Introduction
Endometriosis is a common gynecological disease
associated with severe pelvic pain, affecting 6–10% of women in
their reproductive years and 20–50% of infertile women (1).
The development of novel methods for the diagnosis
or treatment of endometriosis is hampered due to the lack of
knowledge regarding the etiology, pathogenesis and natural
progression of endometriosis. Although there have been several
studies published on this disease, its causative mechanisms have
yet to be identified (2).
Retrograde menstruation is still considered the most prominent
cause among the various theories put forward to explain the
pathogenesis of endometriosis. The majority of women have
retrograde menstruation; however, only 10–15% of women suffer from
endometriosis (3), suggesting
that other factors are involved in the development of
endometriosis.
microRNAs (miRNAs) are small non-coding regulatory
RNAs that regulate the translation of mRNAs by inhibiting ribosomal
functions, decapping the 50 Cap structure, deadenylating the
poly(A) tail and degrading the target mRNA (4). miRNAs are able to regulate the
expression of hundreds of target mRNAs simultaneously, thus
controlling a variety of cellular functions, including cell
proliferation, stem cell maintenance and differentiation (5). Aberrant miRNA expression is
associated with human diseases, such as gynecological diseases,
cancer, inflammatory diseases and cardiovascular disorders.
Emerging data indicate a different molecular environment and
altered miRNA expression in the pathological endometrium, in
contrast to the normal endometrium (6–9).
In this study, a systematic comparison of the miRNAs
in the ectopic, eutopic and normal endometrium was conducted,
leading to the identification of miR-183, which has a regulatory
impact on the development of endometriosis.
Materials and methods
Tissue acquisition
All the endometriotic, eutopic and normal
endometrial tissues were obtained at Nanjing Maternity and Child
Health Care Hospital Affiliated to Nanjing Medical University,
Nanjing, China by laparoscopy and uterine curettage from patients
with or without endometriosis. None of the patients had received
pre-operative hormonal therapy, and all the samples were
histologically confirmed. All the samples were from the
proliferative phase of the menstrual cycle from pre-menopausal
women. The phase of the menstrual cycle was determined by
pre-operative history and a histological evaluation of the
endometrium. The normal samples were obtained from patients with an
average age of 38.9±4.8 years and the endometriosis samples were
otbained from patients with an average age of 36.6±5.1 years. All
patients provided written informed consent prior to participating
in this study. This study was approved by the hospital ethics
committee. Each sample was divided and used for mRNA extraction, as
well as cell isolation.
miRNA microarray
Microarrays were performed by utilizing the miRCURY
LNA™ microRNA Array (v. 14.0; Exiqon, Vedbaek, Denmark). All
procedures were carried out according to the manufacturer’s
instructions. Following RNA measurement on the NanoDrop instrument,
the samples were labeled using the miRCURY™ Hy5™/Hy3™ Power
labeling kit (Exiqon) and hybridized on the miRCURY LNA™ Array (v.
14.0), which contains >1,700 capture probes covering all miRNAs
listed in miRBase v. 14.0. Following the labeling procedure, the
Hy3™-labeled samples and a Hy5™-labeled reference RNA sample were
mixed pairwise and hybridized using the miRCURY LNA™ Array v. 14.0
(Exiqon). The hybridization and subsequent wash steps were
performed according to the miRCURY LNA™ Array manual. The
microarray slides were scanned using the Axon GenePix 4000B
microarray scanner (MDS Analytical Technologies, Silicon Valley,
CA, USA) and image analysis was carried out using GenePix pro v.
6.0 software (MDS Analytical Technologies). The normalized data
were analyzed using the locally weighted scatter plot smoothing
(lowess) regression algorithm [TIGR Microarray Data Analysis System
(MIDAS)]. Following normalization, the differentially expressed
miRNAs were identified through fold change filtering (fold change
>2.0). Hierarchical clustering was performed using MEV software
(v. 4.6, TIGR).
Cell culture and treatment
The endometrial stromal cells (ESCs) from women with
or without endometriosis were cultured according to a previously
described method (10). Following
serum starvation for 12 h, the ESCs (1×105 cells/well)
were treated with 17β-estradiol (E2) (10−8 mol/l),
progesterone (P) (10−8 mol/l), E2 (10−8
mol/l) + P (10−8 mol/l), interleukin-6 (IL-6) (10 ng/ml)
or tumor necrosis factor-α (TNF-α) (10 ng/ml) for 24 h; vehicle
controls were also used (treated with ethanol, 17β-estradiol
solution).
