an M×3005P real-time PCR system
(Agilent Technologies Inc.) using SYBR PreMix ExTaq (Takara Bio
Inc., Shiga, Japan) or Premix Ex Taq (for the use of TaqMan probes;
Takara Bio Inc.). The specific primer and probe sequences for
B-cell CLL/ lymphoma 2 (BCL2)-associated athanogene 3 (BAG3),
B-cell CLL/lymphoma 6 (BCL6), bone morphogenetic protein 2 (BMP2),
connective tissue growth factor (CTGF), DnaJ (Hsp40) homolog,
subfamily B, member 1 (DNAJB1), endothelin 1 (EDN1), early growth
response 1 (ERG1), glyceraldehyde-3-phosphate dehydrogenase
(GAPDH), heat shock 70 kDa protein 1A/B (HSPA1A/B), heat shock
105/110 kDa protein (HSPA1H), heat shock 70 kDa protein 6 (HSP70B’)
(HSPA6), MAX dimerization protein 1 (MXD1) and suppressor of
cytokine signaling 3 (SOCS3) are listed in Table I. GAPDH was used as a control for
normalization.
 | Table I.Nucleotide sequences of primers and
TaqMan probes for target genes. |
Table I.
Nucleotide sequences of primers and
TaqMan probes for target genes.
| Gene | Orientation | Nucleotide sequence
(5′-3′) | GenBank accession
no. |
|---|
| BAG3 | Sense |
CGACCAGGCTACATTCCCAT | NM_004281 |
| Antisense |
TCTGGCTGAGTGGTTTCTGG | |
| BCL6 | Sense |
AATCGTCTCCGGAGTCGAGACA | NM_002357 |
| Antisense |
TGATGTTGCCCTCCCGCAAA | |
| BMP2 | Sense |
ATGCTGTGTCCCGACAGAAC | NM_001200 |
| Antisense |
AACCCTCCACAACCATGTCC | |
| CTGF | Sense |
CTGCAGGCTAGAGAAGCAGA | NM_001901 |
| Antisense |
CCGTCGGTACATACTCCACA | |
| DNAJB1 | Sense |
ACCCGGACAAGAACAAGGAG | NM_006145 |
| Antisense |
GCCACCGAAGAACTCAGCAA | |
| EDN1 | Sense |
CTTGCCAAGGAGCTCCAGAA | NM_001955 |
| Antisense |
GGCTTCCAAGTCCATACGGAA | |
| HSPA1A/B | Sense |
AGGTGCAGGTGAGCTACAAG | NM_005346 |
| Antisense |
ATGATCCGCAGCACGTTGAG | |
| HSPH1 | Sense |
ACCATGCTGCTCCTTTCTCC | NM_006644 |
| Antisense |
CTGGGTTTTCTGGTGGTCTC | |
| HSPA6 | Sense |
GGCCATGACCAAGGACAACA | NM_002155 |
| Antisense |
AACCATCCTCTCCACCTCCT | |
| MXD1 | Sense |
AGACGGGCTCATCTTCGCTT | NM_002357 |
| Antisense |
GCTGAAGCTGGTCGATTTGGTG | |
| SOCS3 | Sense |
TCTGTCGGAAGACCGTCAAC | NM_003955 |
| Antisense |
TAAAGCGGGGCATCGTACTG | |
| EGR1 | Sense |
GGAGCAAAATAAGGAAGAG | NM_001964 |
| Antisense |
GAGAGTTCAGATTTTGTTTTAG | |
| Probe |
FAM-AGCTGAGCTTCGGTTCTCCA-TAMRA | |
| GAPDH | Sense |
AAGGACTCATGACCACAGTCCAT | NM_002046 |
