Introduction
The World Health Organization International Agency
for Research on Cancer reported that approximately 1.29 million
women were diagnosed with breast cancer and >400,000 women
succumbed to the disease in 2008 (1). Thymoquinone is an anti-neoplasic
agent reported to have medicinal potential in the treatment of
breast cancer. The antitumour activity of thymoquinone has been
reported in cells derived from ovarian, breast and colon cancers
(2). The apoptotic activity of
thymoquinone has been reported to induce the total Bax/Bcl-2 ratio
in MCF7 (3), HCT-116 (4) and HL-60 cancer cells (5). These findings were based on flow
cytometry, western blot analysis and multi-colour fluorescence
in situ hybridization (mFISH). Therefore, further
investigation is required to to determine its effects on human
genome expression using cDNA microarray technology.
Materials and methods
Complete medium preparation
RPMI-1640 (Invitrogen, Gibco, Carlsbad, CA, USA)
medium with L-glutamine was used to culture the MCF7 cell line
(ATCC® HTB22™). The medium was supplemented with 10%
heat-inactivated fetal bovine serum (Invitrogen/Gibco) and 1 unit
penicillin/streptomycin (HyClone, Logan, UT, USA).
Thymoquinone solution preparation
The 1 mM stock solution of thymoquinone
(Sigma-Aldrich, Saint-Quentin-Fallavier, France) was prepared with
DMSO (Sigma-Aldrich). The solution was filtered with a 0.02 μm
syringe filter (Hydrophilic Ministart; Sartorius AG, Goettingen,
Germany).
Cell culture and treatment
The MCF7 cells were seeded at 3–4×106
cells/well in 96-well plates. They were cultured at 0.5%
CO2 in a humidified incubator at 37°C (Thermo
Scientific, Waltham, MA, USA) for 24 h. The control cells were
treated with 0.05% dimethyl sulfoxide alone (Sigma-Aldrich). Four
biological replicates from each sample were prepared in separate
culture flasks. The samples were treated with 50 μM thymoquinone
for 24 h. EDTA [0.25% (w/v) + Trypsin/0.53% Mm] solution was used
to detach the cells from the flask surface. The cells were then
centrifuged at 13,000 rpm for 10 min. The supernatant was removed
and the cells were washed with PBS solution prior to centrifugation
at 13,000 rpm for 5 min.
RNA isolation
RNA was isolated from the MCF7 cells using the
RNeasy Plus Mini kit (Qiagen, Valencia, CA, USA) according to the
manufacturer’s instructions. The quantity of RNA was measured using
a spectrophotometer (NanoDrop 2000c; Thermo Scientific). Samples
with the RNA concentration (A260/A280 ≥1.8
ng/μl) and purity (A230/A260 ≥2.0 ng/μl) were
selected. An Agilent 2100 Bioanalyser was used to determine the RNA
integrity number (RIN). The degradation level was identified using
the RNA 6000 Nano LabChip kit (Agilent Technologies, Santa Clara,
CA, USA). The samples with RIN >9.8 were selected for further
analysis.
Two-colour microarray-based gene
expression
The Agilent Low Input Quick Amp Labelling kit (USA)
was used to generate cRNA with a sample input of 200 ng total RNA
in two-color microarray analysis. The RNA Spike-In kit (Agilent
Technologies) was used as an external control (positive control).
It monitors and calibrates the linearity, sensitivity and accuracy
of the microarray workflow. Spike A Mix with cyanine-3 (cye-3) was
used to label the samples (thymoqionone-treated and untreated) and
Spike B Mix with cyanine-5 (cye-5) was used to label the internal
control (Universal Human Reference RNA, Agilent Technologies). T7
RNA polymerase was used to amplify target material.
The array platform used was 8×60K array SurePrint
Technology (Slide Human V1 U252800417900-S01; Interscience,
Rockland, MA, USA) and gasket slides with SureHyb Technology. The
reference design was selected for the study. Four biological
replicates from 50 μM thymoquinone-treated MCF7 and four replicates
from untreated MCF7 cells were hybridised against Human Universal
Reference RNA using a hybridisation kit (Agilent Technologies). The
slide chamber was assembled and placed in rotisserie hybridization
oven and rotated at 10 rpm in 65°C for 17 h. The array slide was
washed and fluorescent imaging system was used to scan the
hybridisation signals (DNA Microarray Scanner, Surescan
High-Resolution Technology, Agilent Technologies).
The results were extracted with Feature Extraction
Project v10.7.3.1 software and analysed using GeneSpring software
v12.1. Statistical analysis was carried out using an unpaired
t-test and a fold change with cut-off value >2 with a P-value
≤0.05 was considered to indicate a statistically significant
difference.
Quantitative reverse transcription PCR
(qRT-PCR)
qRT-PCR was carried otu to validate the results from
microarray analysis and performed in two steps. The high capacity
RNA-to-cDNA kit protocol (Applied Biosystems, Foster City, CA, USA)
was followed to transcribe 2 μg total RNA into single-stranded
cDNA. This was followed by amplification using a thermal cycler
(Eppendrof, Hamburg, Germany). The ΔCT of the
thymoquinone-treated cells was subtracted from the ΔCT
of the untreated cells to determine the differences
(ΔΔCT) and fold change (2−ΔΔCT). The human
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) reference
gene was used as an endogenous control to normalize the
fluorescence signals in the untreated and treated cells. Five
dilution points were prepared in triplicate for each primer to plot
the relative standard curve.
