Introduction
Intervertebral disc degeneration (IDD) is the main
cause of lower back pain, and is a medical condition that places a
heavy burden on the global healthcare system with severe
socioeconomic consequences (1–3).
Due to the nature of the risk factors for IDD, including family
history, lumbar load and workload (4), the incidence of IDD is higher in
developing countries, particularly in China. To date, it is agreed
that IDD is similar to other complex diseases since its etiology
has hereditary and environmental influences, both of which generate
a small overall contribution to the relative risk. The main
pathological changes that occur in IDD involve the excessive
apoptosis of intervertebral disc cells and the components of the
extracellular matrix (ECM). Although a number of studies have
focused on the etiology of IDD, such as genetics (5), mechanical load (6), and environmental factors (7), the underlying pathology is not yet
fully understood.
microRNAs (miRNAs) are a type of small non-coding
RNA molecules of 20–22 nucleotides in length and were first
reported in C. elegans(8,9).
miRNAs function by the partial or completely binding to the
3′-untranslated region (UTR) of their target mRNAs, and thereby
trigger either translation inhibition or mRNA degradation (10,11). A single miRNA is capable of
regulating the expression of several target genes, whereas a single
target gene can also be modulated by several miRNAs (12). On a molecular level, miRNAs have
been shown to act as key regulators in a wide variety of biological
processes, such as cell growth, differentiation, resistance to
chemotherapeutic drugs and organ development (13,14). Furthermore, miRNAs play a central
role in cancer, as well as in inflammatory, neurodegenerative,
pathophysiological and certain reproductive disorders (15,16). In a previous study, it was
demonstrated that the aberrant expression of miRNA-140 was
associated with the degenerative disease, osteoarthritis (OA),
which is characterized by pathological changes similar to those
which occur in IDD (17).
miRNA-140 has also been shown to regulate cartilage development and
homeostasis by targeting insulin-like growth factor binding
protein-5 (IGFBP-5), Smad family member 3 (Smad3) and Adam
metallopeptidase with thrombospondin type 1 motif, 5 (Adamts-5)
(18–20). Several other miRNAs, such as
miR-34a (21), miR-21 (22) and miR-675 (23) have also been found to be
associated with chondrocyte apoptosis, proliferation or cartilage
matrix production. Moreover, miR-155, a well-documented miRNA
involved in various diseases (24), has been reported to promote
Fas-mediated apoptosis by targeting Fas-associated protein with
death domain (FADD) and caspase-3 in human IDD (25). These findings demonstrate the
effectiveness of miRNAs as regulators for determining the
pathogenesis of degenerative disorders, such as IDD.
The aim of the present study was to isolate miRNAs
from patients with IDD and spinal cord injury and subsequently
identify the differential miRNA expression profiles between them.
Bioinformatics analysis was then performed to investigate the
dysregulated miRNA target genes and the signaling pathways
involved, which may enhance our understanding of the molecular
mechanisms leading to IDD.
Materials and methods
Sample collection
The study was approved by the Human Ethics
Committees Review Board at Xi’an Jiaotong University, Xi’an, China
and written informed consent was obtained from each patient prior
to enrollment.
Patients presenting with lumbar intervertebral disc
herniation (LIDH), a medical condition that is representative of
IDD, were selected as the experimental subjects [experimental group
(EG)]. The EG consisted of 20 individuals with severe clinical
symptoms. Disc degeneration was confirmed by both magnetic
resonance imaging (MRI) with an apparent decrease in the
T2-weighted signal, as well as hematoxylin and eosin (H&E)
staining with obvious morphological changes. Disc specimens were
classified as grade IV (IDD group) or grade I (spinal cord injury
group) according to the MRI results (26). Patients with degenerative spinal
stenosis, idiopathic scoliosis, tumors, infections, or previous
lumbar disc surgery were excluded from this study.
The control group (CG) included 20 individuals who
received surgical treatment within 6 h after sustaining injury.
Their medical histories showed no evidence of pre-existing spinal
disorders, disc degeneration or previous spine-related surgeries.
This information was confirmed by both MRI and H&E staining of
the tissue samples. The nucleus pulposus (NP) tissues were
carefully dissected during surgery and subsequently subjected to
various methods of analysis, according to the corresponding
procedures. Briefly, the whole tissues were rinsed with
phosphate-buffered saline (PBS, pH 7.2) and then separated into 2
sections. One half was snap-frozen and stored in liquid nitrogen
within 30 min after removal from the patient, with subsequent
storage at −80ºC, while the remaining half was fixed with
paraformaldehyde.
H&E and TUNEL staining
Standardized H&E staining was used to evaluate
the morphology of NP tissue in the paraffin-embedded samples. To
identify apoptosis in NP cells, TUNEL staining was performed using
the TUNEL apoptosis assay kit according to the manufacturer’s
instructions (Roche, Basel, Switzerland). The results were obtained
using an optical microscope.
RNA extraction and quality
inspection
Total RNA from each sample was individually isolated
using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and the
miRNeasy mini kit (Qiagen, Valencia, CA, USA) according to the
manufacturer’s instructions. This procedure efficiently recovered
all RNA species, including miRNAs. RNA quality and quantity were
measured using a NanoDrop spectrophotometer (ND-1000; NanoDrop
Technologies, Wilmington, DE, USA) and RNA integrity was determined
by gel electrophoresis.
