Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common cancer worldwide and the third highest cause of
cancer-related mortality (500,000 deaths annually) (1). HCC is a primary malignant tumor of
the liver with high prevalence in Asia and Africa (2). There are a number of different
therapies for the treatment of HCC, including percutaneous ethanol
injection therapy (PEIT), transcatheter arterial chemoembolization
(TACE), liver transplantation and surgical intervention. Of these,
surgical intervention has been the most effective therapy for
improving the survival of patients (3). Nevertheless, only a small subset of
HCC patients increase their 5-year survival rate by surgical
resection, primarily due to the high rate of metastasis and the
expression of anti-apoptotic genes associated with HCC (4,5).
Therefore, metastasis remains the major obstacle to the development
of optimal treatment methods for HCC, and novel or adjunct
therapeutic strategies are paramount to overcoming this
obstacle.
Tea is the most widely consumed beverage worldwide;
furthermore, green tea is sold on a large scale, in part, due to
its chemotherapeutic value (6,7).
Catechins constitute approximately 40% of the dry weight of green
tea, and epigallocatechin-3-gallate (EGCG), a polyphenol,
constitutes the highest percentage among the catechins (8–10).
EGCG has immense potential as a therapeutic agent for the treatment
and/or prevention of cancer due to its low cost and high
bioavailability (11). The
anticancer role of EGCG has been investigated epidemiologically, in
in vitro and in vivo models, as well as in clinical
trials (12–14). In vitro studies have
demonstrated the inhibitory effects of EGCG on cancer by
suppressing metastasis (13,15,16). There is also a large body of
evidence demonstrating the effects of EGCG on the migration ability
of several human cancer cell lines by a multifactorial mechanism
involving the downregulation of matrix metalloproteinases (MMPs)
(15,17). Yet, the precise mechanisms of
action of EGCG as an anticancer agent remain unknown.
To our knowledge, the potential effects of EGCG on
HCCLM6, a human HCC metastatic cell line, have not been previously
reported. Thus, in this study, we investigated the effects and
molecular mechanisms of action of EGCG in HCCLM6 cells as a novel
and/or adjunct therapeutic agent in the treatment of HCC.
Materials and methods
Antibodies and reagents
EGCG (≥98% purity),
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
and gelatin were purchased from Sigma (St. Louis, MO, USA). The RNA
PCR kit (AMV) was purchased from the Takara Biotechnology Co., Ltd.
(Dalian, China). DMEM and fetal bovine serum (FBS) were purchased
from Gibco (Grand Island, NY, USA). TRIzol was purchased from
Invitrogen (Camarillo, CA, USA). The Pierce® SilverStain
for Mass Spectrometry kit was purchased from Thermo Scientific
(Rockford, IL, USA). Immobilized pH gradient (IPG) strips and the
2-D Cleanup kit were purchased from Bio-Rad (Hercules, CA, USA).
The BCA protein assay kit was purchased from KeyGen Biotech. Co.
Ltd. (Nanjing, China), and 24-well, double-compartment Transwell
plates were purchased from Corning Inc. (Lowell, MA, USA).
Cells and culture
HCCLM6 cells (ATCC, Manassas, VA, USA) were grown in
DMEM supplemented with 10% heat-inactivated FBS, 0.1% benzyl
penicillin and streptomycin. Cells were maintained at 37°C in a
humidified incubator with an atmosphere of 5% CO2.
Cell viability assay
HCCLM6 cells (6×103 cells/well) were
plated in 96-well plates for 12 h. The cells were treated with
various concentrations (0, 5, 10, 20, 30, 40, 50, 60, 80 or 100
μg/ml) of EGCG and incubated for an additional 24 h. At the end of
the treatment, the culture medium was replaced with fresh complete
medium containing 0.5 mg/ml MTT and incubated at 37°C for 4 h.
Following incubation, the medium was discarded, and DMSO was added
to the wells and gently agitated for 10 min. The absorbance was
then measured at 490 nm. The assay was repeated at least 3 times.
The cell growth inhibition rates were calculated according to the
following formula: inhibition rate (%) = (1 − mean absorbance of
treated group/mean absorbance of untreated group) ×100%.
In vitro migration and invasion
assays
The 24-well Transwell plates and 8.0 μm pore filter
inserts (Corning Inc.) were used for the migration and invasion
assays according to the standard Boyden Chamber protocol. Briefly,
HCCLM6 cells were seeded in a 6-well plate, and at 80% confluency,
the cells were treated with 0, 5 or 10 μg/ml EGCG and incubated for
18 h. The cells were harvested, counted and diluted to
5×105/ml of viable cells in medium without FBS. The cell
suspension (0.2 ml) was placed on top of the filter of the upper
chamber, and 0.6 ml of complete medium was placed in the lower
chamber. The filter insert was placed into the lower chamber and
incubated for 18 h at 37°C. Non-migrated cells were removed from
the top side of the filter by scrubbing with a cotton-tipped swab
moistened with FBS-free medium. Migrated cells on the underside of
the filter were fixed in 20% methanol for 20 min, stained with 0.1%
crystal violet at 37°C for 30 min and then washed with PBS. The
invasion assay was carried out as described above, except that the
top side of the filter membrane was pre-coated with 50 μl of
Matrigel (0.3 mg/50 μl; Bio-Rad) and incubated for 5 h at 37°C to
allow the matrix to form a gel before seeding the cells onto it.