Amplification plots and the standard curve of each
target gene and endogenous control were created to determine the
primer efficiency. Three biological replicates of
thymoquinone-treated samples and three biological replicates of
untreated samples were examined. The StepOnePlus Real-Time PCR
instrument (Applied Biosystems) was used to run this experiment
with a 96-well system. The 5′ nuclease assay (TaqMan probe; Life
Technologies Corp., Carlsbad, CA, USA) was used to detect the
specific PCR product. It was labelled with the reporter
fluorescence dye 6-carboxyfluorescein (6-FAM) at the 5′ end and the
quencher dye MGBNFQ at the 3′ end. The fast mode was selected to
run the experiment for 40 min. The temperature was adjusted to the
one recommended in the manufacturer’s instructions (Applied
Biosystems,).
The comparative CT (ΔΔCT)
method was selected to determine the amount of target nucleic acid
sequence in each sample relative to the untreated samples. The
normalization of the ΔCT value of the
thymoquinone-treated cells to the ΔCT value of the
untreated cells was carried out to determine the ΔΔCT
value.
Results
Quantification and data filtration
Filtering was carried out to minimise errors and
generated 24,971 entities. There were seven entities out of 577
entities with a fold change of >2.0 and and P-value ≤0.05.
Hierarchial clustering
Clustering was carried out to identify entities that
were grouped within a cluster which are co-regulated and
functionally related. Clustering was carried out on both entities
and conditions. This created a two-dimensional dendogram. The
untreated samples had a similar expression profile, which linked to
form a group. A similar pattern was observed in the
thymoquinone-treated samples (Fig.
1).
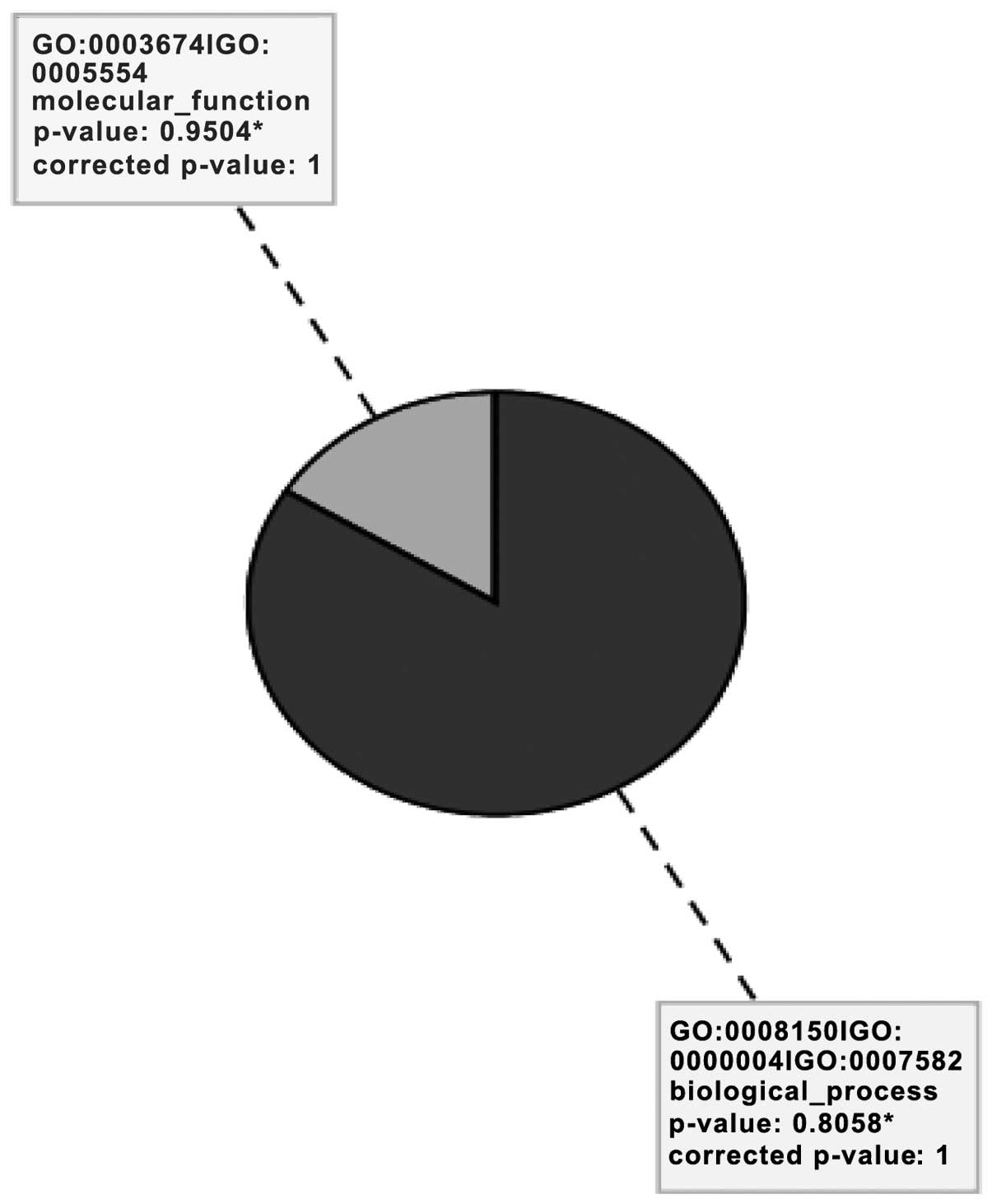
Gene Ontology (GO) analysis
GO describes gene products in terms of their
associated cellular component and molecular function and biological
process (Fig. 2). An analysis
revealed eight significant GO terms related to biological processes
(84%) and molecular functions (16%), as presented in Table I.