| Antisense |
CCATCACGCCACAGTTTCC | |
| Probe |
FAM-CCATCACTGCCACCCAGAAGACTGTG-TAMRA | |
Statistical analysis
Data are presented as the means ± SD. Differences
between pairs of data sets were analyzed using the Student’s
t-test, with values of P<0.05 considered to indicate
statistically significant differences.
Results
Effects of MHT on cell viability and the
cell cycle in cancer cell lines
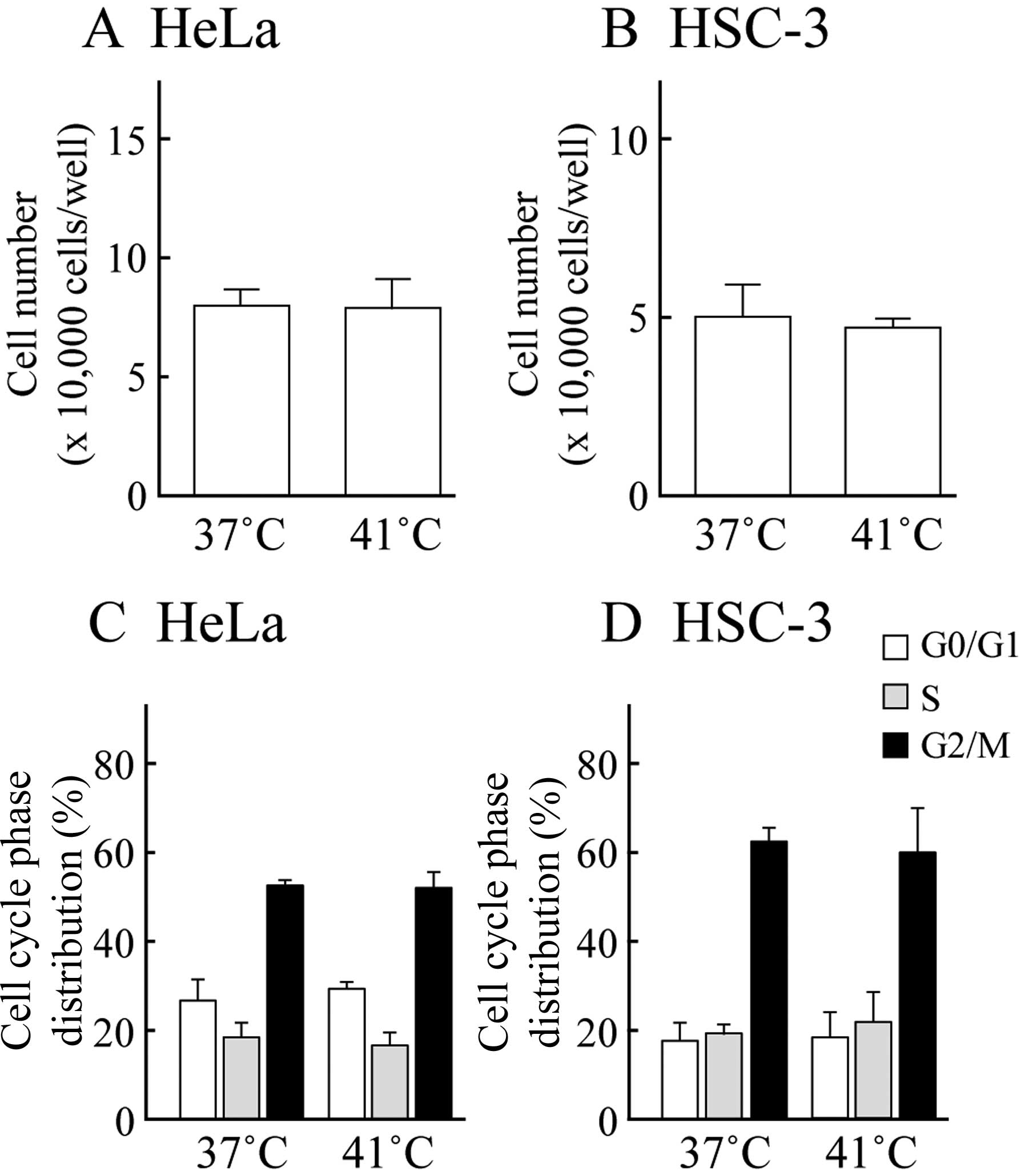
We used a trypan blue dye exclusion test to assess
cell viability. Fig. 1A and B
indicate the number of HeLa and HSC-3 cells following exposure to
MHT at 41°C for 30 min followed by culture at 37°C for 24 h. MHT
did not inhibit cell viability compared to the control (37°C
treatment) in either cell line. Moreover, the cell cycle
distribution was monitored by PI staining with flow cytometry. The
percentages of control HeLa and HSC-3 cells in the G0/G1, S and
G2/M phase were 26.9±4.8, 18.5±3.3 and 52.3±1.2% for the HeLa cells
and 17.5±4.2, 19.3±2.5 and 62.2±3.4% for the HSC-3 cells (means ±
SD), respectively. Changes in cell cycle distribution were not
observed in the cells exposed to MHT (Fig. 1C and D). Exposure to MHT at 41°C
for 30 min did not affect cell viability or the cell cycle
distribution in the other cancer cell lines, MCF-7 and HO-1-N-1
(data not shown). These data demonstrated that the cells exposed to
MHT at 41°C for 30 min did not undergo cell damage and cell cycle
arrest.
Global-scale gene expression
analysis
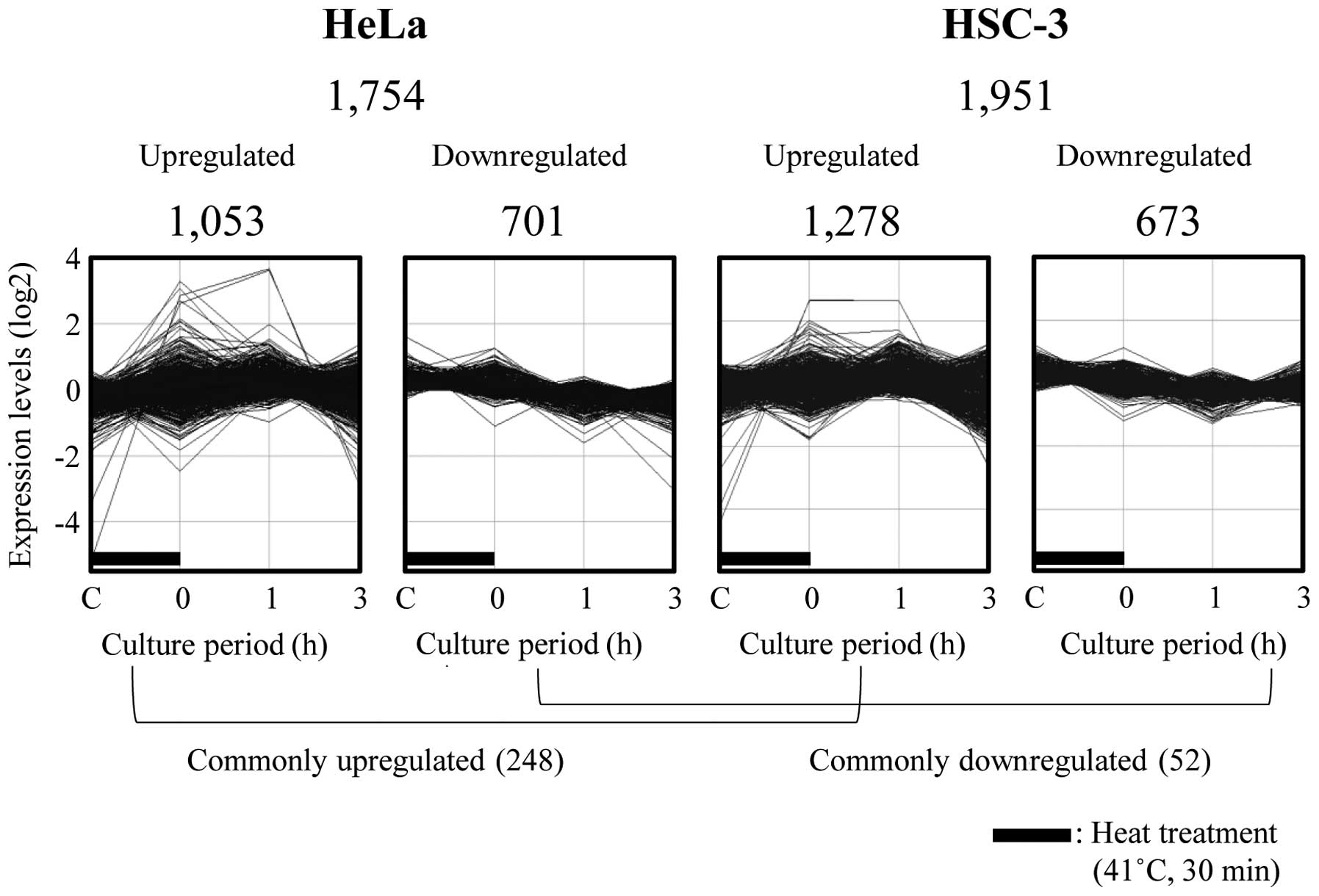
Global-scale gene expression analysis was performed
using a GeneChip microarray system and GeneSpring software. Of the
54,675 probe sets analyzed, many probe sets that were
differentially expressed by ≥1.5-fold were detected. We identified
1,053 and 1,278 upregulated, and 701 and 673 downregulated probe
sets in the HeLa and HSC-3 cells exposed to MHT at 41°C for 30 min,
respectively. Fig. 2 demonstrates
the time course of changes in gene expression. In all, 248 and 52
probe sets were also found to be commonly upregulated and
downregulated in these respective cell lines (Fig. 2). The complete lists of genes from
the HeLa and HSC-3 cell samples have been deposited in the Gene
Expression Omnibus (GEO), a public database (accession numbers:
GSE43701 and GSE43862).