RNA labeling and array hybridization
Following RNA isolation, the miRCURY™ Hy3™/Hy5™
Power labeling kit (Exiqon, Vedbaek, Denmark) was used for miRNA
labeling according to the manufacturer’s instructions. Each 1 μg of
sample was 3′-end-labeled with Hy3 fluorescent label using T4 RNA
ligase. After the labeling procedure was terminated, the
Hy3-labeled samples were hybridized to the miRCURY LNA Array
(v.16.0) (Exiqon) according to the manual provided with the array.
The total mixture with hybridization buffer was hybridized to the
microarray in a 12-Bay Hybridization System (Hybridization System;
NimbleGen Systems, Inc., Madison, WI, USA), which provides an
active mixing action and a constant incubation temperature to
improve hybridization uniformity and enhance the signal. Following
hybridization, the slides were washed several times using the wash
buffer kit (Exiqon), and finally dried by centrifugation. The
slides were then scanned with the Axon GenePix 4000B microarray
scanner (Axon Instruments, Foster City, CA, USA), which contains
>1,891 capture probes annotated in miRBase 16.0 and 66
additional new miRPlus™ human microRNAs that are proprietary and
not found in miRBase. Our microarray data were MIAME compliant and
have been deposited in the MIAME compliant database GEO (Accession
no. GSE45856).
Array data analysis
Scanned images were imported into GenePix Pro 6.0
software (Axon) for grid alignment and data extraction. Replicate
miRNAs were averaged, and miRNAs with intensities ≥50 in all
samples were selected to calculate the normalization factor. The
expressed data were normalized using the median normalization
method. Following normalization, significantly differentially
expressed miRNAs were identified through volcano plot filtering.
Hierarchical clustering was performed using MEV software (v4.6,
TIGR). The miRNA was defined as being differentially expressed
between the compared groups if the P-value was <0.05 and the
fold change above 2.
Quantitative reverse transcription
polymerase chain reaction (qRT-PCR)
Seven miRNAs from the array data analysis were
selected for validation using the SYBR-based qPCR method. The 7
target miRNAs selected belonged to one of the two following
categories: i) miRNAs associated with chondrocyte apoptosis and ECM
degeneration (miR-34a and miR-675*); and ii) miRNAs with
particularly high fold changes in expression according to the
microarray results (miR-10a*, miR-25*,
miR-182, miR-130b* and miR-200c). Total RNA (100 ng) was
reverse transcribed to cDNA using miRNA-specific stem-loop RT
primers in a GeneAmp PCR System 9700 (Applied Biosystems, Foster
City, CA, USA). Quantitative PCR was performed using SYBR-Green
(Invitrogen) according to the manufacturer’s instructions in a
Rotor-Gene 3000 Real-time PCR instrument (Corbett Research,
Brisbane, Australia). The miRNA levels were normalized to U6 as an
internal control. The relative abundance of each miRNA was
calculated using the comparative Ct (2−ΔΔCt) method, and
the results were assessed by a t-test.
Bioinformatics analysis
The 3 most popular databases, TargetScan (27), miRanda (28) and miRDB (29), were used to predict the target
genes of the differentially expressed miRNAs. To reduce the false
positive results, the genes predicted by at least 2 of these 3
databases were selected as differential miRNA targets for further
analysis. Specifically, the target genes for the star form of
miRNAs (miRNA*) were predicted by both the miRanda and
miRDB databases, whereas the target genes for the non-star form of
miRNAs (miRNA) were predicted by the miRanda and TargetScan
databases. For functional annotation analysis, the DAVID database
(30) was used to annotate the
function of target genes in the module. The GO terms with adjusted
P-value <0.05 and count <2 were selected. The target genes
were further put into the KEGG database (31) to identify the enriched pathways.
The count number <2 and P-value >0.05 were selected as the
cut-off criteria.
Statistical analysis
Comparisons of 2-group parameters were performed
using the Student’s t-test. Comparisons of multiple group data were
performed using one-way analysis of variance followed by Turkey’s
post hoc test. A value of P<0.05 was considered to indicate a
statistically significant difference. Statistical analysis was
performed using the SPSS statistical software package (SPSS Inc.,
Chicago, IL, USA).
Results
Basic patient information
Patients with LIDH were selected as the EG. This
group consisted of 10 males and 10 females, ranging from 38 to 68
years in age with an average age of 53.9±8.5 years. In this group,
the degenerative intervertebral disc segments were the L4/5 segment
(12 patients), L5/S1 segment (7 patients), and L3/4 segment (1
patient). According to the MRI results, 14 specimens were
classified as grade V and 6 as grade IV. By contrast, patients with
spinal cord injury were selected as the CG, which included 11 males
and 9 females, ranging from 24 to 55 years in age with an average
age of 41.1±9.0 years. In this group, the degenerative
intervertebral disc segments were the L4/5 segment (8 patients),
L3/4 segment (3 patients), L2/3 segment (3 patients) and the L1/2
segment (6 patients), and 16 were classified as grade II and 4 as
grade I based on an MRI. Three patients from each of these 2 groups
were randomly selected for microarray analysis. The basic
information of these patients is presented in Table I.
 | Table IBasic patient information. |
Table I
Basic patient information.
| Sample ID
(EG/CG) | LIDH patients
(EG) | Spinal cord injury
patients (CG) |
|---|
|
|
|---|
| Age | Gender | Segment | MRI grade | Age | Gender | Segment | MRI grade |
|---|
| PCR2/NPCR2 | 63 | M | L4/5 | V | 56 | F | L4/5 | II |
| PCR4/NPCR4 | 70 | M | L4/5 | V | 51 | M | L3/4 | II |
| PCR6/NPCR6 | 55 | F | L4/5 | IV | 58 | M | L4/5 | II |
Histomorphological analysis of LIDH and
spinal cord injury specimens
The intervertebral disc has a unique structure, with
a gelatinous, amorphic NP surrounded by a highly organized annulus
fibrosus. The ECM, which is produced and maintained by chondrocytic
NP cells, is primarily composed of proteoglycans within a type II
collagen scaffold (32).