Fluorescence-based real-time PCR
(qPCR)
Total RNA was isolated using TRIzol reagent (Takara,
Otsu, Shiga, Japan) and cDNA was synthesized using the
SYBR® PrimeScript™ RT-PCR kit (Takara) on the ABI PRISM
500 Sequence Detection System (Applied Biosystems, Foster City, CA,
USA) according to the manufacturer’s instructions. The housekeeping
gene encoding Hsa-U6 small nuclear RNA (snRNA) was used for
normalization. The primers used were as follows: 5′-CGCG
CGTGAATTACCGAAG-3′ (forward) and 5′-GTGCAGGG TCCGAGGT-3′ (reverse)
for miR-183-3p; 5′-CGCGCTAT GGCACTGGTAG-3′ (forward) and
5′-GTGCAGGGTCC GAGGT-3′ (reverse) for miR-183-5p; and 5′-GCGCGTCGTG
AAGCGTTC-3′ (forward) and 5′-GTGCAGGGTCCGAGGT-3′ (reverse) for
Hsa-U6 snRNA. The conditions for qPCR were as follows: 95°C for 5
min; 45 cycles of 95°C for 15 sec followed by 60°C for 30 sec and
72°C for 30 sec; 95°C for 55 sec, 50°C for 2 min, 95°C for 10 min,
40 cycles of 95°C for 15 sec and 60°C for 1 min. qPCR was carried
out using SYBR-Green JumpStart Taq ReadyMix (Sigma-Aldrich, St.
Louis, MO, USA) and the 7300 Real-Time PCR Detection System (ABI).
The results were analyzed using the comparative threshold cycle
(Ct) method.
miR-183 lentivirus construction and
transduction
The precursor of the miRNA, hsa-miR-183 (GenBank
accession no. MIMAT0000261), and the inhibitor of hsa-miR-183-5p
lentivirus gene transfer vector encoding green fluorescent protein
(GFP) were constructed by Genechem Co., Ltd. (Shanghai, China).
The RNA primers used were: 5′-GAGGATCCCCGGG
TACCAAGGGAGTGGGCAGGCTA-3′ and 5′-ATAAGCTTG
ATATCGTCCCTGCACCCTTGGAAGCA-3′, and were confirmed by sequencing.
The recombinant lentivirus of overexpressed miR-183
(miR-183-lentivirus) and the control lentivirus (GFP-lentivirus)
were prepared and titered to 5.0 E+8 TU/ml (transfection unit).
The sequence of the inhibitor of hsa-miR-183-5p was
TATGGCACTGGTAGAATTCACT, and confirmed by sequencing. The
recombinant lentivirus of miR-183-5p inhibitor
(In-miR-183-lentivirus) and the control lentivirus (GFP-lentivirus)
were prepared and titered to 4.0 E+8 TU/ml (transfection unit).
ESCs from women without endometriosis were plated in
6-well plates (5×104 cells/well) overnight. The
lentiviruses were diluted in 0.2 ml complete medium containing
polybrene (8 mg/ml) and added to the cells for 12 h of incubation
at 37°C, followed by incubation in 0.3 ml of freshly prepared
polybrene-DMEM for another 24 h, which was replaced with fresh DMEM
and the cells were cultured for 3 days. The lentivirus transduction
efficiency of the ESCs was determined by the detection of GFP
signals by fluorescence microscopy at 72 h after transduction. The
miR-183 expression in the stably transduced ESCs was measured by
qPCR. The ESCs transfected with miR-183-lentivirus,
In-miR-183-lentivirus and GFP-lentivirus were kept for further
functional analysis.
Measurement of apoptotic cell death by
flow cytometry
Apoptosis assay was performed according to the
operation manual provided with the BD Annexin V Staining kit (BD
Biosciences, Franklin Lakes, NJ, USA). Briefly, the ESCs were
infected with lentivirus for 5 days under serum-free conditions,
the cells were trypsinized and collected by centrifugation at 1,500
rpm for 5 min, washed once with phosphate-buffered saline (PBS) at
4°C and then resuspend in 1X binding buffer at a concentration of
1×106 cells/ml. Subsequently, 100 μl of the solution
(1×105 cells) were transferred to a 5-ml culture tube.
Annexin V and PI (5 μl/test tube) were then added and the cells
were gently mixed and incubated for 15 min at room temperature in
the dark. This was followed by the addition of 400 μl of 1X binding
buffer to each tube. The cells were then analyzed by flow cytometry
as soon as possible (within 1 h).
Measurement of cell viability by MTT
The ESCs (2.0×103 cells/well) were seeded
into 96-well plates (Corning Costar, Corning, NY, USA) in DMEM
supplemented with 10% fetal bovine serum (FBS) and incubated for
1–5 days. Following incubation, 10 μl of MTT (Sigma-Aldrich)
solution (5 mg/ml in ddH2O) were added to each well. The
plates were incubated for a further 4 h at 37°C. Intracellular
formazan crystals were dissolved by the addition of 100 μl of DMSO
to each well. Cell proliferation was evaluated by measuring the
absorbance at 490 nm.