Stained cells were photographed at ×200 magnification using a Nikon
camera fitted to a Leica microscope (Leica Microsystems, Wetzlar,
Germany).
Gelatin zymography
The HCCLM6 cells (1×105) were seeded in a
6-well plate. At 80% confluency, the cells were treated with 0, 5,
10 or 20 μg/ml EGCG and incubated for 18 h. Following incubation,
the cells and medium were harvested and centrifuged (1,000 rpm for
5 min) to collect the supernatant. The supernatants from the
samples were normalized to the cell number and electrophoresed on a
10% polyacrylamide gel containing 1 mg/ml gelatin at 4°C. Following
electrophoresis, the gels were washed for 15 min in 2.5% Triton
X-100 4 times at room temperature and then incubated for 24 h at
37°C in activation buffer (50 mM Tris-HCl, pH 7.5; 5 mM
CaCl2; 10 mM NaCl; 10 mM ZnCl2). The gels
were stained with 0.2% coomassie brilliant blue (Ameresco Co.,
Framingham, MA, USA) for 3 h and then destained in destaining
buffer (30% methanol, 10% acetic acid) until the gelatinolytic
activity of the MMPs are visible.
RT-PCR
The HCCLM6 cells (3×105) were seeded in
6-well plates until they reached 80% confluency. The cells were
treated with 0, 5, 10 and 20 μg/ml EGCG or 0, 10 and 30 μg/ml EGCG
and incubated for 20 h. Total RNA was extracted with TRIzol
according to the manufacturer’s instructions (Invitrogen).
Semi-quantification and purity assessment were performed by optical
density (OD) measurements at 260 and 280 nm. cDNA was synthesized
from the total RNA using an RNA PCR kit (Takara Biotechnology Co.,
Ltd.). The primers were derived from human sequences (Table I), and the PCR conditions were
optimized until the gene products were within the linear phase of
PCR amplification. The PCR products were resolved on 1% (w/v)
agarose gels containing ethidium bromide. The results were
normalized to β-actin. The PCR conditions for each target gene
(MMP-2, MMP-9 and β-actin) were as follows: 95°C for 5 min; 30
cycles of 95°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec;
72°C for 10 min.
 | Table IPrimer sequences and PCR
conditions. |
Table I
Primer sequences and PCR
conditions.
| Gene | Primer
sequences | PCR cycles (30
cycles) | Amplicon (bp) |
|---|
| MMP-2 | Sense: |
5′-ATGACAGCTGCACCACTGAG-3′ | 95°C, 30 sec | 673 |
| Antisense: |
5′-GCCTCGTATACCGCATCAAT-3′ | 55°C, 30 sec | |
| | | 72°C, 30 sec | |
| MMP-9 | Sense: |
5′-GTGCTGGGCTGCTGCTTTGCTG-3′ | 95°C, 30 sec | 303 |
| Antisense: |
5′-GTCGCCCTCAAAGGTTTGGAAT-3′ | 55°C, 30 sec | |
| | | 72°C, 30 sec | |
| β-actin | Sense: |
5′-GGAGTCCTGTGGCATCCACG-3′ | 95°C, 30 sec | 322 |
| Antisense: |
5′-CTAGAAGCATTTGCGGTGGA-3′ | 55°C, 30 sec | |
| | | 72°C, 30 sec | |
Proteomic analysis of EGCG-treated
cells
The HCCLM6 cells were plated as described above and
treated with EGCG (0 or 20 μg/ml). Total protein was extracted and
pre-treated with the 2-D Cleanup kit (Bio-Rad). Isoelectrofocusing
(IEF) was performed using 7 cm IPG strips (Bio-Rad) with an
immobilized pH gradient from 3–10. The strips were rehydrated at
room temperature for 16 h with 125 μl of swelling buffer (8 mol/l
urea, 2% CHAPS, 2% IPG buffer, 0.3% DTT and a trace of bromophenol
blue), which contained 45 μg of pre-treated proteins from either
the EGCG-treated or untreated cells. IEF was performed at 200 V for
20 min, 450 V for 15 min, 750 V for 15 min and 3,000 V for 2 h.
Following IEF, the strips were immediately equilibrated with the
equilibrium sample buffer (50 mmol/l Tris-HCl, pH 8.8; 6 mol/l
urea; 30% glycerol; 2% SDS; and a trace of bromophenol blue) at
room temperature for 30 min with gentle shaking. SDS-PAGE was
performed using 12% SDS-polyacrylamide gels. The strips were held
in place with 0.5% agarose dissolved in SDS running buffer, and
electrophoresis was then performed (5 mA for 20 min, 15 mA for 20
min, 20 mA for 20 min, and 30 mA in 4°C for 2 h). The gels were
stained with the Silver Stain for Mass Spectrometry kit (Thermo
Scientific) according to the manufacturer’s instructions. Image
analysis was performed using the PDQuest system (Bio-Rad). The
selected spots were excised, and the proteins were purified by an
in-gel digestion with trypsin. The resulting peptides were
subjected to analysis by matrix-assisted laser
desorption/ionization-time of flight mass spectrometry
(MALDI-TOF/MS).