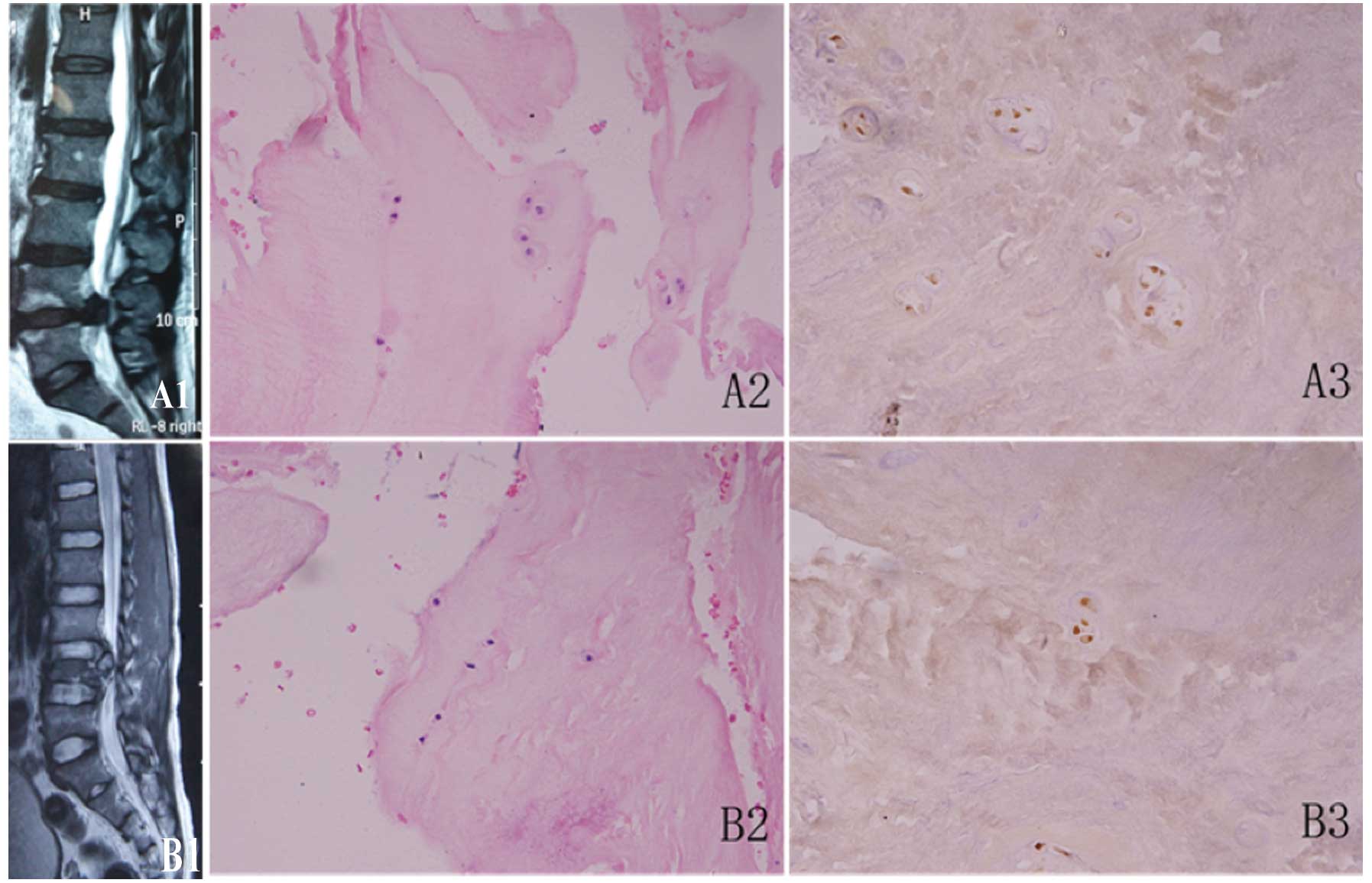
To confirm the intervertebral disc degeneration and
spinal cord injury, the specimens were subjected to H&E
staining. The NP tissue of the lumbar intervertebral disc showed
the following features under light microscopy: the NP cells which
appeared as round, chondrocyte-like cells were the only cellular
structures observed, the cytoplasm was stained red, the nucleus was
stained blue-black and the ECM was stained light red (Fig. 1). In the spinal cord injury group,
there was a higher number of NP cells, most of which were isolated
in the cartilage lacunae; individual NP cells appeared in pairs or
small cell clusters, and only a few empty lacuna (no
chondrocyte-like cells were present) were observed (Fig. 1B2). In the IDD group, the NP cells
whose nuclei had become small were sparse, more cell clusters or
multinucleated giant cells were present and the frequency of empty
lacunae increased (Fig. 1A2).