Invasion (Matrigel) chamber assay
The ESCs (2.5×104) were seeded on a cell
culture Transwell insert coated with extracellular matrix (ECM)
(8-mm pore size, 24-well format; Corning Costar) in 2% FBS medium,
and complete medium (10% FBS) was added to the lower chamber. To
determine the amount of invading cells, the cells were incubated
for 24 h and then removed from the upper chamber using a cotton
swab. The invaded cells on the underside of the insert were fixed
with methanol (2 min). Once fixed, the cells were stained with
crystal violet for 2 min and rinsed with PBS. The undersides of the
membrane were then photographed to compare the number of invaded
cells per insert. The transmigrated cells were counted under a
light microscope. The invaded cells were scored by counting 10
random high-power fields per filter. The counting accuracy was
guaranteed by optical density (OD)570 quantification of the
methanol-solubilized dye.
Statistical analysis
Data represent the means ± SEM of at least 3
independent experiments. The difference between 2 means was
examined by the Student’s t-test, while one-way ANOVA was employed
to compare 3 or more groups. A value of P<0.05 was considered to
indicate a statistically significant difference.
Results
miRNA expression profile
Following the selection of miRNAs by fold change
filtering (fold change >2.0), we found that there were 26
upregulated miRNAs and 19 downregulated miRNAs in the ectopic
endometrium compared with the eutopic endometrium. Compared with
the normal endometrium, 36 downregulated with no upregulated miRNAs
were found in the eutopic endometrium (Table I). Among these differentially
expressed miRNAs, miR-183, miR-215 and miR-363 were found
downregulated both in the ectopic and eutopic tissues, from which
we selected miR-183 as the target miRNA as it was the most
significantly downregulated (Fig.
1).
 | Table IDifferentially expressed miRNAs in
ectopic vs. eutopic endometrium and eutopic vs. normal
endometrium. |
Table I
Differentially expressed miRNAs in
ectopic vs. eutopic endometrium and eutopic vs. normal
endometrium.
| A,
Ectopic/eutopic |
|---|
|
|---|
| miRNAs | Fold-change | P-value |
|---|
| Downregulated |
| hsa-miR-203 | 0.39 | 0.03 |
| hsa-miR-425 | 0.25 | 0.03 |
| hsa-miR-183 | 0.48 | 0.01 |
| hsa-miR-92a | 0.36 | 0.03 |
| hsa-miR-196b | 0.04 | 0.03 |
|
hsa-miR-363* | 0.23 | 0.00 |
| hsa-let-7i | 0.32 | 0.01 |
|
hsa-miRPlus-E1031 | 0.45 | 0.01 |
|
hsa-miRPlus-E1031 | 0.45 | 0.01 |
| hsa-miR-200b | 0.15 | 0.05 |
|
hsa-miRPlus-F1231 | 0.30 | 0.00 |
| hsa-miR-215 | 0.24 | 0.01 |
| hsa-miR-362-3p | 0.31 | 0.04 |
| hsa-miR-342-3p | 0.27 | 0.05 |
| hsa-miR-200c | 0.17 | 0.04 |
| hsa-miR-93 | 0.18 | 0.01 |
|
hsa-miR-24-1* | 0.45 | 0.02 |
| hsa-miR-25 | 0.31 | 0.03 |
|
hsa-miR-106b* | 0.31 | 0.00 |
| Upregulated |
|
hsa-miRPlus-F1038 | 3.21 | 0.02 |
| hsa-miR-1915 | 2.06 | 0.04 |