Statistical analysis
The results of all the experiments are expressed as
the means ± SD. Statistical analyses were performed using SPSS 13.0
software. A P-value <0.05 was considered to indicate a
statistically significant difference.
Results
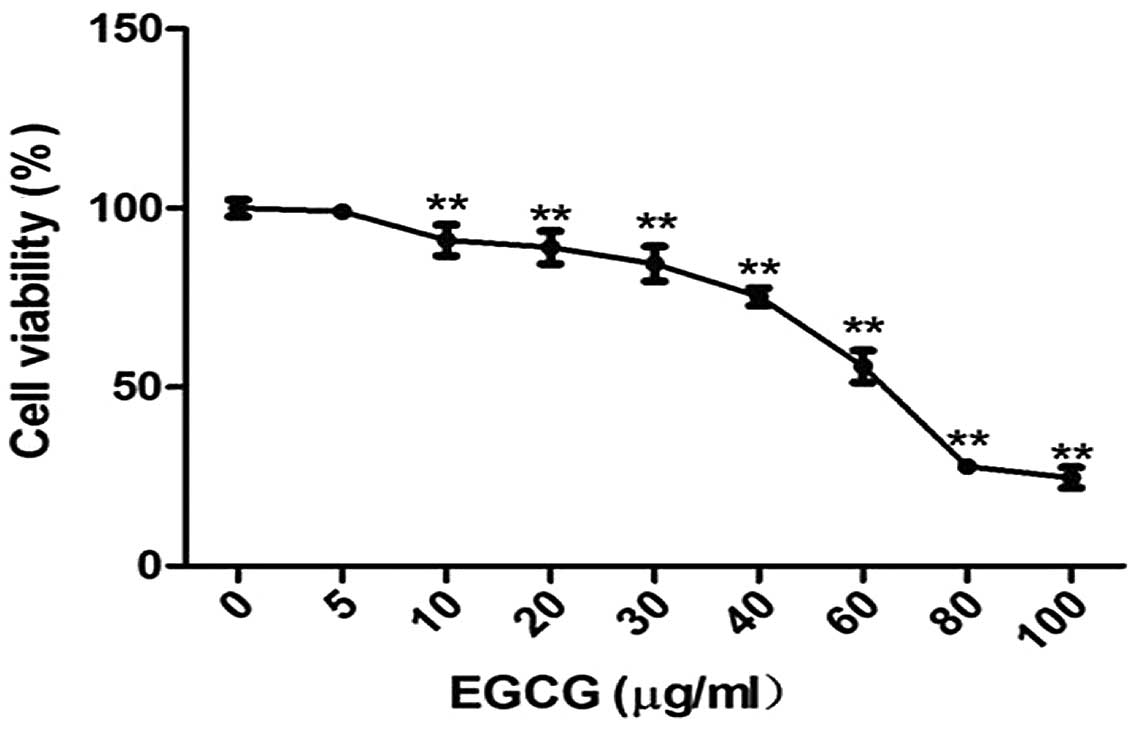
Effects of EGCG on cell viability
To determine the effect of EGCG on HCC, HCCLM6 cells
were treated with EGCG at concentrations varying between 5 and 100
μg/ml. EGCG (10–100 μg/ml) significantly inhibited the growth of
HCCLM6 cells in a dose-dependent manner (Fig. 1) (P<0.01). These results
indicate that EGCG inhibits cell growth (or induces apoptosis) in
the HCC cell line, HCCLM6.
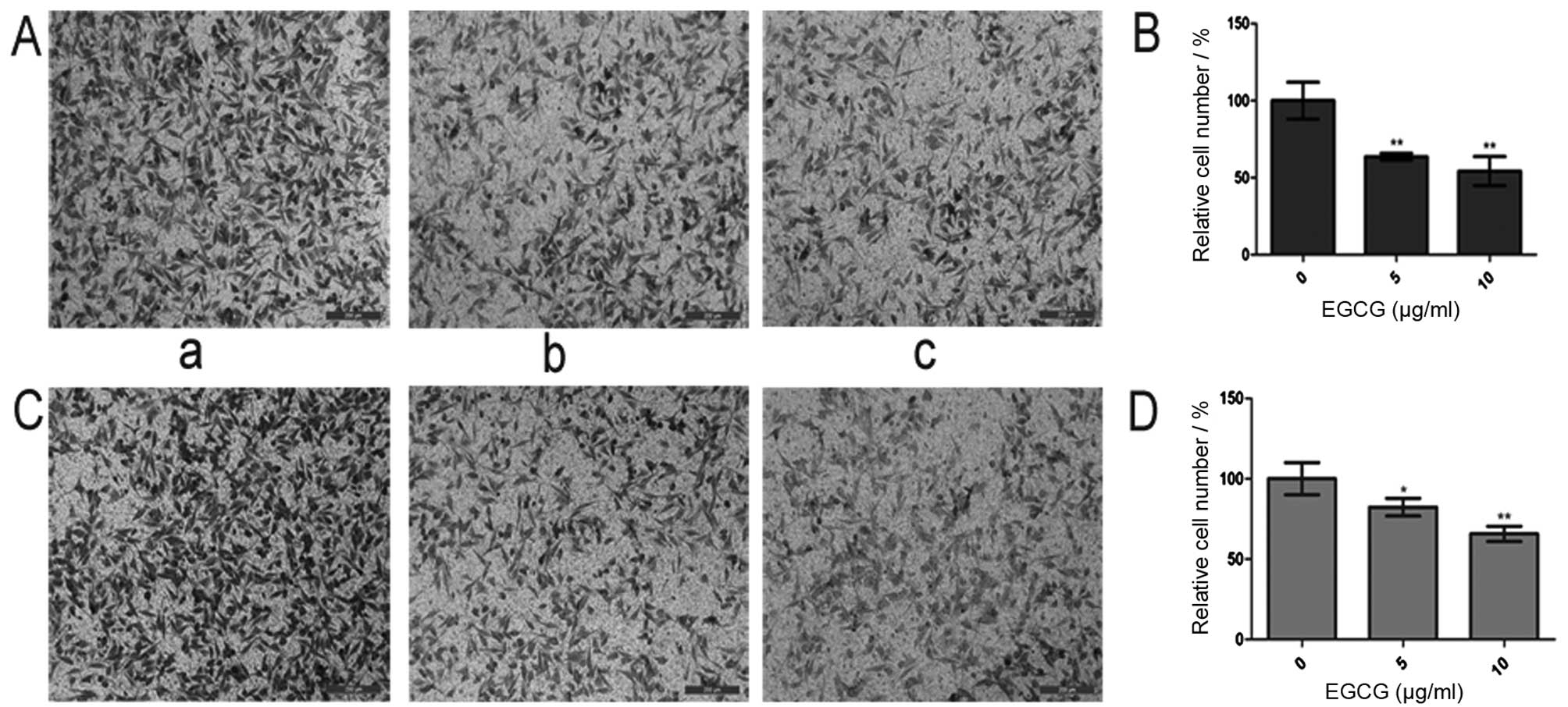
Effects of EGCG on cell migration and
invasion
As indicated in Materials and methods, the 24-well
Transwell membrane inserts had pores large enough to accommodate