TUNEL staining was then performed on the NP tissue
samples from the EG. Fewer chondrocyte-like cells were observed,
and there were many TUNEL-positive cells with brown-stained nuclei
(Fig. 1). These cells also
exhibited changes in nuclear morphology, fragmentation or chromatin
margination (Fig. 1A3). In the
CG, on the other hand, more chondrocyte-like cells and fewer
TUNEL-positive cells were present (Fig. 1B3).
miRNAs are differentially expressed in
IDD
The 6th generation of miRCURY LNA Array (v.16.0)
(Exiqon) employed in this study contained >1,891 capture probes,
covering all human, mouse and rat miRNAs annotated in miRBase 16.0,
as well as all viral miRNAs related to these species. This array
also consisted capture probes for 66 new miRPlus human miRNAs that
are proprietary and not found in miRBase 16.0. After performing
fold change filtering (fold change ≥2) on the differentially
expressed miRNAs, we found that 54 miRNAs were upregulated and 53
miRNAs were downregulated in the IDD group compared with spinal
cord injury group. Volcano plot filtering was then performed to
identify the significantly differentially expressed miRNAs between
these 2 groups. The thresholds for screening differentially
expressed miRNAs were a fold change of ≥2 and a P-value of ≤0.05.
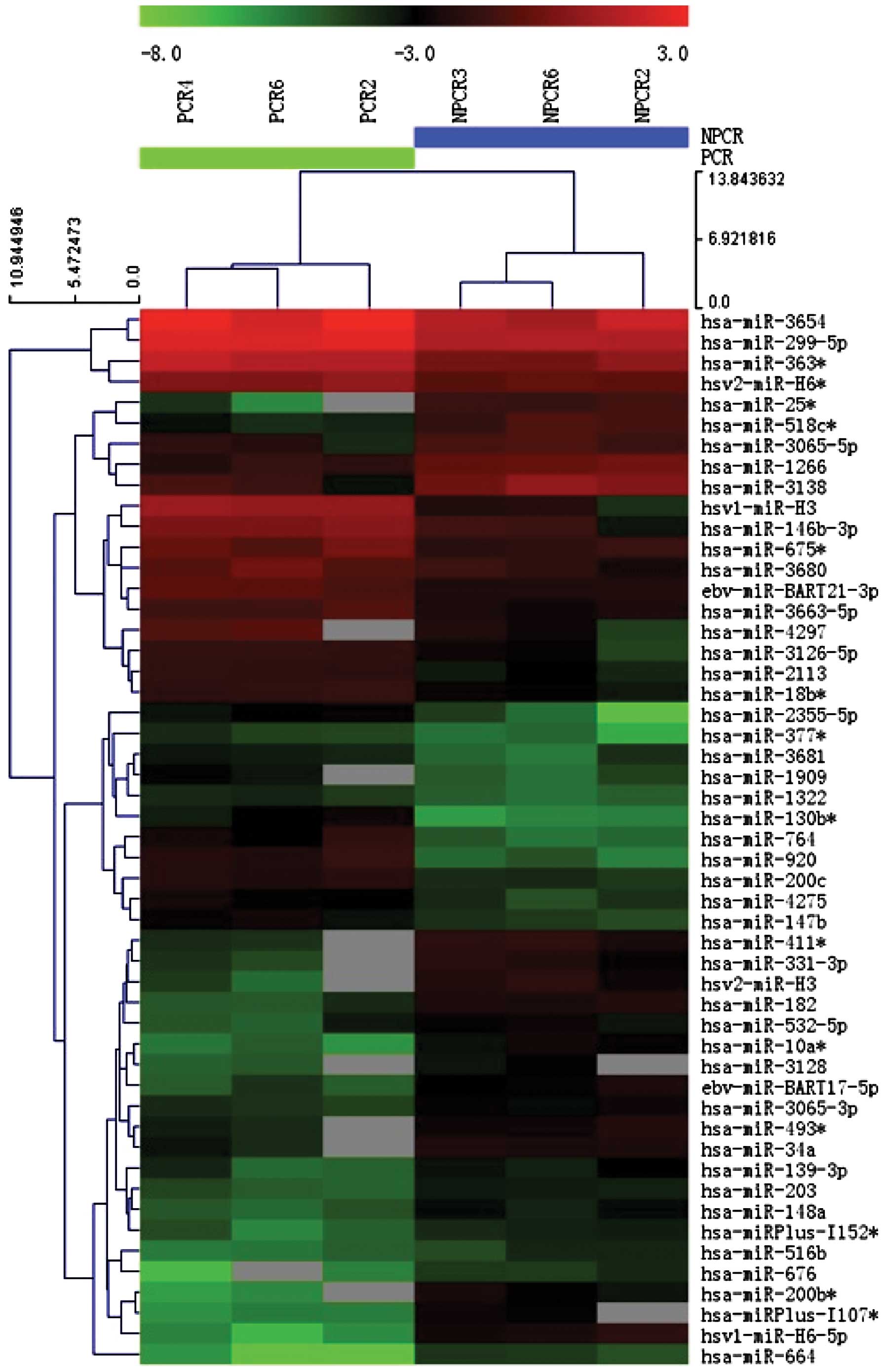
The heat map (Fig. 2) indicated
the results of a two-way hierarchical clustering of miRNAs and
samples. A total of 51 miRNAs exhibited a significant difference in
expression in the IDD group. Amongst these miRNAs, 25 miRNAs showed
an upregulated expression (Table
II), whereas 26 miRNAs showed a downregulated expression
(Table III) in the IDD group
compared with the spinal cord injury group.
 | Table IIUpregulated microRNAs in
intervertebral disc degeneration compared with spinal cord
injury. |
Table II
Upregulated microRNAs in
intervertebral disc degeneration compared with spinal cord
injury.
