Introduction
microRNAs (miRNAs or miRs) have emerged as one of
the important regulators of the interaction network that controls
various cellular processes. They are short non-coding RNAs which
regulate target mRNAs by binding mostly in the 3′ untranslated
region (3′UTR), leading to either translational repression or
degradation of the target. The aberrant expression of miRNAs has
been reported in multiple human cancer types. miRNAs are known to
play an oncogenic or tumor suppressor role and they have been shown
to play key roles in cell survival, proliferation, apoptosis,
migration and invasion; miRNAs also have various other
characteristics which become altered in human cancers (1,2).
Medulloblastoma is a malignant tumor of the
cerebellum. The median age at diagnosis is 5 years, with the age
range extending into young adulthood. Primary management consists
of surgical resection followed by radiation therapy and
chemotherapy. Current therapies have serious short-term and
long-term adverse effects, including post-operative mutism,
neurocognitive deficits, endocrinopathies, sterility and the risk
of secondary high-grade glioma or meningioma (3). Patients with recurrent disease after
primary therapy have a particularly poor prognosis, with a median
survival of <6 months; the 2-year survival rate among these
patients is approximately 9% (4).
Thus, this necessitates the indepth elucidation of the regulation
of pathways for the better understanding of the disease, which may
lead to the development of personalized, more targeted
therapies.
CD164 (endolyn) is a sialomucin, which has been
shown to play roles in regulating the proliferation, adhesion and
differentiation of hematopoietic stem cells (5). In addition, CD164 is highly
expressed in colon cancer sites, and it promotes colon cancer cell
proliferation and metastasis both in vitro and in
vivo (6). It has also been
suggested that CD164 plays a key role in prostate cancer metastasis
(7). The reduction of the CD164
expression level has become a focus of research in colon cancer.
However, to the best of our knowledge, its role in medulloblastoma
has not been reported to date. Moreover, orthodenticle homeobox 2
(OTX2) is a known oncogene for medulloblastoma, and its aberrant
overexpression has been shown to correlate with a poorer survival
in patients with medulloblastoma (8).
In this study, we first demonstrate that miR-219 not
only inhibits proliferation, but also suppresses the invasion and
migration of D283-MED medulloblastoma cells. Moreover, the
knockdown of miR-219 promotes the proliferation, migration and
invasion of D283-MED cells. Secondly, we predicted that miR-219
targets the 3′UTR of CD164 and OTX2 and then confirmed that it
significantly downregulated the protein expression of CD164 and
OTX2 in D283-MED cells. Finally, we demonstrated that the
proliferation, invasion and migration of D283-MED cells were
promoted by the ectopic expression of CD164.
Materials and methods
Cell line and transfection
The D283-MED medulloblastoma cell line was obtained
from the American Type Culture Collection (ATCC; Manassas, VA,
USA). D283-MED cells were cultured in Improved-MEM (Zn Option)
supplemented with 10% FBS, 2 mmol/l L-glutamine, 2 mmol/l
sodiumpyruvate, 100 U/ml penicillin and 100 mg/ml streptomycin. The
cells were maintained in a humidified atmosphere containing 5%
CO2 at 37°C.
Transfection was performed using Lipofectamine 2000
reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer’s instructions. miR-219 mimics, mimic negative
controls (m-NCs), miR-219 inhibitor and inhibitor negative controls
(i-NCs) were purchased from RiboBio Co., Ltd. (Guangzhou,
China).
A final concentration of 50 nM of mimics or 100 nM
of inhibitor and their respective negative controls were used for
each transfection in MTT assay, BrdU incorporation, invasion and
migration expreriments. CD164 expressiion plasmids and empty vector
(pcDNA3.1) were purchased from the National RNAi Core Facility in
Academic Sinica, Taipei, Taiwan. The concentration of the CD164
expression plasmids or the empty vector (pcDNA3.1) used for each
transfection was 10 μg.
miRNA detection
Total RNA from the cultured cells, with efficient
recovery of small RNAs, was isolated using the mirVana miRNA
Isolation kit (Ambion, Austin, TX, USA). The detection of the
mature form of miRNAs was performed using the mirVana qRT-PCR miRNA
Detection kit and qRT-PCR Primer Sets, according to the
manufacturer’s instructions (Ambion). The U6 small nuclear RNA was
used as an internal control.
Examination of cell proliferation by MTT
assay
The examination of cell proliferation by MTT assay
was performed as described in a previous study (9). The effects of miR-219 on cell
proliferation were assessed by 3-(4, 5-dimethylthiazol-2-yl)-2,
5-diphenyltetrazolium (MTT) assay (Sigma, St. Louis, MO, USA)
assay. The absorbance was directly proportional to the number of
surviving cells. The inhibition rate was calculated as follows:
inhibition rate = (1 − A experiment/A control) ×100%.