the migration (and/or the invasion) of single cells. The cells
invading the underside of the 24-well Transwell membrane in these
assays were fixed and stained (Fig.
2A and C). The cell migration analysis of HCCLM6 cells showed
significantly lower (P<0.01) migration in the 5 and 10 μg/ml
EGCG-treated cells compared with the untreated cells (Fig. 2B). The cell invasion analysis of
HCCLM6 cells showed significantly lower (P<0.01) invasion in the
5 and 10 μg/ml EGCG-treated cells compared with the untreated cells
(Fig. 2D).
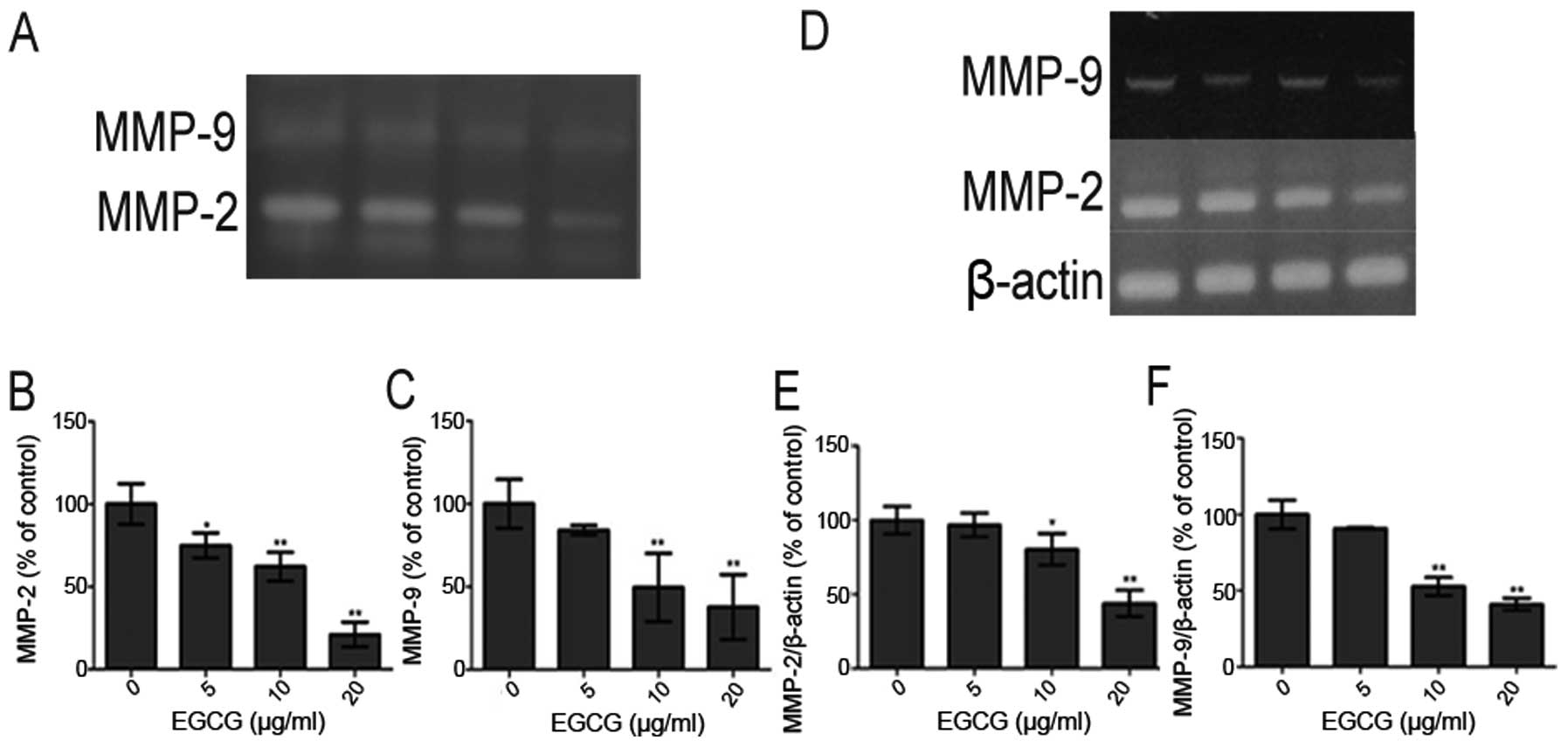
Effect of EGCG on MMP-2 and MMP-9
activity
Gelatin zymography analysis indicated that treatment
with EGCG significantly inhibited the activity of MMP-2 and MMP-9
in the conditioned medium of HCCLM6 cells (Fig. 3A). The activity of MMP-2 and MMP-9
decreased in a dose-dependent manner with the increasing EGCG
concentration (Fig. 3B and C,
respectively).