Identification of biological functions
and gene networks
To identify the biological functions and gene
networks in the differentially expressed probe sets in the HeLa and
HSC-3 cells exposed to MHT, either functional category or gene
network analysis was conducted by use of the Ingenuity Pathways
Knowledge Base. We identified several functionally annotated genes:
494 and 637 upregulated genes, and 303 and 330 down-regulated genes
in the HeLa and HSC-3 cells exposed to MHT, respectively (Table II). The top three biological
functions in each group are summarized in Table II. Biological functions including
cell death and survival, gene expression and cellular function and
maintenance, were observed in the upregulated genes in both cell
lines, and the most prominent functions of commonly upregulated
genes were cellular functions and maintenance (48 genes), as well
as cell death and survival (60 genes). In the downregulated genes
in both cell lines, biological functions including cell cycle, gene
expression and cell death and survival were observed. The specific
biological functions were cell cycle (13 genes) and cell death and
survival (18 genes) in commonly downregulated genes (Table II). Furthermore, we identified two
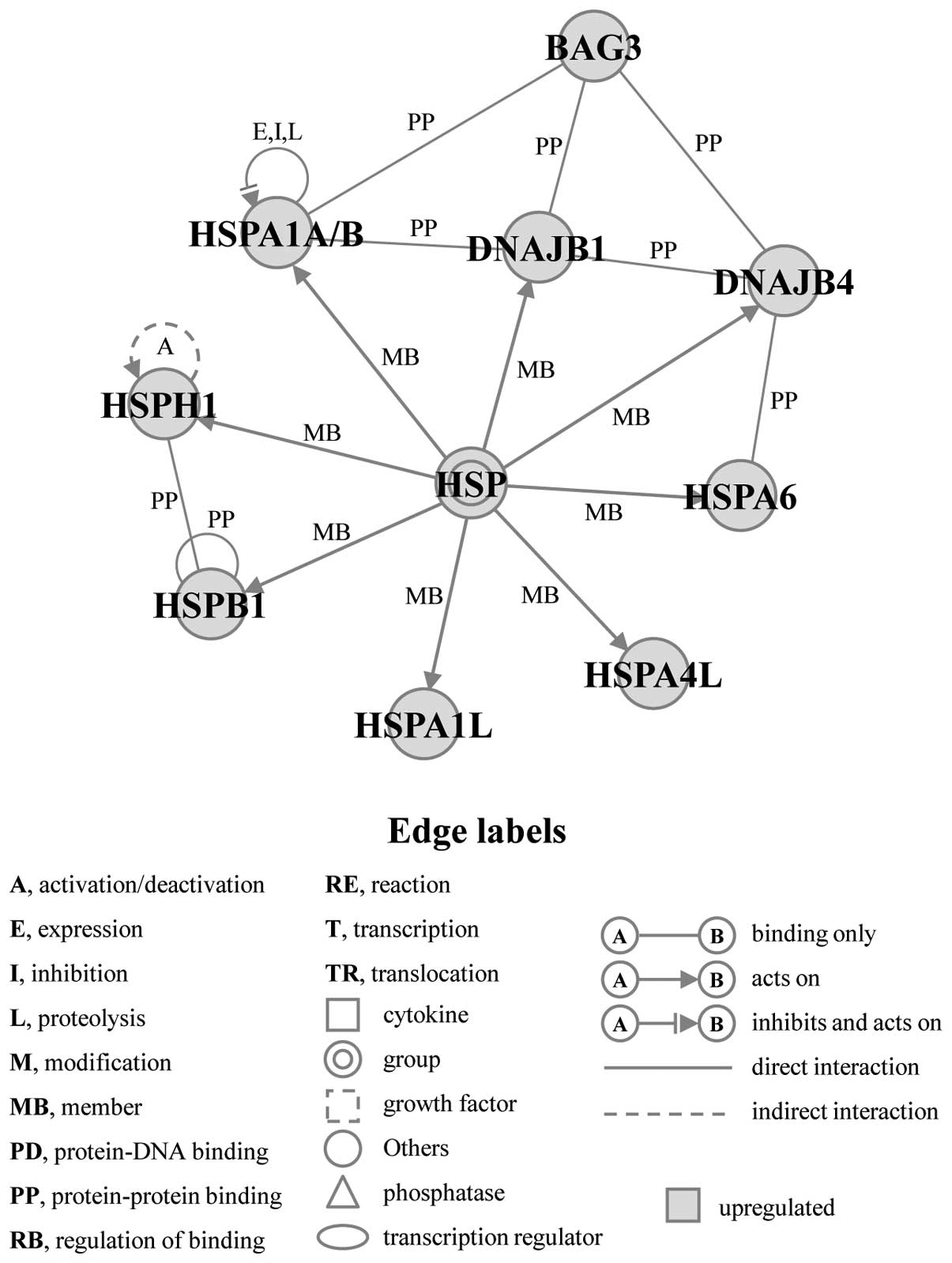
significant gene networks, A and B, that were obtained from
commonly upregulated genes in both cell lines (Figs. 3 and 4). Gene network A included several HSPs,
such as HSPA1A/B, heat shock 70 kDa protein 1-like (HSPA1L), heat
shock protein 4 like (HSPA4L), HSPA6, heat shock 27 kDa protein 1
(HSPB1), HSPH1, DNAJB1, and DnaJ (Hsp40) homolog, subfamily B,
member 4 (DNAJB4), as well as BAG3, and was associated with the
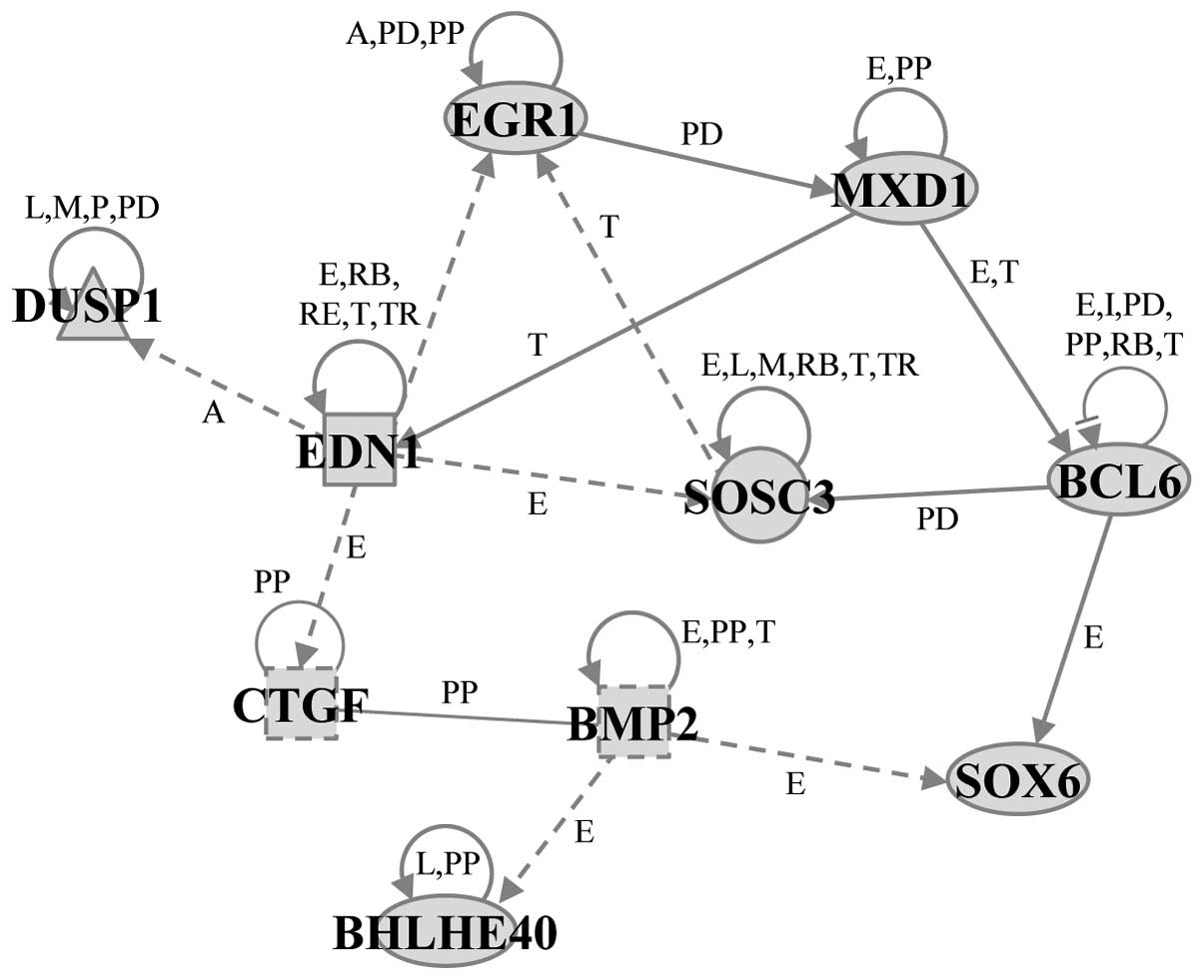
biological functions of cellular function and maintenance (Fig. 3). Gene network B included ten