 | Table IGene Ontology categories of
biological processes and molecular functions. |
Table I
Gene Ontology categories of
biological processes and molecular functions.
| Gene Ontology
categories | Total genes | P-value |
|---|
| Biological
processes |
| Type I
interferon-mediated signaling pathway | 62 | 0.002 |
| Cellular response
to type I interferon | 62 | 0.002 |
| Response to type I
interferon | 63 | 0.002 |
| Xenobiotic
metabolic process | 129 | 0.021 |
| Cellular response
to xenobiotic stimulus | 130 | 0.021 |
| Response to
xenobiotic stimulus | 131 | 0.021 |
| Molecular
functions |
| Androsterone
dehydrogenase activity | 3 | 0.023 |
| Aldo-keto
reductase (NADP) activity | 18 | 0.045 |
Single experiment analysis (SEA)
SEA identifies matching pathways within the
experiment and identifies pathways related to regulated genes. An
analysis of the 577 entities with a P≤0.05 and a fold change of
>2.0 showed 504 pathways. There were three matched entities in
cytochrome p450 (CYP450) (63 entities), phase I and II drug
metabolism (49 entities) and interferon type I (54 entities).
Table II presents
the downregulated pathways with the highest number of matched
entities (16 entities). These were the metapathway
biotransformation and interferon signaling pathway (six entities),
although these were not significant (P>0.05). The most
significant results were observed for the phase I metabolic pathway
(P<0.04). The upregulated pathways with the highest number of
matched entities were also the metapathway biotransformation (four
entities). The most significant results were observed for the GPCR
class B secretion pathway (P=0.009).
 | Table IIRegulated pathways identified by
single experiment analysis. |
Table II
Regulated pathways identified by
single experiment analysis.
| Pathways | P-value | Entities | Matched
entities |
|---|
| Downregulated
pathways |
|
HS_Metapathway_biotransformation | 2.38 | 188 | 16 |
|
HS_Benzo(a)pyrene_metabolism | 1.25 | 9 | 5 |
|
HS_Interferon_α_β_signaling | 2.00 | 26 | 6 |
|
HS_AhR_pathway | 4.32 | 28 | 6 |
|
HS_Estrogen_metabolism | 2.48 | 18 | 4 |
| HS_Oxidative
stress | 7.25 | 30 | 4 |
|
HS_Keap1_Nrf2_pathway | 8.92 | 14 | 3 |
|
HS_Tamoxifen_metabolism | 2.17 | 21 | 3 |
|
HS_Glucuronidation | 0.007 | 26 | 3 |
|
HS_Type_II_interferon_signaling
(IFNG) | 1.68 | 37 | 4 |
|
HS_Iron_metabolism_in_placenta | 0.003 | 12 | 2 |
|
HS_Tryptophan_metabolism | 0.026 | 47 | 3 |
|
HS_Phase_I_-_functionalization_of_compounds | 0.004 | 49 | 3 |
|
HS_Glutathione_metabolism | 0.008 | 20 | 2 |
|
HS_Spingolipid_metabolism | 0.034 | 30 | 2 |
|
HS_Interferon_type_I_ | 0.040 | 54 | 3 |
|
HS_Hypothetical_Network_for_Drug_Addiction | 0.023 | 32 | 2 |
|
HS_Bile_acid_and_bile_salt_metabolism | 0.041 | 24 | 2 |
|
HS_Nicotine_Activity_on_Chromaffin_Cells | 0.029 | 4 | 1 |
|
HS_Cytochrome_P450 | 0.009 | 63 | 3 |
| Upregulated
pathways |
|
Hs_One_Carbon_Metabolism | 0.013 | 27 | 2 |
| HS_GPCRs,
_Class_B_secretin_like | 0.009 | 23 | 2 |
|
HS_miRs_in_Muscles_Cell_Differentiation | 0.015 | 40 | 2 |
|
HS_Metapathway_biotransformation | 0.024 | 188 | 4 |
|
HS_Thyroxine_thyroid_hormone_production | 0.032 | 5 | 1 |
|
HS_Heart_Development | 0.033 | 47 | 2 |
|
HS_Cell_surface_interaction_at_vascular_wall | 0.020 | 39 | 2 |
The most significantly upregulated genes were large
intergenic non-coding RNAs (lincRNAs) and the probe was annotated
with its genomic location (Table
III). The protein tyrosine phosphatase, receptor type, R
(PTPRR) gene was upregulated by 2.2-fold. SEA revealed genes
in the estrogen pathway that were downregulated following treatment
with thymoquinone (Table IV).
The genes that were downregulated were UDP glucuronosyltransferase
1 family, polypeptide A8 (UGT1A8) (by −16.26-fold),
cytochrome P450, family 1, subfamily A, polypeptide 1
(CYP1A1) (by −2.22-fold), cytochrome P450, family 1,
subfamily B, polypeptide 1 (CYP1B1) (by −2.23-fold) and
NAD(P)H dehydrogenase, quinone 1 (NQO1) (by −2.07-fold).