Effect of EGCG on MMP-2 and MMP-9 gene
expression
To determine the effects of EGCG on the mRNA levels
of MMP-2 and MMP-9 genes, the HCCLM6 cells (1×106
cells/ml) were maintained in culture with or without EGCG for 24 h.
Total RNA was isolated for RT-PCR as described above. As shown in
Fig. 3D, the mRNA levels of MMP-2
and MMP-9 were downregulated in a dose-dependent manner by EGCG
(Fig. 3E and F,
respectively).
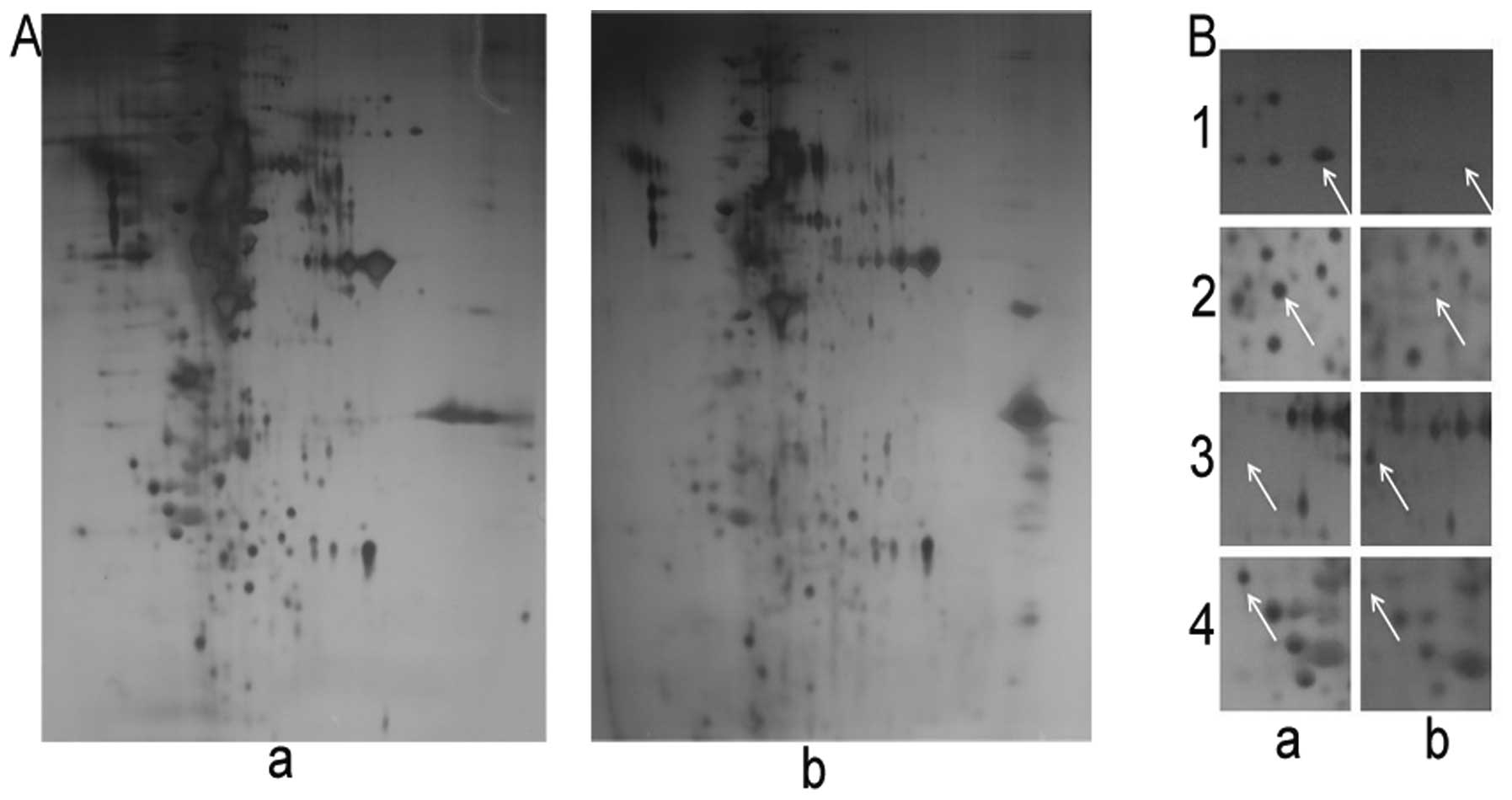
Proteomic analysis of EGCG-treated
cells
The proteins from the EGCG-treated or untreated
HCCLM6 cells were separated by 2-dimensional gel electrophoresis
(2-DE) according to their isoelectric points and molecular weights.