| hsa-miR-637 | 2.35 | 0.03 |
|
hsa-miR-518e* | 2.22 | 0.00 |
|
hsa-miR-519a* | | |
|
hsa-miR-519b-5p | | |
|
hsa-miR-519c-5p | | |
|
hsa-miR-522* | | |
|
hsa-miR-523* | | |
|
hsa-miR-574-5p | 2.00 | 0.04 |
|
hsa-miR-615-3p | 2.14 | 0.02 |
| hsa-miR-1909 | 3.17 | 0.04 |
|
hsa-miR-224* | 2.23 | 0.02 |
| hsa-miR-133b | 2.85 | 0.03 |
| hsa-miR-622 | 2.06 | 0.01 |
|
hsa-miR-628-3p | 2.27 | 0.01 |
|
ebv-miR-BHRF1-2 | 2.78 | 0.03 |
|
hsa-miRPlus-F1215 | 2.28 | 0.01 |
|
hsa-miRPlus-F1221 | 2.32 | 0.01 |
| hsa-miR-1470 | 2.45 | 0.02 |
| hsa-miR-1469 | 2.21 | 0.05 |
|
hsa-miR-520d-5p | 2.34 | 0.00 |
|
hsa-miR-551b* | 2.18 | 0.00 |
|
hsa-miR-361-3p | 2.83 | 0.01 |
| hsa-miR-941 | 3.43 | 0.03 |
|
hsa-miRPlus-F1223 | 2.50 | 0.01 |
| hsa-miR-202 | 12.53 | 0.01 |
| hsa-miR-663b | 2.53 | 0.03 |
|
hsa-miRPlus-F1042 | 2.33 | 0.02 |
| hsa-miR-381 | 2.97 | 0.01 |
| hsa-miR-412 | 3.11 | 0.03 |
|
| B,
Eutopic/normal |
|
| Downregulated | | |
| hsa-miR-921 | 0.49 | 0.01 |
|
hsa-miR-374b* | 0.26 | 0.00 |
|
hsa-miRPlus-E1082 | 0.33 | 0.03 |
|
ebv-miR-BHRF1-3 | 0.28 | 0.02 |
|
hsa-miR-22* | 0.41 | 0.04 |
|
hsa-miRPlus-E1027 | 0.22 | 0.02 |
|
hsa-miR-218-2* | 0.35 | 0.02 |
|
hsa-miR-148b* | 0.17 | 0.00 |
| hsa-miR-183 | 0.38 | 0.00 |
|
hsa-miRPlus-C1100 | 0.27 | 0.02 |
|
hsv1-miR-H5-3p | 0.40 | 0.03 |
|
hsv1-miR-H4* | 0.18 | 0.01 |
| hsa-miR-553 | 0.35 | 0.05 |
| hsa-miR-761 | 0.41 | 0.04 |
| hsa-miR-302e | 0.46 | 0.04 |
|
hsa-miR-9* | 0.38 | 0.01 |
|
hsa-miRPlus-F1212 | 0.26 | 0.03 |
|
hsa-miR-363* | 0.20 | 0.01 |
| hsa-miR-206 | 0.23 | 0.01 |
|
hsa-miRPlus-E1192 | 0.34 | 0.02 |
|
hsa-miRPlus-F1091 | 0.44 | 0.04 |
|
hsa-miRPlus-E1215 | 0.31 | 0.04 |
| hsa-miR-668 | 0.22 | 0.00 |
| hsa-miR-215 | 0.41 | 0.01 |
| hsa-miR-1265 | 0.41 | 0.01 |
| hsa-miR-922 | 0.24 | 0.01 |
|
hsa-miRPlus-E1185 | 0.44 | 0.04 |
|
hsa-miRPlus-E1075 | 0.38 | 0.02 |
|
hsa-miRPlus-E1086 | 0.39 | 0.02 |
| hsa-miR-636 | 0.38 | 0.02 |
|
ebv-miR-BART18-3p | 0.24 | 0.04 |
|
hsa-miRPlus-F1074 | 0.33 | 0.00 |
|
hsa-miRPlus-E1038 | 0.39 | 0.00 |
|
hsa-miRPlus-E1049 | 0.30 | 0.02 |
|
hsa-miRPlus-E1193 | 0.35 | 0.02 |
|
hsa-miRPlus-E1029 | 0.38 | 0.01 |
Quantification of miR-183 expression
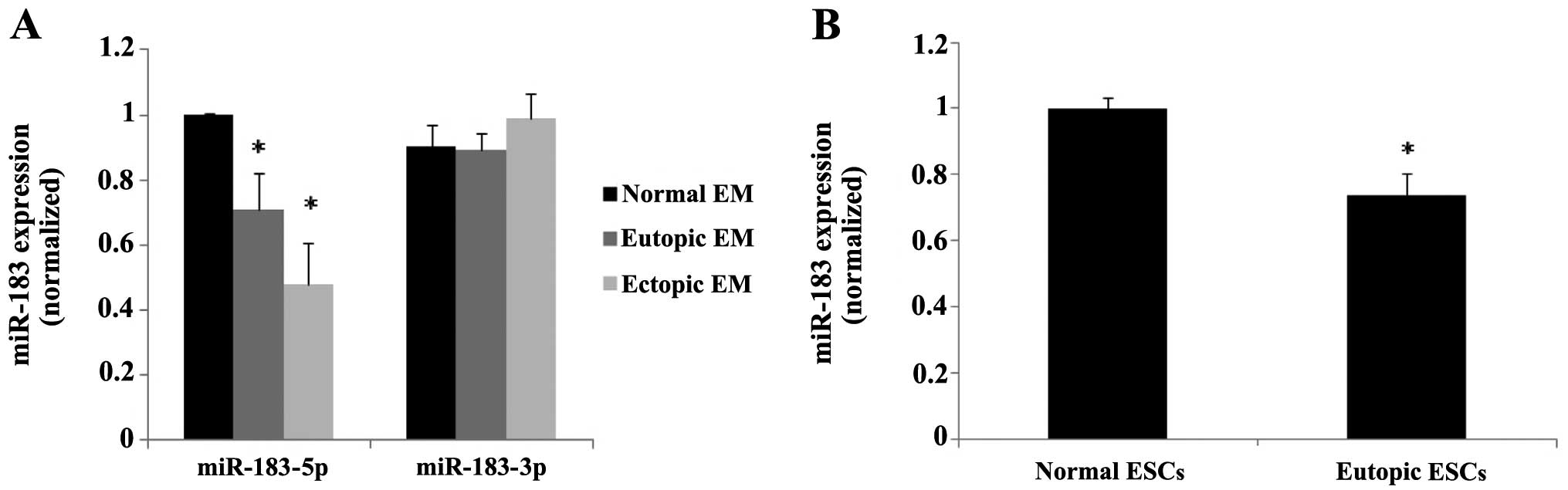
The statistical data indicated that miR-183-5p was
significantly downregulated in the ectopic and eutopic endometrial
tissues from patients with endometriosis compared with those with a
normal endometrium (P=0.011 and P=0.0002, respectively), whereas
miR-183-3p did not show statistically significant differences in
expression (Fig. 2A). Since the
adhesion of endometrial cells to the peritoneal lining is a crucial
step in the early stages of endometriosis, which is dependent on
stromal cells, we further detected the expression of miR-183 in the
ESCs isolated from the ectopic, eutopic and normal endometrium. The
results of cell analysis conformed with the tissue samples
(P<0.05) (Fig. 2B).