Genes in the interferon α and β signaling pathways were also
downregulated following treatment with thymoquinone; these genes
were the interferon-induced protein with tetratricopeptide repeats
(IFIT)1 (by −10.65-fold), IFIT2 (by
−4.20-fold), IFIT3 (by −5.27-fold), interferon, α-inducible
protein (IFI)6 (also known as G1P3) (-by 7.86-fold)
and IFI27 (by −3.19-fold). The genes downregulated in the
type II interferon signaling pathway, included interferon
regulatory factor 9 (IRF9) (−2.01), IFIT2 (−4.20),
2′-5′-oligoadenylate synthetase 1, 40/46 kDa (OAS1) (−3.5)
and IFI6 (−7.86).
 | Table IIIList of genes which were upregulated
following with treatment thymoquinone. |
Table III
List of genes which were upregulated
following with treatment thymoquinone.
| Gene symbol | Description | Fold change |
|---|
|
XLOC001537 | BROAD lincRNAs
version v2 | 6.31 |
| CARD16 | Homo sapiens
caspase recruitment domain family, member 16 | 5.25 |
| C2CD4B | Homo sapiens
C2 calcium-dependent domain containing 4B | 4.31 |
| UBQLNL | Homo sapiens
ubiquilin-like | 4.21 |
| MTR |
Methyltetrahydrofolate-homocysteine
methyltransferase | 3.91 |
| TMEM100 | Protein 100,
transcript variant 2 | 3.70 |
| MOBKL2B | Binder kinase
activator-like 2B | 3.67 |
| RNF17 | (RNF17), transcript
variant 1 | 3.62 |
|
STON1GTF2 | Homo sapiens
ring finger protein 17, transcript variant 1 | 3.60 |
| OR52J3 | Homo sapiens
olfactory receptor, family 52, subfamily J, member 3 | 3.42 |
|
LOC100133130 | Homo sapiens
clone FLB4246 PRO1102 mRNA, complete cds | 3.30 |
|
NCRNA00261 | Homo sapiens
non-protein coding RNA 261, non-coding | 3.28 |
| FBXW2 | Homo sapiens
F-box and WD repeat domain containing 2 | 3.26 |
|
LOC100131763 | Homo sapiens
cDNA FL137602 fis, clone BRCOC2009380 | 3.19 |
| SCIN | Homo sapiens
scinderin, transcript variant 2 | 3.19 |
| PGAP1 | Homo sapiens
post-GPI attachment to protein 1 | 3.17 |
| S100B | Homo sapiens
S100 calcium binding protein B (S100B) | 3.16 |
| SV2A | Homo sapiens
synaptic vesicle glycoprotein 2A | 3.16 |
| C7orf54 | Homo sapiens
chromosome 7 open reading frame 54, non-coding RNA | 3.15 |
| FAM65B | Homo sapiens
family with sequence similarity 65, member B, transcript variant
1 | 3.15 |
 | Table IVList of genes which were
downregulated following treatment with thymoquinone. |
Table IV
List of genes which were
downregulated following treatment with thymoquinone.
| Gene symbol | Description | Fold change |
|---|
| UGT1A8 | Homo sapiens
UDP glucuronosyltransferase 1 family, polypeptide A8 | −16.26 |
| SLC7A11 | Homo sapiens
solute carrier family 7 (anionic amino acid transporter light
chain, xc-system), member 11 | −12.99 |
| IFIT1 | Homo sapiens
interferon-induced protein with tetatricopeptide repeats 1,
transcript variant 2 | −10.65 |
| IF16 | Homo sapiens
interfero, α-inducible protein 6, transcript variant 3 | −7.86 |
|
C17orf64 | Homo sapiens
chromosome 17 open reading frame 64 | −6.20 |
| ALDH3A1 | Homo sapiens
aldehyde dehydrogenase 3 family, member A1, transcript variant
2 | −5.85 |
| UGT1A6 | Homo sapiens
UDP glucuronosyltransferase 1 family, polypeptide A6, transcript