genes: BCL6, basic helix-loop-helix family, member e40 (BHLHE40),
BMP2, CTGF, dual specificity phosphatase 1 (DUSP1), EDN1, EGR1,
MXD1, SOCS3 and SRY (sex determining region Y)-box 6 (SOX6) and was
associated with the biological functions of cell death and survival
(Fig. 4).
 | Table II.Top three biological functions. |
Table II.
Top three biological functions.
| Name | P-value | Numbers of
molecules |
|---|
| HeLa upregulated
genes (494)a | | |
| Cell death and
survival |
3.34E-05-3.39E-02 | 224 |
| Gene
expression |
4.92E-05-3.39E-02 | 169 |
| Cellular
development |
2.89E-04-3.39E-02 | 202 |
| HeLa downregulated
genes (303)a | | |
| Gene
expression |
6.21E-06-3.57E-02 | 140 |
| Cellular assembly
and organization |
1.00E-04-3.57E-02 | 90 |
| Cellular function
and maintenance |
1.00E-04-3.57E-02 | 80 |
| HSC-3 upregulated
genes (637)a | | |
| Cell death and
survival |
2.92E-13-2.85E-03 | 292 |
| Gene
expression |
3.32E-13-2.71E-03 | 248 |
| Cellular growth
and proliferation |
4.54E-10-2.99E-03 | 299 |
| HSC-3 downregulated
genes (330)a | | |
| Cell cycle |
3.44E-06-3.24E-02 | 102 |
| Cell death and
survival |
3.31E-05-3.25E-02 | 148 |
| Cellular
development |
9.25E-05-3.26E-02 | 111 |
| Commonly
upregulated genes (114)a | | |
| Cellular function
and maintenance |
2.21E-07-1.89E-02 | 48 |
| Cell death and
survival |
2.46E-06-2.01E-02 | 60 |
| Cellular
movement |
2.97E-05-2.02E-02 | 40 |
| Commonly
downregulated genes (35)a | | |
| Cell cycle |
9.35E-05-4.86E-02 | 48 |
| Cell death and
survival |
6.09E-04-4.56E-02 | 60 |
| Gene
expression |
6.17E-04-4.86E-02 | 40 |
Verification of differentially expressed
genes by real-time qPCR
To verify the alterations analyzed by the GeneChip
microarray system, we used two cancer cell lines, MCF-7 and
HO-1-N-1, in addition to the HeLa and HSC-3 cell lines.