Transfection efficiency of recombinant
lentivirus
To further investigate the roles of miR-183 in ESCs,
we prepared a lentiviral construct for the overexpression of
miR-183 and anti-miR-183. Fig. 3A
shows that the majority of the ESCs (>80%) expressed GFP and
exhibited a morphology that was similar to that of naive ESCs. qPCR
revealed that the transduction of ESCs with miR-183-lentivirus
increased miR-183 expression by 127-fold, whereas transfection with
In-miR-183-lentivirus increased it by only 0.15-fold (Fig. 3B).
miR-183 decreases the invasive ability of
ESCs
To illustrate the function of miR-183 in the
development of endometriosis, we first examined the effects of
miR-183 overexpression and knockdown in ESCs using an invasion
chamber coated with ECM-Matrigel and found that a significantly
greater number of control ESCs (1.652±0.083) compared with
miR-183-overexpressing ESCs (1.335±0.035) had passed through the
matrix (P<0.05). By contrast, the ESCs in which miR-183
expression was downregulated (1.874±0.099) had greater invasive
potential compared with the control cells (P<0.05) (Fig. 4).
miR-183 induces the apoptosis of ESCs but
has no effect on cell proliferation
The results of Annexin V assay demonstrated that the
overexpression of miR-183 significantly induced the apoptosis of
ESCs (P<0.05), and that the apoptotic rate was decreased when
miR-183 was inhibited (P<0.05) (Fig. 5A). miR-183 did not significantly
alter the cell proliferation rate (Fig. 5B).
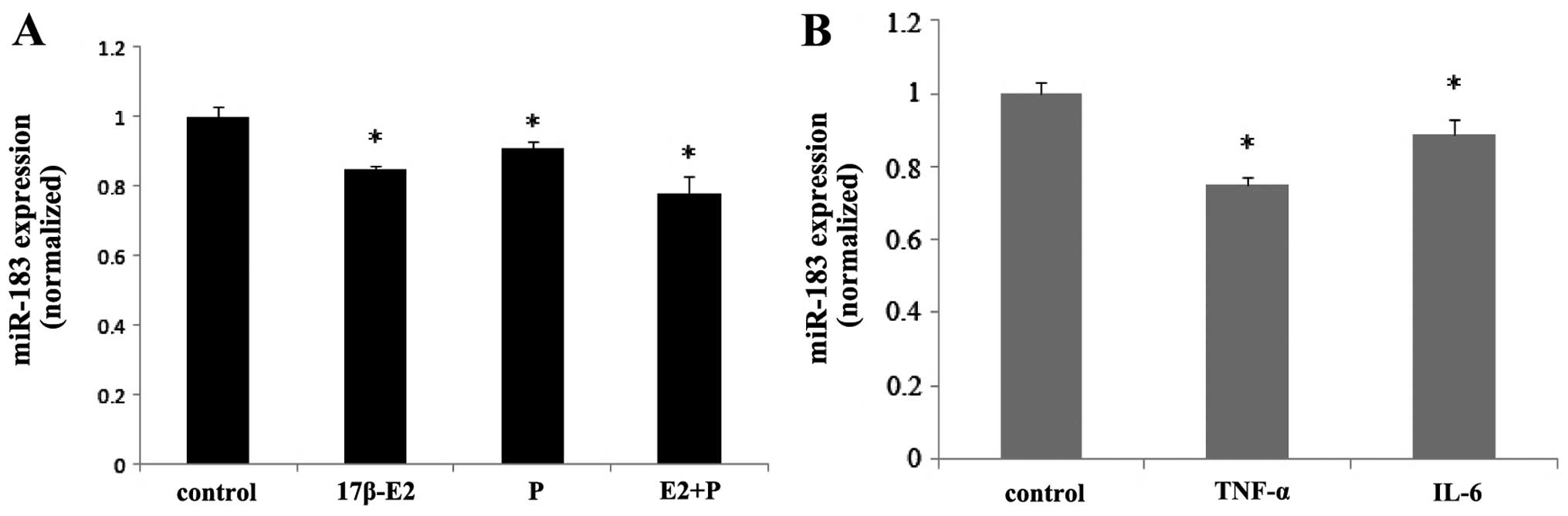
Ovarian steroids and inflammatory factors
inhibit the expression of miR-183
qPCR demonstrated that the treatment of ESCs with
ovarian steroids and inflammatory factors regulated the expression
of miR-183. Both 17β-estradiol and progesterone inhibited the
expression of miR-183 in ESCs (P<0.05), and co-treatment with
these steroids induced synergistic effects (P<0.05) (Fig. 6A). Treatment of the ESCs with
TNF-α and IL-6 also inhibited the expression of miR-183 (P<0.05)
(Fig. 6B).