variant 1 | −5.68 |
| IFIT2 | Homo sapiens
interferon-induced protein with tetratricopeptide repeats 2 | −5.32 |
| IFIT3 | Homo sapiens
interferon-induced protein with tetratricopeptide repeats 3,
transcript variant 1 | −5.27 |
| HMOX1 | Homo sapiens
hemeoxygenase (decycling) 1 | −5.13 |
| DNAH6 | Homo sapiens
dynein, axonemal, heavy chain 6 | −4.58 |
| TPRG1 | Homo sapiens
tumour protein p63 regulated 1 | −4.55 |
| PCLO | Homo sapiens
piccolo (presynaptic cytomatrix protein), transcript variant 1 | −4.33 |
| CHD5 | Homo sapiens
chromodomain helicase DNA binding protein 5 | −4.28 |
| CCR1 | Homo sapiens
chemokine (C-C motif) receptor 1 | −4.26 |
| AKR1C1 | Homo sapiens
aldo-ketoreductase family 1, member C1 (dihydrodiol dehydrogenase
1; 20-α (3-α)-hydroxysteroid dehydrogenase) | −4.21 |
| GSTA5 | Homo sapiens
glutathione S-transferase α 5 | −4.21 |
| AKR1B15 | Homo sapiens
aldo-ketoreductase family 1, member B15 | −4.13 |
|
HEATR7B1 | HEAT repeat
containing 7B1 | −3.98 |
| GPX2 | Homo sapiens
glutathione peroxidase 2 (gastrointestinal) | −3.88 |
Expansion and pathway enrichment
analysis
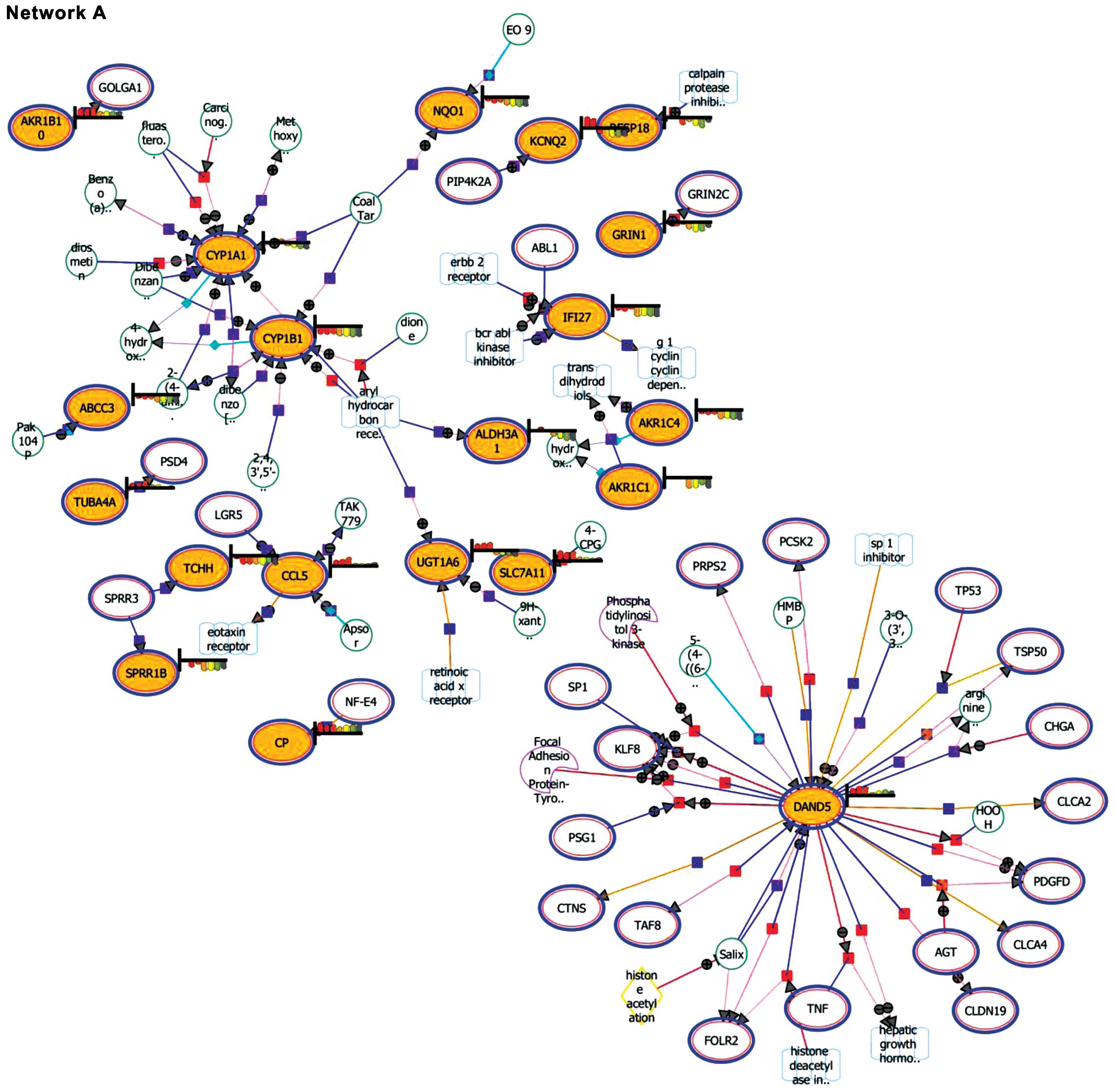
Expansion and pathway enrichment analysis provided
information on the biological functions of the entities. Network A
(downregulated genes) showed the DAN domain family member 5, BMP
antagonist (DAND5) as the focal point among the matched
entities (Fig. 3). In addition,
CYP1A1 and CYP1B1 were also noted to be highlighted
in the interaction. Network B (upregulated genes) highlighted the
involvement of regulator of G-protein signaling 4 (RGS4),
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9
(PSMD9), ubiquitin specific peptidase 9, Y-linked
(USP9Y) and dystrophin (DMD).
Validation of thymoquinone regulation of
gene expression by qRT-PCR
The efficiency of each primer was determined using
the relative standard curve method. The optimal concentration based
on the efficiency of the primers was selected for relative
quantification (RQ). The ΔΔCT value of CYP1A1,
solute carrier family 7 (anionic amino acid transporter light
chain, xc-system), member 11 (SLC7A11) and UGT1A8
showed a downregulation of 43-, 15-, and 11-fold, respectively.
However, the ΔΔCT value of the protein tyrosine
phosphatase, receptor type, R (PTPRR) and caveolin 1,
caveolae protein, 22 kDa (CAV1) genes showed an upregulation
of 2.2- and 1.5-fold, respectively.
Discussion
Almost 75% of breast cancer patients are estrogen
receptor-positive (6). Tamoxifen
is a selective estrogen-receptor modulator. It has been shown to
reduce invasive breast cancer risk by 50% (7,8).
Despite advances in treatment, there is a need to investigate the
use of new drugs to modulate the expression of the estrogen
receptor in estrogen receptor-positive breast cancer cells.
Furthermore, recurrence develops after certain durations of
treatment. In this study, cDNA microarray technology was used to
measure the expression levels of all genes within the human genome.
Microarray data analysis provided information on the genes and
pathways targeted by 50 μM thymoquinone following 24 h of
treatment.