Analysis of cell proliferation by BrdU
incorporation
Cell proliferation was assessed using a colorimetric
BrdU cell proliferation kit according to the manufacturer’s
instructions (Cat. no. 11647229001; Roche, Indianapolis, IN, USA).
The transfected cells were labeled with BrdU for 3 to 4 h. The
genomic DNA was fixed and denatured, followed by incubation with
peroxidase-conjugated anti-BrdU antibody for 90 min. A substrate
for the conjugated peroxidase was then added and the reaction
product was quantified by measuring the absorbance. The results
were then normalized by the number of total viable cells.
Migration and invasion assay
For Transwell migration assays, 2.5×104
to 5×104 cells were plated in the top chamber with the
non-coated membrane (24-well insert; pore size, 8 mm; BD
Biosciences, San Jose, CA, USA). For invasion assays,
1.25×105 cells were plated in the top chamber with
Matrigel-coated membrane (24-well insert; pore size, 8 mm; BD
Biosciences). For both assays, the cells were plated in medium
without serum, and medium supplemented with serum was used as a
chemo-attractant in the lower chamber. The cells were incubated for
24 h and cells that did not migrate or invade through the pores
were removed by a cotton swab. Cells on the lower surface of the
membrane were stained with the Diff-Quick Stain Set (Dade Behring,
Inc., Newark, DE, USA) and counted.
Reverse transcription-polymerase chain
reaction
Total RNA was isolated from the cells using TRIzol
reagent (Invitrogen). cDNA was synthesized from 1 μg of total RNA
in a 20 μl reverse transcription (RT) system followed by PCR
amplification in a 50-μl PCR system performed using an RT-PCR kit
(Promega, Madison, WI, USA). The housekeeping gene,
glyceraldehyde-3-phosphate dehydrogenase (GAPDH), was used as an
RNA loading control. The PCR primer sequences were as follows:
CD164 forward, 5′-GTGCTGTCCGCGG ACAAGAAC-3′ and reverse,
5′-TGTGAACAATAGCTCTCATC-3′; OTX2 forward,
5′-TCTTATCTAAAGCAACCGCCTTACGCAGTC-3′ and reverse,
5′-GCACCCTGGATTCTGGCAAGTTGATTTTCA-3′; and GAPDH forward,
5′-TGTCATCAACGGGAAGCCCA-3′ and reverse,
5′-TTGTCATGGATGACCTTGGC-3′.
PCR was conducted according to manufacturer’s
instructions and the PCR products were analyzed by agarose gel
electrophoresis. Gels were photographed and the densities of the
bands were determined using a computerized image analysis system
(Alpha Innotech, San Leandro, CA, USA). The area of each band was
calculated as the integrated density value (IDV). Mean values were
calculated from 3 separate experiments. The IDV ratios of CD164 to
GAPDH and OTX2 to GAPDH were calculated for each sample.
Western blot analysis
Western blot analysis was performed as previously
described (9). The antibodies
used are listed in Table I.
Briefly, following incubation with the primary antibody (Table I) overnight at 4°C,
IRDye™-800-conjugated anti-rabbit secondary antibodies (Li-COR
Biosciences, Lincoln, NE, USA) were used for 30 min at room
temperature. The specific proteins were visualized using an
Odyssey™ Infrared Imaging System (Gene Company).
 | Table IAntibodies used in western blot
analysis. |
Table I
Antibodies used in western blot
analysis.
| Primary antibody | Secondary
antibody |
|---|
| Anti-CD164 (1:500;
Abcam) | Anti-rabbit
secondary |
| Anti-OTX2 (1:500;
Abcam) | antibodies
(Li-COR) |
| Anti-ISL1 (1:500;
Abcam) | |
| Anti-EYA1 (1:500;
Abcam) | |
| Anti-RECK (1:500;
Abcam) | |
| Anti-UBE2N (1:500;
Abcam) | |
| Anti-β-actin (1:500;
Abcam) | |
Microarray analysis
The preparation of RNA from the cells and analysis
of the Affymetrix GeneChip microarray data (Affymetrix, Santa
Clara, CA, USA) were carried out as previously described (10). Total RNA was prepared using TRIzol
reagent (Invitrogen) according to the manufacturer’s instructions.