Time-dependent changes in the expression levels of selected genes
that belonged to gene network A or B were measured using real-time
qPCR. Among the genes in gene network A, the expression levels of
HSPA1A/B, HSPA6, HSPH1, DNAJB1 and BAG3 were markedly elevated
immediately following exposure to MHT in all four cell lines. The
expression levels of BCL6, BMP2, CTGF, EDN1 and MXD1 in gene
network B were significantly increased at 1 h following exposure to
MHT in all four cell lines. The expression levels of EGR1 and SOCS3
were significantly increased at 0, 1 and 3 h following exposure to
MHT in all four cell lines. These results are in very close
agreement with those of the GeneChip® analysis with the
HeLa and HSC-3 cells (Fig.
5).
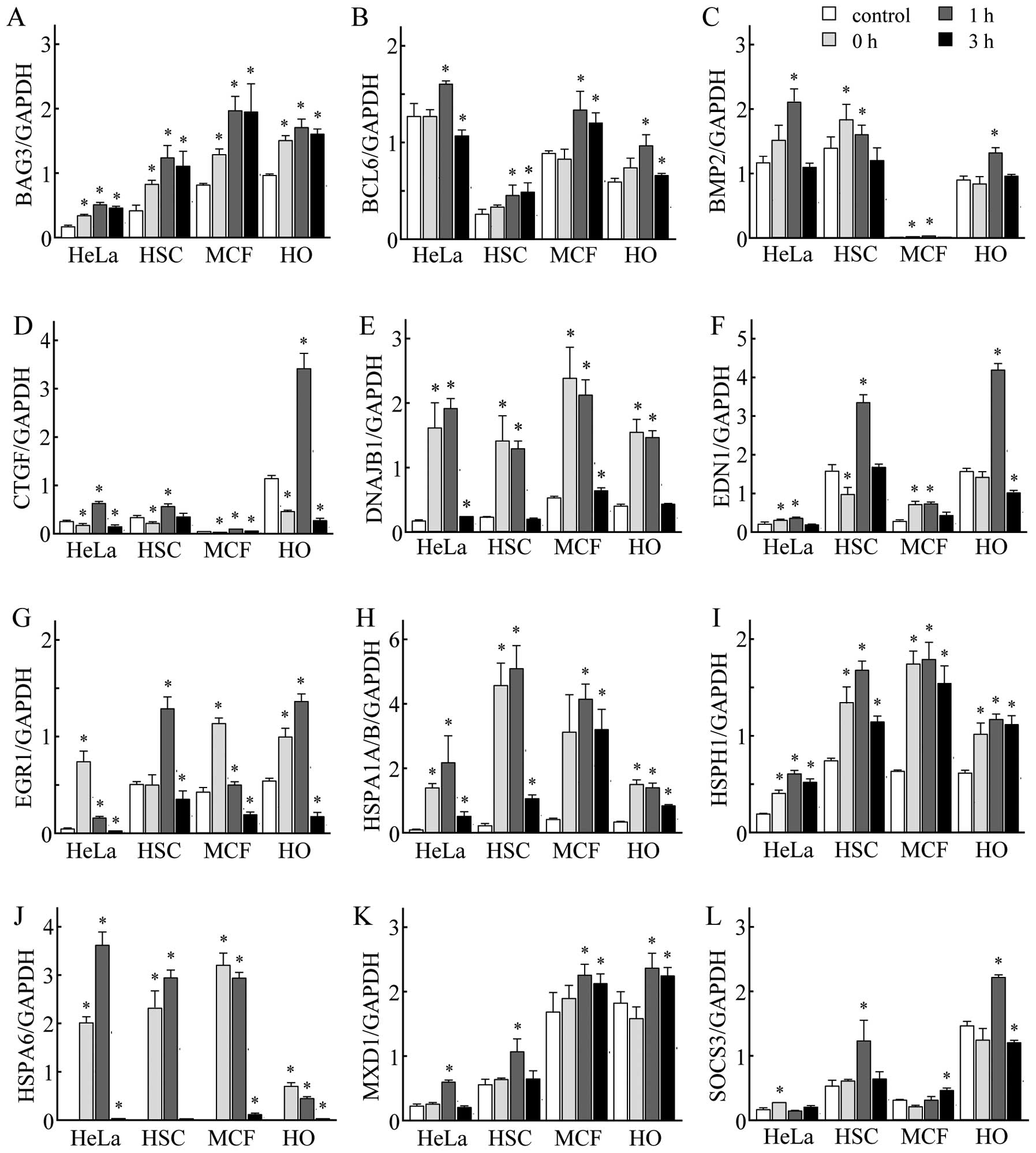 | Figure 5.Verification of GeneChip®
microarray results by real-time quantitative polymerase chain
reaction (qPCR). The cancer cell lines, HeLa, HSC-3 (HSC), MCF-7
(MCF) and HO-1-N-1 (HO), were incubated at 41°C for 30 min and then
cultured at 37°C for 0, 1 and 3 h. Real-time qPCR was performed.
(A) BCL2-associated athanogene 3 (BAG3), (B) B-cell CLL/lymphoma 6
(BCL6), (C) bone morphogenetic protein 2 (BMP2), (D) connective
tissue growth factor (CTGF), (E) DnaJ (Hsp40) homolog, subfamily B,
member 1 (DNAJB1), (F) endothelin 1 (EDN1), (G) early growth
response 1 (EGR1), (H) heat shock 70 kDa protein 1A/B (HSPA1A/B),
(I) HSPH1, (J) heat shock 70 kDa protein 6 (HSP70B’) (HSPA6), (K)
MAX dimerization protein 1 (MXD1) and (L) suppressor of cytokine
signaling 3 (SOCS3). Each expression level was normalized to the
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression level.
Data are presented as the means ± SD (n=3–4). *P<0.05
vs. control (non-treated cells). |
Discussion
HT is a well-established treatment modality that is
used alone or in combination with radiotherapy and/or chemotherapy
for the treatment of a variety of tumors (1). However, due to the complexity of the
heat stress signal transduction, the elucidation of the molecular
mechanisms involved in the cellular responses to heat stress has
remained a major challenge in studies of HT. In this study, we used
global-scale microarray analysis, in conjunction with
bioinformatics tools, and identified unique common gene networks in
response to MHT in cancer cells.