Steroid hormones play a main role in the maintenance
of normal female function and are normally produced by ovaries
(9). Estradiol has been known to
be the main and most active endogenous estrogen in pre-menopausal
women. It is present in the form of estrone in post-menopausal
women (10) and in pregnant women
it is in the form of estriol (11). It has been reported that elevated
circulating estrogen levels cause increased breast cancer risk
(12).
CYP450 enzymes are haem-containing enzymes which
catalyse phase 1 drug metabolism (13). CYP1A1 is expressed in
extrahepatic tissues, such as mammary glands, the prostate,
ovaries, uterus, colon, testis, adrenal, thymus and lungs (14). The overexpression of CYP1B1
has been reported in mammary tumours and breast cancer (15–17). The enzymes are present in the
estrogen metabolic pathway and are known as 17β-estradiol (E2)
hydroxylases (18). The
conversion of estradiol to 2-hydroxyestradiol (2-OHE2)
and 4-hydroxyestradiol (4-OHE2) is influenced by the
CYP1A1 and CYP1B1 genes, respectively (19,20). 4-OHE2 is known as the
most carcinogenic metabolite of estrogen (21,22). It has been reported to be a vital
factor in hormonal carcinogenesis (23). The CYPIAI gene regulates
the metabolism of estradiol into 2-hydroxyestradiol. It is also
involved in the metabolism of estrone to 4-hydroxyestrone
(4-OHE1). 4-OHE1 is known as one of the most
potent carcinogenic estrogen metabolites (20). Our findings revealed a
downregulation in the expression of the CYP1A1 and
CYP1B1 genes. Further validation with qRT-PCR revealed a
significant downregulation of the CYPIAI gene by 43-fold.
The downregulation of CYP450 enzymes has been reported to suppress
estradiol-2,3-quinone (E2-2,3-Q) and
estradiol-3,4-quinone (E2-3,4-Q) activity (22,24).
Previous studies have reported the anticancer and
anti-angiogenic effects of 2- and 4-methoxyestradiol (2- and
4-MeOE2) (20,25). Increased levels of the methoxy
form of estrone have been shown to have anticancer and
anti-angiogenic potential in humans (20). The formation of 2- and
4-MeOE2 occurs through the methylation of
2-OHE2 and 4-OHE2. The oral administration of
2-MeOE2 in phase I and phase II clinical trials has been
shown to be well tolerated by patients. Its anti-proliferative
effects have been demonstrated in nasopharngeal carcinoma and
tumor-derived uterine leiomyoma cell lines (26,27). The phase I enzyme UGT1A8 plays a
key role in drug metabolism. In this study, we observed a
significant downregulation of the UGT1A8 gene by 16.26-fold.
The validation using real-time PCR on relative gene expression
revealed a downregulation of 11-fold. The downregulation of this
gene would retain 2-MeOE2 levels.
Multidrug resistance is the most important mechanism
by which cancer chemotherapeutic drugs fail to take effect and
resistance develops against anticancer drugs (28,29). The IFN-related DNA damage
resistance signature (IRDS) is associated with resistance to
chemotherapy or radiation in various cancer cell lines. Some of the
IRDS genes are STAT1, ISG15 and IFIT1. They
are associated with the interferon signaling pathway (30). The upregulation of these genes has
been reported in different types of cancer. In our study, whole
genome microarray data analysis revealed that the IFIT1 gene
was downregulated by 10.65-fold. In addition, the IFIT2 and
IFIT3 genes were downregulated by 5.32- and 5.27-fold,
respectively. The overexpression of the IFIT3 and
IFI27 genes has been reported to induced tumour
proliferation, angiogenesis and chemoresistance in pancreatic
carcinoma cells (32). The
findings of our study revealed the downregulation of the
IFI27 and IFI6 genes by 3.19- and 7.86-fold,
respectively following treatment with thymoquinone. The
downregulation of the IFI6 or GIP3 gene has been
shown to reduce MCF-7 cell growth (33).
Interferon-stimulated gene factor 3 (ISGF3)
is regulated by interferon α. ISGF3 genes consist of
STAT1, STAT2 and IRF9. It has been grouped as
ISGF3-α and ISGF3-γ(34,35). ISGF3-γ functions through
type I interferons (IFN-α and IFN-β) (31). The overexpression of IRF9
has been observed in almost half of breast and uterine tumours. It
may be connected to downstream mediators of interferon signaling to
drug resistance (31). In this
study, thymoquinone induced the downregulation of ISGF3
(IRF9) by 2.01-fold. A previous study using cDNA microarray
analysis demonstrated a high expression of the STAT1 and
STAT2 genes in MCF-7 cells that overexpressed
IRF9(31). The
downregulation of IRF9 gene expression may induce the lower
expression of interferon-inducible genes (31). This finding further supports the
chemopreventive effects of thymoquinone. STAT1 is commonly
overexpressed in breast cancer. It is found to be related with
increased resistance to radiation and chemotherapy (36). In breast cancer, the increase in
estrogen levels leads to a higher expression of STAT1 gene
(36).
Glutathione (GSH) is known as a compound that
promotes drug-resistance through the removal of free radicals
(37). The SLC7A11 gene
functions in chemoresistance through the maintenance of
intracellular GSH levels (28).
In this study, we observed a downregulation of SLC7A11 gene
expression by 12.99-fold. Validation using real-time PCR of
relative gene expression revealed a downregulation of 15-fold.