RNA was further purified using RNeasy columns and treatment with
RNase-free DNase I (Qiagen, Chatsworth, CA, USA). Total RNA was
used to generate cRNA, which was labeled with biotin as recommended
by Affymetrix. cRNA was then hybridized to Affymetrix Hu95A
GeneChips, which contain approximately 12,000 human oligonucleotide
probe sets. After washing, the chips were scanned and analyzed
using MicroArray Suite 5.0 (Affymetrix). The average intensities
for each GeneChip were globally scaled to a target intensity of
150. Further analysis was performed using GeneSpring software
version 5.0.1 to obtain expression level information, fold change,
and P-values for each gene relative to the control, as previously
described (10). Genes with
similar expression profiles were grouped together using
hierarchical clustering and the resulting gene tree is shown. The
magnitude of fold-induction or -repression for each miRNA (relative
to the median of its expression across all experimental samples) is
indicated by the colour bar. Data shown are based on 3 replicate
studies.
Bioinformatics analysis
The analysis of potential microRNA target sites was
carried out using 3 commonly used prediction algorithms: miRanda
(http://www.microrna.org/), TargetScan (http://www.targetscan.org) and PicTar (http://pictar.bio.nyu.edu), as previously described
(11).
Results
Overexpression of miR-219 inhibits the
proliferation, migration and invasion of D283-MED cells
In an attempt to identify the role of miR-219 in
regulating the proliferation of medulloblastoma cells, D283-MED
cells were transfected with miR-219 mimics. Following stable
transfection, miR-219 expression was detected by RT-PCR and the
proliferation rates of the D283-MED cells were determined by MTT
assay. The results revealed that the exogenous miR-219 stably
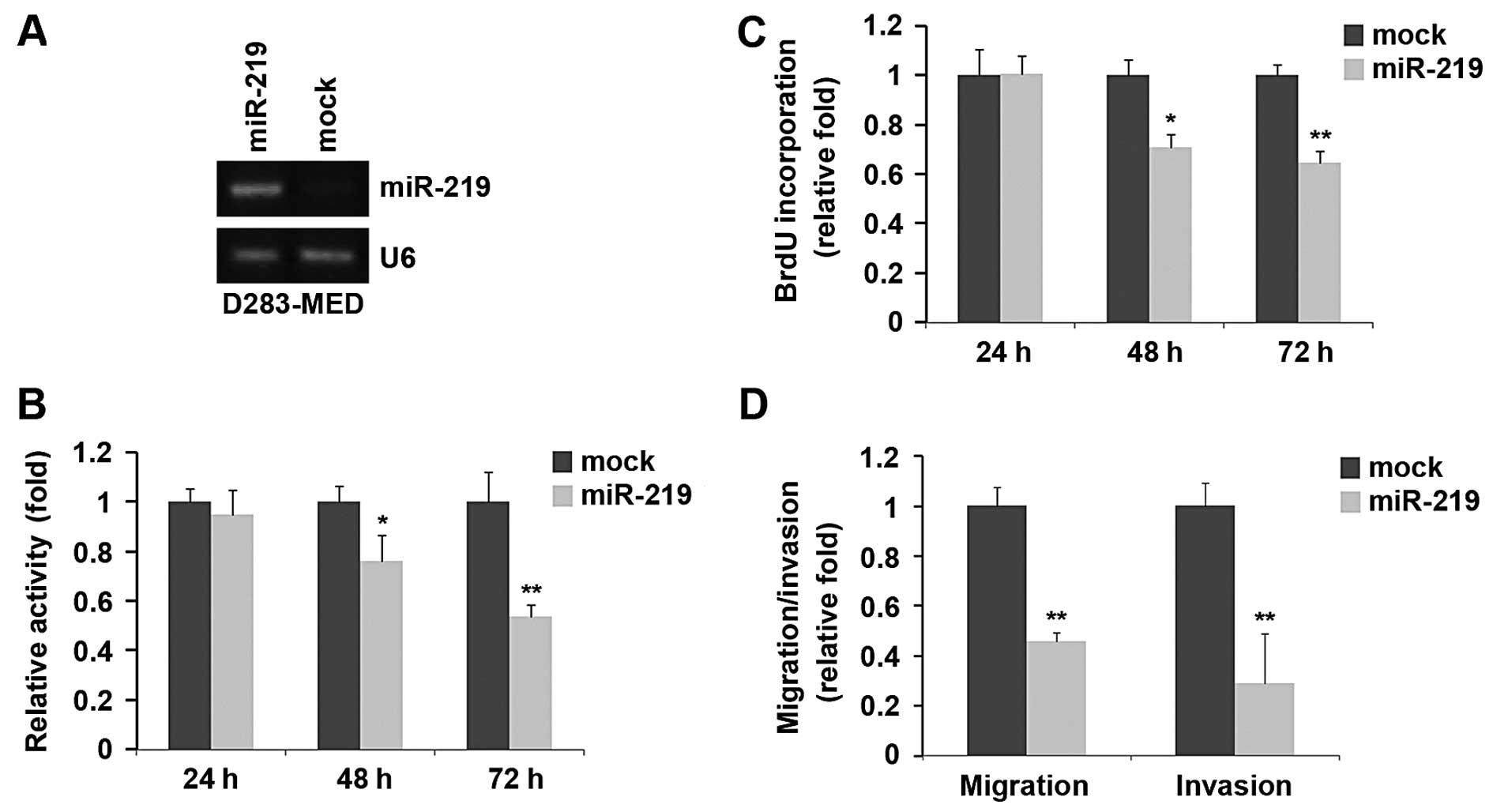
increased its expression in D283-MED cells (Fig. 1A). The overexpression of miR-219
significantly reduced the proliferation rate of the D283-MED cells
after 48 and 72 h of transfection and the inhibition of cell
proliferation was time-dependent (Fig. 1B). This was further revealed by
BrdU incorporation assay, showing that transfection with miR-219
resulted in reduced DNA synthesis activity per viable cell in the
D283-MED cells (Fig. 1C).