It is well known that a temperature of 42.5°C for
cancer cells is the inflection point of HT. That is, cancer cells
exposed to temperatures >42.5°C undergo cell death; however, the
exposure of cells to a temperature of up to 42.5°C induces little
or no cytotoxicity (2). In the
present study, the exposure of HeLa and HSC-3 human SCC cells to
MHT at 41°C, below the inflection point, did not affect cell
viability or the cell cycle, as has also been shown in our previous
studies (24–27). By contrast, several genes that
were upregulated or downregulated by ≥1.5-fold were identified in
both cancer cell lines following exposure to MHT. These results
were comparable to those from previous studies on MHT conditions
using cancer or normal cell lines (17,23–27). Previous studies have indicated
that heat induces a significant decrease in mRNA and protein
synthesis in order to increase the transcription of heat-responsive
genes, such as HSPs, and that this overall reprogramming of gene
expression permits the selective synthesis of HSPs (31,32). Therefore, in this study, we
focused on the common and upregulated genes elicited by MHT. We
successfully identified two significant gene networks, A and B,
derived from commonly upregulated genes in HeLa and HSC-3 cells
exposed to MHT. In addition, the expression levels of 12 out of 19
genes selected were increased in the four cancer cell lines. Gene
network A included several HSPs, as well as BAG3, and was
specifically associated with the biological functions of cellular
function and maintenance. Several microarray studies have
demonstrated that the induction of HSPs is a common response to
heat stress with or without cell death (15–27). HSPs protect cells both by limiting
the effects of protein-damaging agents through protein chaperoning
and refolding and by directly blocking the pathway of cell death,
such as apoptosis or necrosis (12). In gene network A, HSPA1A/B
(33,34), HSPB1 (35), HSPH1 (36) and DNAJB1 (33) were found to be associated with
cytoprotection. BAG3 is a family of co-chaperons that interact with
Hsp70 (37,38). The interaction between BAG3 and
DNAJB1 or DNAJB4 has also been observed in human cells (39). The siRNA-mediated inhibition of
BAG3 expression has been shown to increase apoptosis, indicating
that BAG3 plays a role in the prevention of cell death (37,38).
Furthermore, Ingenuity Pathway Analysis allowed us
to identify the significant gene network B, which was associated
with the biological functions of cell death and survival. Seven
genes in network B, BCL6 (40),
BMP2 (41), CTGF (42), EDN1 (43), EGR1 (44), MXD1 (45) and SOCS3 (46), have been described as anti-cell
death molecules in several types of cells, including cancer cells.
Of note, we elucidated the interactions among the genes in gene
network B: that is, EDN1 is involved in the upregulation of mRNAs
for EGR1 (47), CTGF (48) and SOCS3 (49). It has also been reported that the
binding of MXD1 DNA and EGR1 protein (50), as well as the protein-protein
interaction between CTGF and BMP2 (51), occur in mammalian cells. Genes
having cytoprotective or anti-cell death activity in gene networks
A and B may play a role in protecting cancer cells from MHT
stress.
Although HT is an important modality in cancer
treatment, the acquisition of thermoresistance in cancer cells due
to the elevation of HSPs makes HT less effective (13,14). The induction of HSPs is regulated
mainly at the transcriptional level by the activation of heat shock
transcription factor 1 (HSF1) (52,53). In addition, the induction of BAG3
is mediated, at least in part, by the activation of HSF1, as in the
case of HSPs (54). Of note, the
inhibition of the functions of HSF1 (55–57) or Hsp70 (58) by the use of gene targeting has
been shown to sensitize HT-induced cell death in cancer cells. The
targeting of heat shock response pathways, including these
molecules in combination with HT may become a promising therapeutic
approach for the treatment of cancer. In this study, we identified
two gene networks comprising of several cytoprotective genes in
four cancer cell lines exposed to MHT. Thus, the findings presented
in this study may prove useful towards the design of more effective
therapeutic strategies involving MHT or HT for the treatment of
cancer.
Abbreviations:
|
BAG3
|
BCL2-associated athanogene 3;
|
|
BCL2
|
B-cell CLL/lymphoma 2;
|
|
BCL6
|
B-cell CLL/lymphoma 6;
|
|
BHLHE40
|
basic helix-loop-helix family, member
e40;
|
|
BMP2
|
bone morphogenetic protein 2;
|
|
CTGF
|
connective tissue growth factor;
|
|
DMEM
|
Dulbecco’s modified Eagle’s
medium;
|
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B,
member 1;
|
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B,
member 4;
|
|
DUSP1
|
dual specificity phosphatase 1;
|
|
EDN1
|
endothelin 1;
|
|
ERG1
|
early growth response 1;
|
|
FBS
|
fetal bovine serum;
|
|
GAPDH
|
glyceraldehyde-3-phosphate
dehydrogenase;
|
|
HSF1
|
heat shock transcription factor 1;
|
|
HSP
|
heat shock protein;
|
|
HSPA1A/B
|
heat shock 70 kDa protein 1A/B;
|
|
HSPA1H
|
heat shock 105/110 kDa protein;
|
|
HSPA1L
|
heat shock 70 kDa protein 1-like;
|
|
HSPA4L
|
heat shock protein 4 like;
|
|
HSPA6
|
heat shock 70 kDa protein 6
(HSP70B’);
|
|
HSPB1
|
heat shock 27 kDa protein 1;
|
|
HT
|
hyperthermia;
|
|
MHT
|
mild hyperthermia;
|
|
MXD1
|
MAX dimerization protein 1;
|
|
PI
|
propidium iodide;
|
|
qPCR
|
quantitative polymerase chain
reaction;
|
|
SCC
|
squamous cell carcinoma;
|
|
SOCS3
|
suppressor of cytokine signaling
3;
|
|
SOX6
|
SRY (sex determining region Y)-box
6
|
Acknowledgements
The present study was supported in
part by a Grant-in-Aid for Challenging Exploratory Research
(23650303) from the Japan Society for the Promotion of Science, and
by research grants from the University of Toyama.
References
|
1.
|
Hall EJ: Hyperthermia. Radiobiology for
the Radiologist. 5th edition. Hall EJ: Lippincott Williams &
Wilkins; pp. 495–520. 2000
|
|
2.
|
Dewey WC: Arrhenius relationships from the
molecule and cell to the clinic. Int J Hyperthermia. 25:3–20. 2009.