Discussion
Endometriosis is a prevalent gynecological disease
characterized by the growth of endometriotic tissue outside the
uterine cavity. miRNAs are naturally occurring post-transcriptional
regulatory molecules that potentially play a role in endometriotic
lesion development (11). In
recent years, emerging evidence suggests that the dysregulation of
miRNA expression is involved in endometriosis. Previously, Ohlsson
Teague et al(12) screened
miRNA expression by microarray analysis in paired ectopic and
eutopic endometrial tissues and identified 14 upregulated (miR-145,
miR-143, miR-99a, miR-99b, miR-126, miR-100, miR-125b, miR-150,
miR-125a, miR-223, miR-194, miR-365, miR-29c and miR-1) and 8
downregulated (miR-200a, miR-141, miR-200b, miR-142-3p, miR-424,
miR-34c, miR-20a and miR-196b) miRNAs. More recently, Hawkins et
al(13) also found 10
upregulated (miR-202, 193a-3p, 29c, 708, 509-3-5p, 574-3p, 193a-5p,
485-3p, 100 and 720) and 12 downregulated (miR-504, 141, 429, 203,
10a, 200b, 873, 200c, 200a, 449b, 375 and 34c-5p) miRNAs in
endometriomas compared with the normal endometrium using
next-generation sequencing technology. Many of the identified
miRNAs, such as miR-199a, miR-126 and miR-10b were subsequently
investigated in recent studies (14–16), further indicating that miRNAs play
a role in the development of endometriosis.
Published studies have identified differentially
expressed miRNAs in endometriotic tissues. However, to the best of
our knowledge, the differences in miRNA expression in endometrial
tissue from women with or without endometriosis have not yet been
validated. It is well known that eutopic endometrial cells may
function differently in women with endometriosis compared with a
normal endometrium in disease-free women. These cells have more
chances of survival outside the uterine cavity, which lead to the
development of well-documented changes at the peritoneum and other
ectopic sites (17). A heritable
or acquired molecular aberration within the endometrium may have an
impact on selective survival advantage to refluxed endometrial
tissue in women predisposed to the development of endometriosis.
The identification of molecular differences in the eutopic
endometrium of women with endometriosis is an important step toward
understanding the pathogenesis of this condition and toward
developing effective strategies for the treatment of associated
infertility and pain (17). In
this study, we detected the miRNA expression profiles in
endometrial tissue from endometriosis-free women, as well as in
tissue from eutopic endometrium from women with surgically
confirmed endometriosis and the tissue from ectopic endometrium.
The results revealed 26 upregulated miRNAs and 19 downregulated
miRNAs in the ectopic endometrium compared with the eutopic
endometrium. Compared with the normal endometrium, 36 downregulated
with no upregulated miRNAs were found in the eutopic endometrium.
Among these differentially expressed miRNAs, miR-183, miR-215 and
miR-363 were found to be downregulated in both the ectopic and
eutopic tissues, from which we selected miR-183 as the target miRNA
as it was the most significantly downregulated. The differential
expression of miR-183 was further confirmed by qPCR. However,
little is known about the role of miR-183 in the development of
endometriosis.
The adhesion of endometrial cells to the peritoneal
lining is a crucial step during the early stages of endometriosis,
which is dependent on stromal cells, as no cell adhesion in the
endometrial epithelium is identified during the first 24–48 h
(18). In our study, we therefore
selected ESCs as the target cells. The results of qPCR further
confirmed that miR-183 expression in the eutopic endometrium was
higher than that in the control group.
miR-183 is an miRNA involved in the regulation of
cell growth, cell differentiation, apoptosis, cell motility, cell
adhesion and cell invasion (19–21). It has been reported that miR-183
is upregulated in colorectal cancer, prostate cancer and
hepatocellular carcinomas, while it is downregulated in ovarian
cancer, breast cancer stem cells and osteosarcomas (5,20,22–25), suggesting that its roles vary
depending on the cellular context.
In this study, we used ESCs as an in vitro
model of endometriosis to examine the functional impact of miR-183
overexpression and inhibition on endometriotic cell behavior.