Given that miR-219 markedly inhibited D283-MED cell
proliferation, we then sought to determine whether miR-219 has an
effect on the migration and invasion of D283-MED cells. The cell
migration and invasion assay of D283-MED cells revealed that the
overexpression of miR-219 not only inhibited the migration of
D283-MED cells, but also suppressed their invasion (Fig. 1D).
Knockdown of miR-219 promotes the
proliferation, migration and invasion of D283-MED cells
To provide further evidence that miR-219 is involved
in D283-MED cell proliferation, migration and invasion, we examined
the effects of an inhibitor of miR-219. Following transfection,
miR-219 expression was detected by RT-PCR and the proliferation
rates of the D283-MED cells were examined by MTT assay. The results
revealed that miR-219 inhibitior decreased miR-219 expression in
the D283-MED cells. In addition, the proliferation of D283-MED
cells transfected with miR-219 inhibitor was found to be higher
than that of the cells transfected with i-NCs (Fig. 2A and B). Consistent with the
results of MTT assay, BrdU incorporation assay demonstrated that
DNA synthesis was increased by miR-219 inhibitor in the cells
(Fig. 2C). Finally, we found that
miR-219 inhibitor also markedly increased the migration and
invasion of the D283-MED cells (Fig.
2D). miR-219 inhibitors play an opposite role to miR-219 in
regulating the proliferation, migration and invasion of D283-MED
cells.
CD164 and OTX2 protein but not mRNA
expression is suppressed by miR-219 in D283-MED cells
We then performed an analysis of potential microRNA
target sites using 3 commonly used prediction algorithms, miRanda
(http://www.microrna.org/), TargetScan (http://www.targetscan.org) and PicTar (http://pictar.bio.nyu.edu) (11). All 3 algorithms predicted the
CD164, eyes absent homolog 1 (Drosophila) (EYA1), ISL LIM
homeobox 1 (ISL1), lysosomal protein transmembrane 4 alpha
(LAPTM4A), OTX2, reversion-inducing-cysteine-rich protein with
kazal motifs (RECK), T, Brachyury homolog (mouse) (T),
thyrotropin-releasing hormone degrading enzyme (TRHDE) and
ubiquitin-conjugating enzyme E2N (UBE2N) genes to be targets of
miR-219 (Fig. 3A).