The PTPRR gene is known as an inhibitor of
MAPK (38). We found that
PTPRR was upregulated by 2.20-fold and the myocyte enhancer
factor 2C (MEF2C) gene by 2.22-fold, which promotes
apoptosis. A previous study demonstrated that the PTPRR gene
exerts an inhibitory effect on p44/42 MAPK signaling and
transcription factor AP1 expression in cervical cancer
(39). In this study, we found
that the caspase-10 gene, which is known as one of the
initiator caspases, was upregulated by 1.57-fold. Previously,
caspase cascade activation has been shown to involve caspase-3, -7
and -9, and not caspase-8 in MCF7/DOX cells (40).
In conclusion, our findings suggest that
thymoquinone acts in a synergistic manner involving the estrogen
metabolic and interferon pathways as indicated by our microarray
findings. The co-adminstration of thymoquinone and tamoxifen
requires further investigation in order to fully elucidate the
treatment outcomes.
Acknowledgements
This study was funded by Universiti Sains Malaysia
short-term grant 304/CIPPT/6311033 and Advanced Medical and Dental
Postgraduate Fund. The authors would like to extend their
appreciation to the statistician.
References
|
1
|
IARC. World Cancer Report. Geneva: World
Health Organization; Lyon: 2008, http://www.iarc.fr/en/publications/pdfs-online/wcr/2008/index.php.
|
|
2
|
Banerjee S, Azmi AS, Padhye S, et al:
Structure-activity studies on therapeutic potential of Thymoquinone
analogs in pancreatic cancer. Pharm Res. 27:1146–1158. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Woo C, Loo SY, Gee V, et al: Anticancer
activity of thymoquinone in breast cancer cells: possible
involvement of PPAR-gamma pathway. Biochem Pharmacol. 82:464–475.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gali-Muhtasib H, Diab-Assaf M, Boltze C,
Al-Hmaira J, Hartig R, Roessner A and Schneider-Stock R:
Thymoquinone extracted from black seed triggers apoptotic cell
death in human colorectal cancer cells via a p53-dependent
mechanism. Int J Oncol. 25:857–866. 2004.PubMed/NCBI
|
|
5
|
El-Mahdy MA, Zhu Q, Wang QE, Wani G and
Wani AA: Thymoquinone induces apoptosis through activation of
caspase-8 and mitochondrial events in p53-null myoloblastic
leukemia HL-60 cells. Int J Cancer. 117:409–417. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rugo HS: The breast cancer continuum in
hormone-receptor-positive breast cancer in postmenopausal women:
evolving management options focusing on aromatase inhibitors. Ann
Oncol. 19:16–27. 2007. View Article : Google Scholar
|
|
7
|
Fisher B, Costantino JP, Wickerham DL, et
al: Tamoifen for prevention of breast cancer: report of the
National Surgical Adjuvant Breast and Bowel Project P-1 Study. J
Natl Cancer Inst. 90:1371–1388. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jordan VC: Chemoprevention of breast
cancer with selective oestrogen-receptor modulators. Nat Rev.
7:46–53. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Duursen MB: Estrogen metabolizing enzymes:
biomarkers of exposure, effect and susceptibility for
carcinogenesis (Dissertation). Proefschrift Universiteit Utrecht
Press; 2005
|
|
10
|
de Padua Mansur A, Silva TC, Takada JY, et
al: Long-term prospective study of the influence of estrone levels
on events in postmenopausal women with or at high risk for coronary
artery disease. ScientificWorldJournal. 2012:3635952012.PubMed/NCBI
|
|
11
|
Zhou J and Seidel E: Estrogens induce
visfatin expression in 3T3-L1 cells. Peptides. 31:271–274. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abrahamsson A, Morad V, Saarinen N and
Dabrosin C: Estradiol, tamoxifen, and flaxseed alter IL-1beta and
IL-1Ra levels in normal human breast tissue in vivo. J Clin
Endocrinol Metab. 97:E2044–E2054. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Androutsopoulos VP, Tsatsakis AM and
Spandidos DA: Cytochrome P450 CYP1A1: wider roles in cancer
progression and prevention. BMC Cancer. 9:1872009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Furukawa M, Nishimura M, Ogino D, et al:
Cytochrome p450 gene expression levels in peripheral blood
mononuclear cells in comparison with the liver. Cancer Sci.
95:520–529. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fu J, Weise A, Falany J, et al: Expression
of estrogenicity genes in a lineage cell culture model of human
breast cancer progression. Breast Cancer Res Treat. 120:35–45.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao YN, Zhang W, Chen YC, Fang F and Liu
XQ: Relative imbalances in the expression of
catechol-O-methyltransferase and cytochrome P450 in breast
cancer tissue and their association with breast carcinoma.
Maturitas. 72:139–145. 2012.PubMed/NCBI
|
|
17
|
Gajjar KL, Martin-hirsch PL and Martin FL:
CYP1B1 and hormone-induced cancer. Cancer Lett. 324:13–30. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mikstack R, Rimando AM, Dutkiewicz Z,
Stefanski T and Sobiak S: Design, synthesis and evaluation of the
inhibitory selectivity of novel transresveratrol analogues on human
recombinant CYP1A1, CYP1A2 and CYP1B1. Bioorg Med Chem.
20:5117–6126. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ashley-Martin J, Vanleeuwen J, Cribb A,
Andreou P and Guernsey JR: Breast cancer risk, fungicide exposure
and CYP1A1*2A gene-environment interactions in a
province-wide case control study in Prince Edward Island, Canada.