View Article : Google Scholar
|
|
3.
|
van der Zee J, González González D, van
Rhoon GC, van Dijk JD, van Putten WL and Hart AA: Comparison of
radiotherapy alone with radiotherapy plus hyperthermia in locally
advanced pelvic tumours: a prospective, randomised, multicentre
trial. Dutch Deep Hyperthermia Group Lancet. 355:1119–1125.
2000.PubMed/NCBI
|
|
4.
|
Wust P, Hildebrandt B, Sreenivasa G, et
al: Hyperthermia in combined treatment of cancer. Lancet Oncol.
3:487–497. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Issels RD: Hyperthermia adds to
chemotherapy. Eur J Cancer. 44:2546–2554. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Issels RD, Lindner LH, Verweij J, et al:
Neo-adjuvant chemotherapy alone or with regional hyperthermia for
localised high-risk soft-tissue sarcoma: a randomised phase 3
multicentre study. Lancet Oncol. 11:561–570. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Westermann A, Mella O, Van Der Zee J, et
al: Long-term survival data of triple modality treatment of stage
IIB-III-IVA cervical cancer with the combination of radiotherapy,
chemotherapy and hyperthermia - an update. Int J Hyperthermia.
28:549–553. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Beachy SH and Repasky EA: Toward
establishment of temperature thresholds for immunological impact of
heat exposure in humans. Int J Hyperthermia. 27:344–352. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Frey B, Weiss EM, Rubner Y, et al: Old and
new facts about hyperthermia-induced modulations of the immune
system. Int J Hyperthermia. 28:528–542. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Lindquist S: The heat-shock response. Annu
Rev Biochem. 55:1151–1191. 1986. View Article : Google Scholar
|
|
11.
|
Richter K, Haslbeck M and Buchner J: The
heat shock response: life on the verge of death. Mol Cell.
40:253–266. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Beere HM: ‘The stress of dying’: the role
of heat shock proteins in the regulation of apoptosis. J Cell Sci.
117:2641–2651. 2004.
|
|
13.
|
Li GC and Werb Z: Correlation between
synthesis of heat shock proteins and development of thermotolerance
in Chinese hamster fibroblasts. Proc Natl Acad Sci USA.
79:3218–3222. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Nollen EA, Brunsting JF, Roelofsen H,
Weber LA and Kampinga HH: In vivo chaperone activity of heat shock
protein 70 and thermotolerance. Mol Cell Biol. 19:2069–2079.
1999.PubMed/NCBI
|
|
15.
|
Dinh HK, Zhao B, Schuschereba ST, Merrill
G and Bowman PD: Gene expression profiling of the response to
thermal injury in human cells. Physiol Genomics. 7:3–13.
2001.PubMed/NCBI
|
|
16.
|
Sonna LA, Gaffin SL, Pratt RE, Cullivan
ML, Angel KC and Lilly CM: Effect of acute heat shock on gene
expression by human peripheral blood mononuclear cells. J Appl
Physiol. 92:2208–2220. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Murray JI, Whitfield ML, Trinklein ND,
Myers RM, Brown PO and Botstein D: Diverse and specific gene
expression responses to stresses in cultured human cells. Mol Biol
Cell. 15:2361–2374. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Zhou M, Zhang A, Lin B, Liu J and Xu LX:
Study of heat shock response of human umbilical vein endothelial
cells (HUVECs) using cDNA microarray. Int J Hyperthermia.
23:225–258. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Wong HR, Odoms K and Sakthivel B:
Divergence of canonical danger signals: the genome-level expression
patterns of human mononuclear cells subjected to heat shock or
lipopolysaccharide. BMC Immunol. 9:242008. View Article : Google Scholar
|
|
20.
|
Narita N, Noda I, Ohtsubo T, et al:
Analysis of heat-shock related gene expression in head-and-neck
cancer using cDNA arrays. Int J Radiat Oncol Biol Phys. 53:190–196.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Kato N, Kobayashi T and Honda H: Screening
of stress enhancer based on analysis of gene expression profiles:
enhancement of hyperthermia-induced tumor necrosis by an MMP-3
inhibitor. Cancer Sci. 94:644–649. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Borkamo ED, Dahl O, Bruland O and Fluge O:
Global gene expression analyses reveal changes in biological
processes after hyperthermia in a rat glioma model. Int J
Hyperthermia. 24:425–441. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Laramie JM, Chung TP, Brownstein B, Stormo
GD and Cobb JP: Transcriptional profiles of human epithelial cells
in response to heat: computational evidence for novel heat shock
proteins. Shock. 29:623–630. 2008.PubMed/NCBI
|
|
24.
|
Tabuchi Y, Takasaki I, Wada S, et al:
Genes and genetic networks responsive to mild hyperthermia in human
lymphoma U937 cells. Int J Hyperthermia. 24:613–622. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Furusawa Y, Tabuchi Y, Wada S, Takasaki I,
Ohtsuka K and Kondo T: Identification of biological functions and
gene networks regulated by heat stress in U937 human lymphoma
cells. Int J Mol Med. 28:143–151. 2011.PubMed/NCBI
|
|
26.
|
Tabuchi Y, Wada S, Furusawa Y, Ohtsuka K
and Kondo T: Gene networks related to the cell death elicited by
hyperthermia in human oral squamous cell carcinoma HSC-3 cells. Int
J Mol Med. 29:380–386. 2012.PubMed/NCBI
|
|
27.
|
Tabuchi Y, Furusawa Y, Kariya A, Wada S,
Ohtsuka K and Kondo T: Common gene expression patterns responsive
to mild temperature hyperthermia in normal human fibroblastic
cells. Int J Hyperthermia. 29:38–50. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Furusawa Y, Iizumi T, Fujiwara Y, et al:
Inhibition of checkpoint kinase 1 abrogates G2/M checkpoint
activation and promotes apoptosis under heat stress. Apoptosis.
17:102–112. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Tabuchi Y, Takasaki I, Doi T, Ishii Y,
Sakai H and Kondo T: Genetic networks responsive to sodium butyrate
in colonic epithelial cells. FEBS Lett. 580:3035–3041. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Ahmed K, Furusawa Y, Tabuchi Y, et al:
Chemical inducers of heat shock proteins derived from medicinal
plants and cytoprotective genes response. Int J Hyperthermia.