The 2-DE gels represent the pattern of all the proteins in the
untreated and EGCG-treated HCCLM6 cells (Fig. 4A). Of the proteins whose
expression was significantly altered by EGCG treatment, 10 were
selected (data not shown) and analyzed by MALDI-TOF/MS. Using
bioinformatics mining with the MASCOT search engine (http://www.matrixscience.com/) and NCBI BLASTP
(http://www.ncbi.nlm.nih.gov/blast),
we determined significant functional differences (Table II) in 4 of these proteins
(Fig. 4B).
 | Table IICharacteristics of the 4 proteins
with significant functional differences and their relative
expression levels in HCCLM6 cells treated with EGCG. |
Table II
Characteristics of the 4 proteins
with significant functional differences and their relative
expression levels in HCCLM6 cells treated with EGCG.
| Spot no. | Protein(s)
identified | Accession no. | Molecular mass | pI | Score
intensity | Spot |
|---|
| 1 | Far upstream
element binding protein 1 (FUBP1) | Q96AE4 | 67690 | 7.18 | 96 | −a |
| 2 | Heat shock protein
beta 1 (HSPB1) | P04792 | 22826 | 5.98 | 70 | −a |
| 3 | Nucleophosmin
(NPM) | P06748 | 32726 | 4.64 | 83 | +a |
| 4 | Heat shock 60 kDa
protein 1 (chaperonin) (CH60) | P10809 | 61187 | 5.70 | 82 | −a |
Discussion
In this study, EGCG exhibited anticancer effects by
clearly inhibiting the metastatic potential (migration and
invasion) of HCCLM6 cells. MMP-2 and MMP-9 expression and enzyme
activity correlated with the EGCG inhibition of metastasis,
suggesting that EGCG prevents metastasis by inhibiting these
enzymes (18). EGCG has been
shown to inhibit tumor invasion and migration associated with MMPs
in human breast cancer and pancreatic cancer cells (15,19). Metastasis, the spread of cancer in
the body, is a major cause of mortality (19). Collagenase type IV (containing
MMP-2 and MMP-9) is a key enzyme involved in tumor invasion and
migration, as demonstrated by the anti-metastatic effects of
several collagenase inhibitors; some inhibitors have already been
used in clinical trials (20). In
our study, EGCG significantly inhibited HCCLM6 cell metastasis in a
dose-dependent manner at both the mRNA expression and protein
(enzyme) activity levels (Figs. 3
and 4). These results suggest
that EGCG inhibits the metastasis of HCCLM6 cells through the
downregulation of MMPs.
To further understand the effects of EGCG on HCCLM6
cells and to identify potential novel therapeutic targets for HCC,
the protein profiles of EGCG-treated and untreated cells were
analyzed. We identified 4 proteins associated with cell growth and
proliferation: far upstream element binding protein 1 (FUBP1), heat
shock protein (HSP)B1, heat shock 60 kDa protein 1 (chaperonin)
(CH60) and nucleophosmin (NPM) (Table II). FUBP1 is a DNA binding
protein that activates the far upstream element of c-Myc to
stimulate its expression in HCC (21,22). FUBP1 functions as an ATP-dependent
DNA helicase, which is overexpressed in soft tissue metastasis, and
has been shown to be an important element in the progression of
breast cancer metastasis (23).
Therefore, FUBP1 may be an important component in the overall
metastatic process of HCC. Panel 1 of Fig. 4B shows the decreased expression of
FUBP1 protein following treatment with EGCG. EGCG anti-metastatic
activity in HCCLM6 and in HCC, in general, may involve FUBP1
through a molecular mechanism similar to that proposed in other
soft tissues (23).
HSPB1, also known as HSP27, belongs to the
ubiquitous family of small HSPs. In panel 2 of Fig. 4B, the enlarged 2-DE gel image
shows a significant reduction in HSPB1 expression following
treatment with EGCG. In MDA-MB-231 breast cancer cells, increased
HSPB1 expression has been shown to enhance metastasis through the
upregulation of MMP-9 (24,25). HSP27 is a potent therapeutic
target in breast cancer bone metastasis; the anti-metastatic agent,
midazolium trans-imidazole dimethyl sulfoxide tetrachlororuthenate
(NAMI-A), has been shown to decrease HSP27 protein expression
(26). In the present study, we
hypothesized that treatment with EGCG may have decreased HSP27
expression by downregulating MMP-9 and reducing HCCLM6 cancer cell
metastasis (Table II and
Fig. 3).