Functional analysis indicated that miR-183 plays a promotional role
in ESC apoptosis and exhibits a negative regulatory impact on the
invasive ability of the cells; however, it has no effect on ESC
proliferation. The cause of endometriosis has been attributed to
the attachment and invasion of the retrograded endometrial
fragments into the peritoneum, where they establish a blood supply,
triggering a suboptimal immune response that does not adequately
clear the implants, which induces their continued survival and
growth. This effect of miR-183 can be expected in vivo to
enhance the implantation and establishment of the ectopic
lesion.
Hormonal alterations may influence the ability of
endometrial cells to proliferate, attach to the mesothelium and
evade immune-mediated clearance. In addition to the concept of
endometriosis as an estrogen-dependent disorder, there is
increasing evidence to suggest that an incomplete transition of the
endometrium from the proliferative to the secretory phase has
significant molecular implications toward enhancing the survival
and implantation of the refluxed endometrium. On the one hand,
miRNAs have emerged as a major regulatory system of steroid hormone
responses in the female reproductive tract (26); on the other hand, several miRNAs
are possibly influenced by the hormonal milieu (27,28). The inflammatory environment within
the pelvis may also contribute to the pathophysiology of
endometriosis. In our study, compared with the disease-free
controls, the eutopic endometrium from women with endometriosis
showed an increased basal production of IL-6 and TNF-α, which both
play a prominent role in a number of chronic inflammatory
conditions (29). To determine
wether ovarian steroids and inflammatory factors exert regulatory
effects on miR-183, we detected miR-183 expression in ESCs treated
with 17β-estradiol, progesterone, TNF-α and IL-6 by qPCR. Our
results provide evidence that ovarian steroids and inflammatory
factors decrease the expression of miR-183 in ESCs; however,
further studies are required to examine the molecular mechanisms
involved.
In conclusion, we identified several differentially
expressed miRNAs, including miR-183 in the normal, eutopic and
ectopic endometrium by miRNA microarray screening analysis. The
downregulation of the expression of miR-183 enhanced the invasive
potential and inhibited the apoptosis of ESCs. The targets of this
miRNA related to cellular functions are still being investigated.
miR-183 gene expression can be inhibited by the stimulation of
ovarian steroids and inflammatory factors; such a regulatory
function may further manifest the growth and invasive capacity of
ESCs by altering the expression of miR-183. These findings suggest
that the aberrant miR-183 expression is involved in the development
and progression of endometriosis as part of epigenetic
mechanisms.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (no. 81100405), the Key Program
of Nanjing Medical Science and Technology Development Foundation,
Nanjing Department of Health (no. ZKX10019) (no. QRX11109) and
Nanjing Medical University (no. 2002NJMU212).
References
|
1
|
Nyholt DR, Low SK, Anderson CA, Painter
JN, Uno S, Morris AP, MacGregor S, Gordon SD, Henders AK, Martin
NG, Attia J, Holliday EG, McEvoy M, Scott RJ, Kennedy SH, Treloar
SA, Missmer SA, Adachi S, Tanaka K, Nakamura Y, Zondervan KT,
Zembutsu H and Montgomery GW: Genome-wide association meta-analysis
identifies new endometriosis risk loci. Nat Genet. 44:1355–1359.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bulun SE: Endometriosis. N Engl J Med.
360:268–279. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cramer DW and Missmer SA: The epidemiology
of endometriosis. Ann NY Acad Sci. 955:11–22. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shimono Y, Zabala M, Cho RW, Lobo N,
Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, Dirbas FM,
Somlo G, Pera RA, Lao K and Clarke MF: Downregulation of miRNA-200c
links breast cancer stem cells with normal stem cells. Cell.
138:592–603. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gilabert-Estelles J, Braza-Boils A, Ramon
LA, Zorio E, Medina P, Espana F and Estelles A: Role of microRNAs
in gynecological pathology. Curr Med Chem. 19:2406–2413. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancer. Adv Exp Med Biol. 774:1–20. 2013.
View Article : Google Scholar
|
|
8
|
Goettsch C, Hutcheson JD and Aikawa E:
MicroRNA in cardiovascular calcification: focus on targets and
extracellular vesicle delivery mechanisms. Circ Res. 112:1073–1084.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wei Y, Nazari-Jahantigh M, Chan L, Zhu M,
Heyll K, Corbalán-Campos J, Hartmann P, Thiemann A, Weber C and
Schober A: The microRNA-342-5p fosters inflammatory macrophage
activation through an Akt1- and microRNA-155-dependent pathway
during atherosclerosis. Circulation. 127:1609–1619. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shi YL, Luo XZ, Zhu XY, Hua KQ, Zhu Y and
Li DJ: Effects of combined 17beta-estradiol with TCDD on secretion
of chemokine IL-8 and expression of its receptor CXCR1 in
endometriotic focus-associated cells in co-culture. Hum Reprod.