| ID | Name | Fold change | P-value |
|---|
| 147900 | hsv2-miR-H6* | 2.012 | 0.011 |
| 148599 | has-miR-3680 | 2.335 | 0.047 |
| 17411 | hsa-miR-147b | 2.491 | 0.010 |
| 42932 | hsa-miR-920 | 8.215 | 0.005 |
| 145990 |
ebv-miR-BART21-3p | 2.702 | 0.003 |
| 146058 | hsv1-miR-H3 | 9.582 | 0.000 |
| 42787 | hsa-miR-130b* | 6.266 | 0.005 |
| 147743 | hsa-miR-4275 | 2.374 | 0.009 |
| 148633 | hsa-miR-299-5p | 2.155 | 0.000 |
| 42461 |
hsa-miR-146b-3p | 3.798 | 0.003 |
| 146096 | hsa-miR-764 | 6.153 | 0.037 |
| 148597 |
hsa-miR-3663-5p | 2.497 | 0.006 |
| 145826 | hsa-miR-18b* | 2.391 | 0.002 |
| 42899 | hsa-miR-377* | 2.496 | 0.021 |
| 147800 |
hsa-miR-2355-5p | 4.078 | 0.005 |
| 148353 | hsa-miR-3681 | 2.228 | 0.031 |
| 42859 | hsa-miR-675* | 2.462 | 0.025 |
| 147925 |
hsa-miR-3126-5p | 2.662 | 0.005 |
| 148379 | hsa-miR-3654 | 2.258 | 0.041 |
| 17427 | hsa-miR-200c | 3.330 | 0.001 |
| 27544 | hsa-miR-363* | 2.668 | 0.005 |
| 146179 | hsa-miR-2113 | 2.877 | 0.000 |
| 146180 | hsa-miR-1909 | 2.861 | 0.007 |
| 147632 | hsa-miR-4297 | 3.904 | 0.007 |
| 46408 | hsa-miR-1322 | 2.186 | 0.010 |
 | Table IIIDownregulated microRNAs in
intervertebral disc degeneration compared with spinal cord
injury. |
Table III
Downregulated microRNAs in
intervertebral disc degeneration compared with spinal cord
injury.
| ID | Name | Fold change | P-value |
|---|
| 28019 |
hsa-miR-10a* | 0.194 | 0.003 |
| 146077 | hsv2-miR-H3 | 0.204 | 0.041 |
| 147654 | hsa-miR-3138 | 0.260 | 0.017 |
| 145973 | hsa-miR-664 | 0.176 | 0.003 |
| 46818 |
ebv-miR-BART17-5p | 0.298 | 0.008 |
| 17624 | hsa-miR-532-5p | 0.431 | 0.045 |
| 145821 |
hsa-miR-518c* | 0.206 | 0.005 |
| 10975 | hsa-miR-182 | 0.248 | 0.001 |
| 148471 |
hsa-miRPlus-l152* | 0.400 | 0.006 |
| 11004 | hsa-miR-203 | 0.424 | 0.001 |
| 145974 |
hsa-miR-200b* | 0.121 | 0.027 |
| 42451 | hsa-miR-139-3p | 0.448 | 0.047 |
| 42929 |
hsa-miR-25* | 0.110 | 0.003 |
| 147682 | hsa-miR-H6-5p | 0.080 | 0.004 |
| 148393 | hsa-miR-676 | 0.242 | 0.016 |
| 147536 |
hsa-miRPlus-l107* | 0.150 | 0.004 |
| 10955 | hsa-miR-148a | 0.360 | 0.004 |
| 46517 | hsa-miR-1266 | 0.370 | 0.004 |
| 147903 |
hsa-miR-3065-3p | 0.456 | 0.004 |
| 11125 |
has-miR-493* | 0.449 | 0.042 |
| 42784 |
hsa-miR-411* | 0.275 | 0.028 |
| 42887 | hsa-miR-331-3p | 0.275 | 0.040 |
| 148033 |
hsa-miR-3065-5p | 0.443 | 0.030 |
| 27217 | hsa-miR-34a | 0.455 | 0.005 |
| 11151 | hsa-miR-516b | 0.420 | 0.029 |
| 147545 | hsa-miR-3128 | 0.307 | 0.029 |
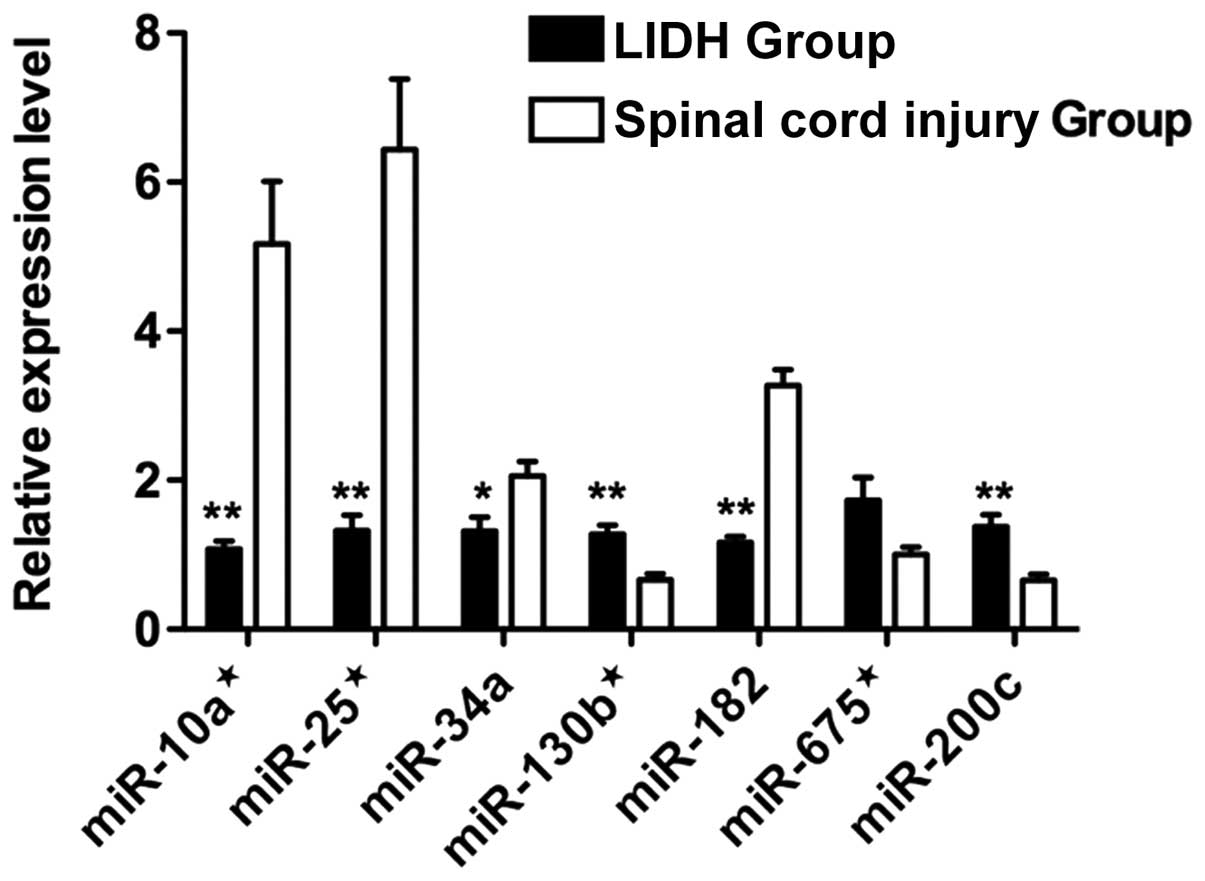
Based on their expression levels and fold difference
in expression, 3 upregulated miRNAs (miR-130b*,
miR-675* and miR-200c) and 4 downregulated miRNAs
(miR-10a*, miR-25*, miR-34a and miR-182) were
selected for validation by qRT-PCR. All of the miRNAs examined,
with the exception of miR-675*, showed a statistically
significant difference in expression in a manner consistent with
the data from microarray analysis (P<0.05) (Fig. 3).
Bioinformatics analysis
Since miRNAs function by targeting mRNAs, we
retrieved the putative target genes of differentially expressed
miRNAs from 3 databases and selected the target genes retrieved by
at least 2 databases. The target genes were then subjected pathway
enrichment analysis using KEGG pathways to find the canonical
pathways controlled by the identified miRNAs. Among the top 10