NPM is a multifunctional protein that shuttles
between the nucleoli and the cytoplasm, functioning as a chaperone
for the nuclear export of ribosomal subunits (27,28). NPM shifts its location from the
nucleolus to the nucleoplasm (NPM translocation) and accumulates if
cells are exposed to actinomycin D, doxorubicin, or other DNA
damaging agents (29,30). In our study, treatment with EGCG
significantly increased NPM expression in HCCLM6 cells (Fig. 4B, panel 3). HeLa cells exposed to
long-term and/or to high doses of actinomycin D (or other
antibiotics with anticancer activity) have shown a significant
accumulation of NPM in the nucleoplasm, which antagonizes both cell
growth and RNA synthesis (29).
EGCG may impair RNA synthesis, RNA processing and cell growth due
to a loss of NPM binding targets in the nucleolus, which may cause
it to accumulate in the nucleoplasm. The resulting loss of
ribosomal assembly integrity may inhibit cellular growth (and/or
metastasis) in HCCLM6 cells. In a recent study, increased NPM
expression levels were suggested to enhance cellular
transformation, antagonize the repression of cell adhesion genes
and inhibit apoptosis mediated by the Myc-Miz1 complex if the
alternate reading frame (Arf) tumor suppressor protein is present
(31). These contradicting
observations of NPM expression in cancer require further
investigation to clearly define the ‘antagonistic’ function of this
protein, either enhancing or reducing carcinogenesis and/or tumor
progression.
CH60 has been associated with tumor metastasis by
regulating tumor immunity (32,33). The overexpression of HSP60 has
been shown to increase the migration and invasive potential of
human pharyngeal squamous carcinoma cells (FADU cells) in
vitro and in vivo (34). The enlargement of the 2-DE gel in
panel 4 of Fig. 4B shows a
significant loss of CH60 expression in the HCCLM6 cells following
treatment with EGCG. CH60 may be a critical factor in the
anti-metastatic activity of EGCG in HCCLM6 cells.
The present study demonstrates that EGCG inhibits
HCCLM6 cell metastasis by inhibiting MMP-2, MMP-9, FUBP1, HSPB1 and
CH60 expression and increasing NPM expression. However, further
studies are required to investigate the specific anti-metastatic
mechanisms of action of EGCG and its effects on FUBP1, HSPB1, NPM
and CH60 expression in cells in vitro and in vivo.
Nevertheless, the data from the present study suggest that EGCG has
potential as an anticancer agent in the treatment of HCC.
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
EGCG
|
epigallocatechin-3-gallate
|
|
2-DE
|
2-dimensional gel electrophoresis
|
|
MTT
|
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
|
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Estimating the world cancer burden: Globocan 2000. Int J Cancer.
94:153–156. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
El-Serag HB and Mason AC: Rising incidence
of hepatocellular carcinoma in the United States. N Engl J Med.
340:745–750. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ikai I, Yamaoka Y, Yamamoto Y, et al:
Surgical intervention for patients with stage IV-A hepatocellular
carcinoma without lymph node metastasis: proposal as a standard
therapy. Ann Surg. 227:433–439. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee JS, Chu IS, Heo J, Calvisi DF, et al:
Classification and prediction of survival in hepatocellular
carcinoma by gene expression profiling. Hepatology. 40:667–676.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Song HY, Liu YK, Feng JT, et al: Proteomic
analysis on metastasis-associated proteins of human hepatocellular
carcinoma tissues. J Cancer Res Clin Oncol. 132:92–98. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shukla Y: Tea and cancer chemoprevention:
a comprehensive review. Asian Pacific J Cancer Prev. 8:155–165.
2007.PubMed/NCBI
|
|
7
|
Yang CS, Wang X, Lu G and Picinich SC:
Cancer prevention by tea: animal studies, molecular mechanisms and
human relevance. Nat Rev Cancer. 9:429–439. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang CS and Wang ZY: Tea and cancer. J
Natl Cancer Inst. 85:1038–1049. 1993. View Article : Google Scholar
|
|
9
|
Balentine DA, Wiseman SA and Bouwens LCM:
The chemistry of tea flavonoids. Crit Rev Food Sci Nutr.
37:693–704. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mukhtar H and Ahmad N: Green tea in
chemoprevention of cancer. Toxicol Sci. 52:111–117. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Singh BN, Shankar S and Srivastava RK:
Green tea catechin, epigallocatechin-3-gallate (EGCG): mechanisms,
perspectives and clinical applications. Biochem Pharmacol.
82:1807–1821. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Thawonsuwan J, Kiron V, Satoh S, et al:
Epigallocatechin-3-gallate (EGCG) affects the antioxidant and
immune defense of the rainbow trout, Oncorhynchus mykiss.
Fish Physiol Biochem. 36:687–697. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mukhtar H and Ahmad N: Tea polyphenols:
prevention of cancer and optimizing health. Am J Clin Nutr.