28:1–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Mariner PD, Walters RD, Espinoza CA, et
al: Human Alu RNA is a modular transacting repressor of mRNA
transcription during heat shock. Mol Cell. 29:499–509. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Spriggs KA, Bushell M and Willis AE:
Translational regulation of gene expression during conditions of
cell stress. Mol Cell. 40:228–237. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Zhou H, Li SH and Li XJ: Chaperone
suppression of cellular toxicity of huntingtin is independent of
polyglutamine aggregation. J Biol Chem. 276:48417–48424.
2001.PubMed/NCBI
|
|
34.
|
Hageman J, van Waarde MA, Zylicz A,
Walerych D and Kampinga HH: The diverse members of the mammalian
HSP70 machine show distinct chaperone-like activities. Biochem J.
435:127–142. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Lee YJ, Lee DH, Cho CK, et al: HSP25
inhibits radiation-induced apoptosis through reduction of
PKCdelta-mediated ROS production. Oncogene. 24:3715–3725. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Yamagishi N, Goto K, Nakagawa S, Saito Y
and Hatayama T: Hsp105 reduces the protein aggregation and
cytotoxicity by expanded-polyglutamine proteins through the
induction of Hsp70. Exp Cell Res. 316:2424–2433. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Rosati A, Ammirante M, Gentilella A, et
al: Apoptosis inhibition in cancer cells: a novel molecular pathway
that involves BAG3 protein. Int J Biochem Cell Biol. 39:1337–1342.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Jacobs AT and Marnett LJ: HSF1-mediated
BAG3 expression attenuates apoptosis in 4-hydroxynonenal-treated
colon cancer cells via stabilization of anti-apoptotic Bcl-2
proteins. J Biol Chem. 284:9176–9183. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Behrends C, Sowa ME, Gygi SP and Harper
JW: Network organization of the human autophagy system. Nature.
466:68–76. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Kumagai T, Miki T, Kikuchi M, et al: The
proto-oncogene Bc16 inhibits apoptotic cell death in
differentiation-induced mouse myogenic cells. Oncogene. 18:467–475.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Chen S, Guttridge DC, Tang E, Shi S, Guan
K and Wang CY: Suppression of tumor necrosis factor-mediated
apoptosis by nuclear factor kappaB-independent bone morphogenetic
protein/Smad signaling. J Biol Chem. 276:39259–39263. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Wang MY, Chen PS, Prakash E, et al:
Connective tissue growth factor confers drug resistance in breast
cancer through concomitant up-regulation of Bcl-xL and cIAP1.
Cancer Res. 69:3482–3491. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Raymond MN, Bole-Feysot C, Banno Y, Tanfin
Z and Robin P: Endothelin-1 inhibits apoptosis through a
sphingosine kinase 1-dependent mechanism in uterine leiomyoma ELT3
cells. Endocrinology. 147:5873–5882. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Weisz L, Zalcenstein A, Stambolsky P, et
al: Transactivation of the EGR1 gene contributes to mutant p53 gain
of function. Cancer Res. 64:8318–8327. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Lüscher B: Function and regulation of the
transcription factors of the Myc/Max/Mad network. Gene. 277:1–14.
2001.
|
|
46.
|
Puhr M, Santer FR, Neuwirt H, et al:
Down-regulation of suppressor of cytokine signaling-3 causes
prostate cancer cell death through activation of the extrinsic and
intrinsic apoptosis pathways. Cancer Res. 69:7375–7384. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Bruneau BG, Piazza LA and de Bold AJ: BNP
gene expression is specifically modulated by stretch and ET-1 in a
new model of isolated rat atria. Am J Physiol. 273:H2678–H2686.
1997.PubMed/NCBI
|
|
48.
|
Koitabashi N, Arai M, Kogure S, et al:
Increased connective tissue growth factor relative to brain
natriuretic peptide as a determinant of myocardial fibrosis.
Hypertension. 49:1120–1127. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Chang HH, Huang YM, Wu CP, et al:
Endothelin-1 stimulates suppressor of cytokine signaling-3 gene
expression in adipocytes. Gen Comp Endocrinol. 178:450–458. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
50.
|
Virolle T, Krones-Herzig A, Baron V, De
Gregorio G, Adamson ED and Mercola D: Egr1 promotes growth and
survival of prostate cancer cells. Identification of novel Egr1
target genes. J Biol Chem. 278:11802–11810. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
51.
|
Maeda S: An impact of CCN2-BMP-2 complex
upon chondrocyte biology: evoking a signalling pathway bypasses ERK
and Smads? J Biochem. 150:219–221. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52.
|
Morimoto RI: Regulation of the heat shock
transcriptional response: cross talk between a family of heat shock
factors, molecular chaperones, and negative regulators. Genes Dev.
12:3788–3796. 1998. View Article : Google Scholar
|
|
53.
|
Akerfelt M, Morimoto RI and Sistonen L:
Heat shock factors: integrators of cell stress, development and
lifespan. Nat Rev Mol Cell Biol. 11:545–555. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54.
|
Franceschelli S, Rosati A, Lerose R, De
Nicola S, Turco MC and Pascale M: Bag3 gene expression is regulated
by heat shock factor 1. J Cell Physiol. 215:575–577. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
55.
|
Rossi A, Ciafrè S, Balsamo M, Pierimarchi
P and Santoro MG: Targeting the heat shock factor 1 by RNA
interference: a potent tool to enhance hyperthermochemotherapy
efficacy in cervical cancer. Cancer Res. 66:7678–7685. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
56.
|
Nakamura Y, Fujimoto M, Hayashida N, Takii
R, Nakai A and Muto M: Silencing HSF1 by short hairpin RNA
decreases cell proliferation and enhances sensitivity to
hyperthermia in human melanoma cell lines. J Dermatol Sci.
60:187–192. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57.
|
Tabuchi Y, Furusawa Y, Wada S, Ohtsuka K
and Kondo T: Silencing heat shock transcription factor 1 using
small interfering RNA enhances mild hyperthermia and hyperthermia
sensitivity in human oral squamous cell carcinoma cells. Thermal
Med. 27:99–108. 2011. View Article : Google Scholar
|
|
58.
|
Gabai VL, Budagova KR and Sherman MY:
Increased expression of the major heat shock protein Hsp72 in human
prostate carcinoma cells is dispensable for their viability but
confers resistance to a variety of anticancer agents. Oncogene.
24:3328–3338. 2005. View Article : Google Scholar
|