signaling pathways mostly likely to be regulated by the miRNAs were
the phosphoinositide 3-kinase (PI3K)-Akt, mitogen-activated protein
kinase (MAPK), epidermal growth factor receptor (EGFR; ErbB) and
Wnt pathways (Fig. 4A). The
network between the miRNAs and signaling pathways is illustrated in
Fig. 4B. The function of the
target genes was then predicted by the GO enrichment analysis. The
predicted target genes were principally enriched for GO terms
related to processes, such as protein binding and anatomical
structure (Fig. 5A). The network
miRNAs involved were presented in Fig. 5B.
Discussion
In the present study, 3 samples were selected from
each of the LIDH and spinal cord injury groups. Their miRNA
expression patterns were quantified and we then evaluated the
differences in their respective miRNA expression profiles by
microarray analysis. We demonstrated that 25 miRNAs were
upregulated and 26 miRNAs were downregulated in the IDD group
compared with the spinal cord injury group. Given the potential for
false positives with microarray technology, as well as its inherent
limitations as regards sensitivity and quantification, qRT-PCR was
then performed to validate the microarray data. However, microarray
expression analysis is still a powerful, high-throughput and
versatile tool for the study of genome-wide miRNA expression
profiles. Therefore, microarray technology was used in this study
to preliminarily screen the differentially expressed miRNAs,
potentially shedding light on the regulatory mechanisms of miRNAs
in IDD.
Since NP tissue cannot be separated from completely
normal living bodies to serve as normal controls, the controls are
usually selected from the following 2 groups when investigating
differential miRNA expression profiles in patients with IDD: i)
cases in which the NP tissue is detached from the body immediately
following accidental death; ii) patients with spinal disease
requiring discectomy, such as those with congenital scoliosis or
severe spinal injury. For the former group, NP tissue may be
obtained through osteotomy, whereas in the latter group, NP tissue
can be acquired via discectomy and fusion fixation. In all selected
cases, it is imperative to exclude other spinal diseases and other
underlying diseases that may cause changes in gene expression. In
this study, we selected patients with spina cord injury as the CG
for the following reasons: i) separating NP tissue from accidental
death victims is very difficult as it is not easy to verify whether
the intervertebral disc has degraded, and there are various changes
in gene expression following accidental death; ii) congenital
scoliosis is a congenital disorder, and given that the local lesion
was stimulated by abnormal stress for a long period of time, it is
likely that its gene expression differs from that in normal healthy
individuals; iii) there were a number of severe spinal injuries
needing discectomy and fusion fixation in our clinic, and thus it
was convenient to obtain the NP tissue from these patients. Changes
due to local inflammation and associated post-trauma reactions in
the intervertebral disc should be taken into consideration when
analyzing the results. It is noted that we cannot entirely exclude
the possibility that the LIDH and spinal cord injury groups had
different genetic backgrounds.