Int J Environ Res Publ Health. 9:1846–1858. 2012.PubMed/NCBI
|
|
20
|
Williams-Brown MY, Salih SM and Xu X: The
effect of tamoxifen and raloxifene on estrogen metabolism and
endometrial cancer risk. J Steroid Biochem Mol Biol. 126:78–86.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Scornaienchi ML, Thornton C, Willett KL
and Wilson JY: Cytochrome P450-mediated 17b-estradiol metabolism in
zebrafish (Danio rerio). J Endocrinol. 206:317–325. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han EH, Kim HG, Hwang YP, Song GY and
Jeong HG: Prostaglandin E2 induces CYP1B1 expression via
ligand-independent activation of the ERalpha pathway in human
breast cancer cells. Toxicol Sci. 114:204–216. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guengerich PF, Chun Y, Kim D, Gillam E and
Shimada T: Cytochrome P450 1B1: a target for inhibition in
anticarcinogenesis strategies. Mutat Res. 523–524:173–182.
2003.PubMed/NCBI
|
|
24
|
Zahid M, Kohli E, Saeed M, Rogan E and
Cavalieri E: The greater reactivity of estradiol-3,4-quinone vs
estradiol-2,3-quinone with DNA in the formation of depurinating
adducts: implications for tumor-initiating activity. Chem Res
Toxicol. 19:164–172. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lakhani NJ, Sarkar MA, Venitz J and Figg
WD: 2-Methoxyestradiol, a promising anticancer agent.
Pharmacotherapy. 23:165–172. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Newman SP, Foster PA, Stengel C, et al:
STX140 is efficacious in vitro and in vivo in taxane-resistant
breast carcinoma cells. Clin Cancer Res. 14:597–606. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Verenich S and Gerk PM: Therapeutic
promises of 2-methoxyestradiol and its drug disposition challenges.
Mol Pharm. 7:2030–2039. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang Y, Dai Z, Barbacioru C and Sadee W:
Cystine-glutamate transporter SLC7A11 in cancer chemosensitivity
and chemoresistance. Cancer Res. 65:7446–7454. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Krishna R and Mayer LD: Multidrug
resistance (MDR) in cancer: mechanisms, reversal using modulators
of MDR and the role of MDR modulators in influencing the
pharmacokinetics of anticancer drugs. Eur J Pharm Sci. 11:265–283.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Weichselbaum RR, Ishwaran HYT, Nuyten DSA,
et al: An interferon-related gene signature for DNA damage
resistance is a predictive marker for chemotherapy and radiation
for breast cancer. Proc Natl Acad Sci USA. 105:18490–18495. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Luker KE, Pica CM, Schreiber RD and
Piwnica-Worms SD: Overexpression of IRF9 confers resistance to
antimicrotubule agents in breast cancer cells. Cancer Res.
61:6540–6547. 2001.PubMed/NCBI
|
|
32
|
Camaj P, Ischenko I, Seeliger H, Arnold G,
Jauch KW and Bruns CJ: Overexpression of the gene IFIT3 enhances
tumor growth, angiogenesis, metastasing and chemoresistance of the
pancreas carcinoma cells. In: Chirurgisches Forum und DGAV Forum;
2009; 38. Springer; Berlin Heidelberg: pp. 17–18. 2009
|
|
33
|
Cheriyath V, Kuhns MA, Jacobs BS, et al:
G1P3, an interferon- and estrogen-induced survival protein
contributes to hyperplasia, tamoxifen resistance and poor outcomes
in breast cancer. Oncogene. 31:2222–2236. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Veals SA, Schindler C, Leonard D, Fu XY,
Aebersold R, Darnell JE Jr and Levy DE: Subunit of an
alpha-interferon-responsive transcription factor is related to
interferon regulatory factor and Myb families of DNA-binding
proteins. Mol Cell Biol. 12:3315–3324. 1992.
|
|
35
|
Wong LH, Hatzinisiriou I, Devenish RJ and
Ralph SJ: IFN-γ priming up-regulates IFN-stimulated gene factor 3
(ISGF3) components, augmenting responsiveness of IFN-resistant
melanoma cells to type I IFNs. J Immunol. 160:5475–5484. 1998.
|
|
36
|
Englert NA, Spink BC and Spink DC:
Persistent and non-persistent changes in gene expression result
from long-term estrogen exposure of MCF-7 breast cancer cells. J
Steroid Biochem Mol Biol. 123:140–150. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pham AN, Bower PE, Alvardo O, Ravula R,
Gout PW and Huang Y: Pharmacogenomic approach reveals a role for
the x(c)- cystine/glutamate antiporter in growth and celastrol
resistance of glioma cell lines. J Pharmacol Exp Ther. 323:949–958.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lin G, Aranda V, Muthuswamy SK and Tonks
NK: Identification of PTPN23 as a novel regulator of cell invasion
in mammary epithelial cells from a loss-of-function screen of the
‘PTP-ome’. Genes Dev. 25:1412–1425. 2011.PubMed/NCBI
|
|
39
|
Su PH, Lin YW, Huang RL, et al: Epigenetic
silencing of PTPRR activates MAPK signaling, promotes metastasis
and serves as a biomarker of invasive cervical cancer. Oncogene.
32:15–26. 2012.PubMed/NCBI
|
|
40
|
Arafa el-SA, Zhu Q, Shah ZI, Wani G,
Barakat BM, Racoma I, El-Mahdy MA and Wani AA: Thymoquinone
up-regulates PTEN expression and induces apoptosis in
doxorubicin-resistant human breast cancer cells. Mutat Res.
706:28–35. 2011. View Article : Google Scholar : PubMed/NCBI
|