71(Suppl 6): S1698–S1702. 2000.PubMed/NCBI
|
|
14
|
Thangapazham RL, Singh AK, Sharma A, et
al: Green tea polyphenols and its constituent epigallocatechin
gallate inhibits proliferation of human breast cancer cells in
vitro and in vivo. Cancer Lett. 245:232–241. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shankar S, Ganapathy S, Hingorani SR and
Srivastava RK: EGCG inhibits growth, invasion, angiogenesis and
metastasis of pancreatic cancer. Front Biosci. 13:440–452. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stuart EC, Scandlyn MJ and Rosengren RJ:
Role of epigallocatechin gallate (EGCG) in the treatment of breast
and prostate cancer. Life Sci. 79:2329–2336. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sen T, Moulik S, Dutta A, et al:
Multifunctional effect of epigallocatechin-3-gallate (EGCG) in
downregulation of gelatinase-A (MMP-2) in human breast cancer cell
line MCF-7. Life Sci. 84:194–204. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee SJ, Lee KW, Hur HJ, Chun JY, Kim SY
and Lee HJ: Phenolic phytochemicals derived from red pine (Pinus
densiflora) inhibit the invasion and migration of SK-Hep-1
human hepatocellular carcinoma cells. Ann NY Acad Sci.
1095:536–544. 2007.PubMed/NCBI
|
|
19
|
Woodhouse EC, Chuaqui RF and Liotta LC:
General mechanisms of metastasis. Cancer. 80(Suppl 8): S1529–S1537.
1997. View Article : Google Scholar
|
|
20
|
Hidalgo M and Eckhardt SG: Development of
matrix metalloproteinase inhibitors in cancer therapy. J Natl
Cancer Inst. 93:178–193. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Duncan R, Bazar L, Michelotti G, Tomonaga
T, Krutzsch H, Avigan M and Levens D: A sequence-specific,
single-strand binding protein activates the far upstream element of
c-myc and defines a new DNA-binding motif. Genes Dev. 8:465–480.
1994. View Article : Google Scholar
|
|
22
|
Zubaidah RM, Tan GS, Tan SB, Lim SG, Lin Q
and Chung MC: 2-D DIGE profiling of hepatocellular carcinoma
tissues identified isoforms of far upstream binding protein (FUBP)
as novel candidates in liver carcinogenesis. Proteomics.
8:5086–5096. 2008. View Article : Google Scholar
|
|
23
|
Sanz R, Aragüés R, Stresing V, Martín B,
Landemaine T, Oliva B, et al: Functional pathways shared by liver
and lung metastases: a mitochondrial chaperone machine is
up-regulated in soft-tissue breast cancer metastasis. Clin Exp
Metastasis. 24:673–683. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hansen RK, Parra I, Hilsenbeck SG,
Himelstein B and Fuqua SA: Hsp27-induced MMP-9 expression in
influenced by the Src tyrosine protein kinase yes. Biochem Biophys
Res Commun. 282:186–193. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gibert B, Eckel B, Gonin V, Goldschneider
D, Fombonne J, Deux B, et al: Targeting heat shock protein 27
(HspB1) interferes with bone metastasis and tumour formation in
vivo. Br J Cancer. 107:63–70. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sanna B, Debidda M, Pintus G, et al: The
anti-metastatic agent imidazolium
trans-imidazoledimethylsulfoxide-tetrachlororuthenate induces
endothelial cell apoptosis by inhibiting the mitogen-activated
protein kinase/extracellular signal-regulated kinase signaling
pathway. Arch Biochem Biophys. 403:209–218. 2002. View Article : Google Scholar
|
|
27
|
Borer RA, Lehner CF, Eppenberger HM and
Nigg EA: Major nucleolar proteins shuttle between nucleus and
cytoplasm. Cell. 56:379–390. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Maggi LB Jr, Kuchenruether M, Dadey DY,
Schwope RM, Grisendi S, Townsend RR, et al: Nucleophosmin serves as
a rate-limiting nuclear export chaperone for the Mammalian
ribosome. Mol Cell Biol. 28:7050–7065. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yung BY, Bor AM and Chan PK: Short
exposure to actinomycin D induces ‘reversible’ translocation of
protein B23 as well as ‘reversible’ inhibition of cell growth and
RNA synthesis in HeLa cells. Cancer Res. 50:5987–5991. 1990.
|
|
30
|
Chan PK and Chan FY: A study of
correlation between NPM-translocation and apoptosis in cells
induced by daunomycin. Biochem Pharmacol. 57:1265–1273. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Herkert B, Dwertmann A, Herold S, Abed M,
Naud JF, Finkernagel F, et al: The Arf tumor suppressor protein
inhibits Miz1 to suppress cell adhesion and induce apoptosis. J
Cell Biol. 188:905–918. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Feng Y, Tian ZM, Wan MX and Zheng ZB:
Protein profile of human hepatocarcinoma cell line SMMC-7721:
identification and functional analysis. World J Gastroenterol.
13:2608–2614. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiang D, Ying W, Lu Y, Wan J, Zhai Y, Liu
W, et al: Identification of metastasis-associated proteins by
proteomic analysis and functional exploration of interleukin-18 in
metastasis. Proteomics. 3:724–737. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tsai YP, Yang MH, Huang CH, et al:
Interaction between HSP60 and beta-catenin promotes metastasis.
Carcinogenesis. 30:1049–1057. 2009. View Article : Google Scholar : PubMed/NCBI
|