21:870–879. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Teague EM, Print CG and Hull ML: The role
of microRNAs in endometriosis and associated reproductive
conditions. Hum Reprod Update. 16:142–165. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ohlsson Teague EM, Van der Hoek KH, Van
der Hoek MB, Perry N, Wagaarachchi P, Robertson SA, Print CG and
Hull LM: MicroRNA-regulated pathways associated with endometriosis.
Mol Endocrinol. 23:265–275. 2009.PubMed/NCBI
|
|
13
|
Hawkins SM, Creighton CJ, Han DY, Zariff
A, Anderson ML, Gunaratne PH and Matzuk MM: Functional microRNA
involved in endometriosis. Mol Endocrinol. 25:821–832. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dai L, Gu L and Di W: MiR-199a attenuates
endometrial stromal cell invasiveness through suppression of the
IKKβ/NF-κB pathway and reduced interleukin-8 expression. Mol Hum
Reprod. 18:136–145. 2012.PubMed/NCBI
|
|
15
|
Liu S, Gao S, Wang XY and Wang DB:
Expression of miR-126 and Crk in endometriosis: miR-126 may affect
the progression of endometriosis by regulating Crk expression. Arch
Gynecol Obstet. 285:1065–1072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schneider C, Kässens N, Greve B, Hassan H,
Schüring AN, Starzinski-Powitz A, Kiesel L, Seidler DG and Götte M:
Targeting of syndecan-1 by micro-ribonucleic acid miR-10b modulates
invasiveness of endometriotic cells via dysregulation of the
proteolytic milieu and interleukin-6 secretion. Fertil Steril.
99:871–881. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu H and Lang JH: Is abnormal eutopic
endometrium the cause of endometriosis? The role of eutopic
endometrium in pathogenesis of endometriosis. Med Sci Monit.
17:RA92–RA99. 2011.PubMed/NCBI
|
|
18
|
Witz CA: Current concepts in the
pathogenesis of endometriosis. Clin Obstet Gynnecol. 42:566–585.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang G, Mao W and Zheng S: MicroRNA-183
regulates Ezrin expression in lung cancer cells. FEBS Lett.
582:3663–3668. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu J, Feng Y, Ke Z, Yang Z, Zhou J, Huang
X and Wang L: Down-regulation of miR-183 promotes migration and
invasion of osteosarcoma by targeting Ezrin. Am J Pathol.
180:2440–2451. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lodrini M, Oehme I, Schroeder C, Milde T,
Schier MC, Kopp-Schneider A, Schulte JH, Fischer M, De Preter K,
Pattyn F, Castoldi M, Muckenthaler MU, Kulozik AE, Westermann F,
Witt O and Deubzer HE: MYCN and HDAC2 cooperate to repress miR-183
signaling in neuroblastoma. Nucleic Acids Res. 41:6018–6033. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bandrés E, Cubedo E, Agirre X, Malumbres
R, Zárate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M and
García-Foncillas J: Identification by Real-time PCR of 13 mature
microRNAs differentially expressed in colorectal cancer and
non-tumoral tissues. Mol Cancer. 5:292006.PubMed/NCBI
|
|
23
|
Schaefer A, Jung A, Mollenkopf HJ, Wagner
I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G and Jung
K: Diagnostic and prognostic implications of microRNA profiling in
prostate carcinoma. Int J Cancer. 126:1166–1176. 2010.PubMed/NCBI
|
|
24
|
Li J, Fu H, Xu C, Tie Y, Xing R, Zhu J,
Qin Y, Sun Z and Zheng X: miR-183 inhibits TGF-beta1-induced
apoptosis by downregulation of PDCD4 expression in human
hepatocellular carcinoma cells. BMC Cancer. 10:3542010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dahiya N, Sherman-Baust CA, Wang TL,
Davidson B, Shih IeM, Zhang Y, Wood W III, Becker KG and Morin PJ:
MicroRNA expression and identification of putative miRNA targets in
ovarian cancer. PLoS One. 3:e24362008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Panda H, Pelakh L, Chuang TD, Luo X,
Bukulmez O and Chegini N: Endometrial miR-200c is altered during
transformation into cancerous states and targets the expression of
ZEBs, VEGFA, FLT1, IKKβ, KLF9, and FBLN5. Reprod Sci. 19:786–796.
2012.PubMed/NCBI
|
|
27
|
Lam EW, Shah K and Brosens JJ: The
diversity of sex steroid action: the role of micro-RNAs and FOXO
transcription factors in cycling endometrium and cancer. J
Endocrinol. 212:13–25. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nothnick WB and Healy C: Estrogen induces
distinct patterns of microRNA expression within the mouse uterus.
Reprod Sci. 17:987–994. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Burney RO and Giudice LC: Pathogenesis and
pathophysiology of endometriosis. Fertil Steril. 98:511–519. 2012.
View Article : Google Scholar : PubMed/NCBI
|