Susceptibility to IDD is greater in individuals with
particular alleles, such as the Trp2 allele of COL9A2 (33) or different genetic polymorphisms
in vitamin D receptor (VDR) (34). Following gene sequence alignment
and target gene prediction using the 3 popular databases (PicTar,
TargetScan and miRanda), the COL9A2 gene was found to be a putative
target gene of miR-146b-3p, which was upregulated by 3.80-fold in
the degenerative disc (P=0.003). Moreover, miR-146 has been
reported to target several other mRNAs, including apoptosis-related
genes, such as FADD (35),
inflammation-associated genes, such as interleukin (IL)-1β, IL-6
and tumor necrosis factor (TNF) (36), as well as metabolism-related
genes, such as matrix metalloproteinase (MMP)-16 (37). Given that the abovementioned
biological processes and genes are involved in the pathological
changes observed in IDD, we predicted that miR-146 may play an
important role in the occurrence and development of IDD.
A previous study demonstrated that silencing miR-34a
can effectively reduce IL-1β-induced apoptosis in rat chondrocytes
(21). In this study, miR-34a was
found to be downregulated by 0.45-fold (P<0.05) in degenerative
NP compared with the spinal cord injury group. This result was
further confirmed by qRT-PCR (0.64-fold, P<0.05). One probable
explanation is that spinal cord injury may induce the apoptosis of
intervertebral disc cells through a caspase-dependent pathway.
Thoracolumbar fractures can induce early caspase-dependent
apoptosis in disc cells of the affected intervertebral disc, in
part by downregulating the anti-apoptotic protein, Bcl-2, as well
as signaling via the death receptor complex [TNF receptor (TNFR) I
and Fas receptor (FasR)] (38).
As previously demonstrated, compared with degenerative
intervertebral discs, traumatic thoracolumbar intervertebral discs
have an increased number of TUNEL-positive cells, which is evidence
of apoptosis involving both receptor-mediated and
mitochondrial-dependent pathways (39). In this study, the spinal cord
injury group samples were obtained upon surgery performed on
patients with spinal cord injury that took place within 6 h
following injury. The morphological changes in our spinal cord
injury group samples were the same as those observed in the normal
NP tissue. Based on these data we hypothesized the following: soon
after spinal injury, the expression of miR-34a is upregulated to a
level even higher than that observed during degeneration, resulting
in a large number of cells undergoing apoptosis and further
promoting intervertebral disc degeneration.
In the present study, the differentially expressed
miRNAs were predicted to control several pathways relevant for the
regulation of IDD. It has been demonstrated that the
transcriptional activation of the PI3K-Akt pathway is involved in
lumbar disc degeneration (40).
As shown in a previous study, hyperbaric oxygen treatment
suppresses the MAPK signaling pathway in degenerated human
intervertebral disc cells (41).
In addition, investigators from The Netherlands have used a canine
model of IDD to examine the biochemical changes associated with
chondroid metaplasia, and found a downreguation of Wnt signaling
and caveolin-1 expression (42).
Our results suggest that miRNAs are important regulators of IDD
through the modulation of several signaling pathways.
In conclusion, our results demonstrated that 25
miRNAs were upregulated and 26 were downregulated in the NP tissue
of LIDH patients compared with the patients spinal cord injury.
Bioinformatics analysis predicted the target genes and signaling
pathways of these miRNAs, which may enhance our understanding of
the involvement of miRNAs in the occurrence and development of IDD.
Further studies on miRNA functions and target gene verification
would provide an experimental basis for the diagnosis and treatment
of IDD, which remains an important area for future
investigation.
Acknowledgements
We thank Yong Fan and Min-jie Ma for their
assistance in sample collection; Hui Yang, Xu Chen and Ke Xu for
their experimental assistance; and Kang Chen, Biotech (Shanghai,
China) for their skillful assistance with the microRNA
microarrays.
References
|
1
|
Juniper M, Le TK and Mladsi D: The
epidemiology, economic burden, and pharmacological treatment of
chronic low back pain in France, Germany, Italy, Spain and the UK:
a literature-based review. Expert Opin Pharmacother. 10:2581–2592.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Phillips C, Main C, Buck R, Aylward M,
Wynne-Jones G and Farr A: Prioritising pain in policy making: the
need for a whole systems perspective. Health Policy. 88:166–175.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Waddell G: Low back pain: a twentieth
century health care enigma. Spine (Phila Pa 1976). 21:2820–2825.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang YG, Sun Z, Zhang Z, Liu J and Guo X:
Risk factors for lumbar intervertebral disc herniation in Chinese
population: a case-control study. Spine (Phila Pa 1976).
34:E918–E922. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kalichman L and Hunter DJ: The genetics of
intervertebral disc degeneration. Associated genes. Joint Bone
Spine. 75:388–396. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Walter BA, Korecki CL, Purmessur D,
Roughley PJ, Michalek AJ and Iatridis JC: Complex loading affects
intervertebral disc mechanics and biology. Osteoarthritis
Cartilage. 19:1011–1018. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Battie MC and Videman T: Lumbar disc
degeneration: epidemiology and genetics. J Bone Joint Surg Am.
88(Suppl 2): S3–S9. 2006. View Article : Google Scholar
|
|
8
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
|
|
9
|
Wightman B, Ha I and Ruvkun G:
Posttranscriptional regulation of the heterochronic gene lin-14 by
lin-4 mediates temporal pattern formation in C. elegans.
Cell. 75:855–862. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang F, Niu G, Chen X and Cao F: Molecular
imaging of microRNAs. Eur J Nucl Med Mol Imaging. 38:1572–1579.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Selbach M, Schwanhausser B, Thierfelder N,
Fang Z, Khanin R and Rajewsky N: Widespread changes in protein
synthesis induced by microRNAs. Nature. 455:58–63. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Griffiths-Jones S: The microRNA Registry.
Nucleic Acids Res. 32:D109–D111. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang F, Song X, Li X, et al: Noninvasive
visualization of microRNA-16 in the chemoresistance of gastric
cancer using a dual reporter gene imaging system. PLoS One.
8:e617922013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Teague EM, Print CG and Hull ML: The role
of microRNAs in endometriosis and associated reproductive
conditions. Hum Reprod Update. 16:142–165. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Miyaki S, Nakasa T, Otsuki S, et al:
MicroRNA-140 is expressed in differentiated human articular
chondrocytes and modulates interleukin-1 responses. Arthritis
Rheum. 60:2723–2730. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Miyaki S, Sato T, Inoue A, et al:
MicroRNA-140 plays dual roles in both cartilage development and
homeostasis. Genes Dev. 24:1173–1185. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pais H, Nicolas FE, Soond SM, et al:
Analyzing mRNA expression identifies Smad3 as a microRNA-140 target
regulated only at protein level. RNA. 16:489–494. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tardif G, Hum D, Pelletier JP, Duval N and
Martel-Pelletier J: Regulation of the IGFBP-5 and MMP-13 genes by
the microRNAs miR-140 and miR-27a in human osteoarthritic
chondrocytes. BMC Musculoskelet Disord. 10:1482009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Abouheif MM, Nakasa T, Shibuya H, Niimoto
T, Kongcharoensombat W and Ochi M: Silencing microRNA-34a inhibits
chondrocyte apoptosis in a rat osteoarthritis model in vitro.
Rheumatology (Oxford). 49:2054–2060. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kongcharoensombat W, Nakasa T, Ishikawa M,
et al: The effect of microRNA-21 on proliferation and matrix
synthesis of chondrocytes embedded in atelocollagen gel. Knee Surg
Sports Traumatol Arthrosc. 18:1679–1684. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dudek KA, Lafont JE, Martinez-Sanchez A
and Murphy CL: Type II collagen expression is regulated by
tissue-specific miR-675 in human articular chondrocytes. J Biol
Chem. 285:24381–24387. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Teng G and Papavasiliou FN: Shhh!
Silencing by microRNA-155. Philos Trans R Soc Lond B Biol Sci.
364:631–637. 2009.
|
|
25
|
Wang HQ, Yu XD, Liu ZH, et al: Deregulated
miR-155 promotes Fas-mediated apoptosis in human intervertebral
disc degeneration by targeting FADD and caspase-3. J Pathol.
225:232–242. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pfirrmann CW, Metzdorf A, Zanetti M,
Hodler J and Boos N: Magnetic resonance classification of lumbar
intervertebral disc degeneration. Spine (Phila Pa 1976).
26:1873–1878. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar
|
|
29
|
Wang X: miRDB: a microRNA target
prediction and functional annotation database with a wiki
interface. RNA. 14:1012–1017. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
31
|
Kanehisa M and Goto S: KEGG: kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hadjipavlou AG, Tzermiadianos MN, Bogduk N
and Zindrick MR: The pathophysiology of disc degeneration: a
critical review. J Bone Joint Surg Br. 90:1261–1270. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Aladin DM, Cheung KM, Chan D, et al:
Expression of the Trp2 allele of COL9A2 is associated with
alterations in the mechanical properties of human intervertebral
discs. Spine (Phila Pa 1976). 32:2820–2826. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Eser B, Cora T, Eser O, et al: Association
of the polymorphisms of vitamin D receptor and aggrecan genes with
degenerative disc disease. Genet Test Mol Biomarkers. 14:313–317.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Curtale G, Citarella F, Carissimi C, et
al: An emerging player in the adaptive immune response:
microRNA-146a is a modulator of IL-2 expression and
activation-induced cell death in T lymphocytes. Blood. 115:265–273.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xie YF, Shu R, Jiang SY, Liu DL, Ni J and
Zhang XL: MicroRNA-146 inhibits pro-inflammatory cytokine secretion
through IL-1 receptor-associated kinase 1 in human gingival
fibroblasts. J Inflamm (Lond). 10:202013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xia H, Qi Y, Ng SS, et al: microRNA-146b
inhibits glioma cell migration and invasion by targeting MMPs.
Brain Res. 1269:158–165. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Heyde CE, Tschoeke SK, Hellmuth M,
Hostmann A, Ertel W and Oberholzer A: Trauma induces apoptosis in
human thoracolumbar intervertebral discs. BMC Clin Pathol. 6:52006.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tschoeke SK, Hellmuth M, Hostmann A, et
al: Apoptosis of human intervertebral discs after trauma compares
to degenerated discs involving both receptor-mediated and
mitochondrial-dependent pathways. J Orthop Res. 26:999–1006. 2008.
View Article : Google Scholar
|
|
40
|
Pasku D, Soufla G, Katonis P, Tsarouhas A,
Vakis A and Spandidos DA: Akt/PKB isoforms expression in the human
lumbar herniated disc: correlation with clinical and MRI findings.
Eur Spine J. 20:1676–1683. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Niu CC, Lin SS, Yuan LJ, et al: Hyperbaric
oxygen treatment suppresses MAPK signaling and mitochondrial
apoptotic pathway in degenerated human intervertebral disc cells. J
Orthop Res. 31:204–209. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Erwin M: Canonical Wnt signaling and
caveolae play a role in intervertebral disc degeneration; the
continuing saga of the mysterious notochordal cell. Arthritis Res
Ther. 15:1132013. View
Article : Google Scholar : PubMed/NCBI
|