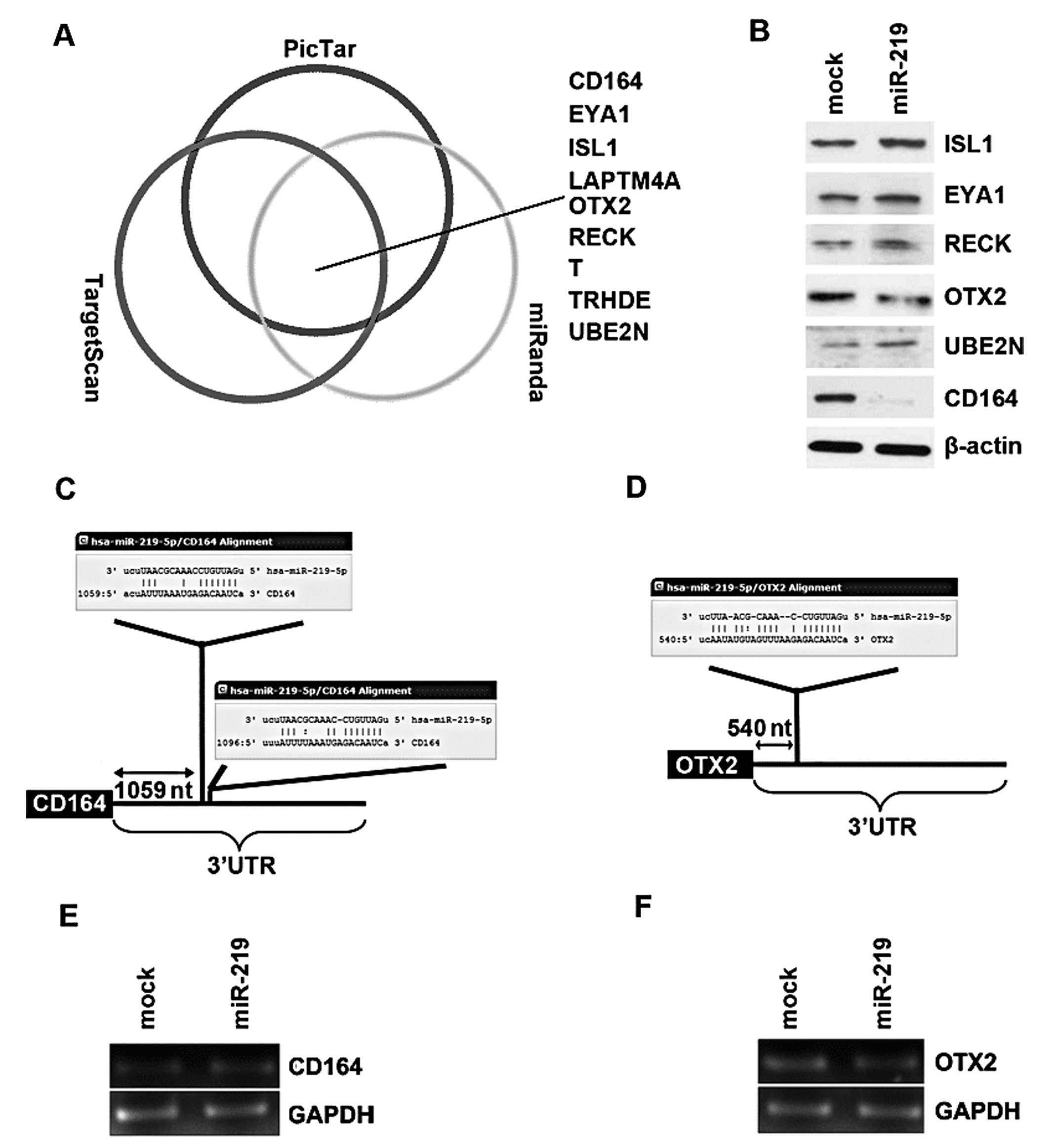 | Figure 3microRNA-219 (miR-219) reduces CD164
and OXT2 expression in D283-MED cells. (A) Diagram showing the
predicted target genes of miR-219 from databases (miRanda,
TargetScan and PicTar). (B) Western blot analysis results of ISL1,
EYA1, RECK, OTX2, UBE2N and CD164 protein expression in D283-MED
cells transfected with miR-219 mimics or mimic negative controls
(m-NCs; mock) at 48 h after transfection, n=3. (C) miRanda,
TargetScan and PicTar predicted that miR-219 targets the 3′UTR of
CD164. (D) miRanda, TargetScan and PicTar predicted that miR-219
targets the 3′UTR of OTX2. (E) Expression of miR-219 reduced CD164
protein expression, but not mRNA levels. D283-MED cells were
transfected with miR-219 mimics or m-NCs and then subjected to
RT-PCR. (F) Expression of miR-219 reduced OTX2 protein expression,
but not mRNA levels. D283-MED cells were transfected with miR-219
mimics or m-NCs and then subjected to RT-PCR. |
In an attempt to further identify the role of
miR-219 in regulating gene expression in medulloblastoma cells, the
D283-MED cells were transfected with miR-219, and then western blot
analysis was performed. Due to the lack of LAPTM4A, T and TRHDE
antibodies, we only detected CD164, OTX2, ISL1, EYA1, RECK and
UBE2N protein expression. We found that miR-219 markedly inhibited
the expression of 2 out of the 6 genes, CD164 and OXT2 in the
D283-MED cells (Fig. 3B). A total
of 2 miR-219 target sites (Fig.
3C) were found in the 3′UTR of CD164 and 1 target site
(Fig. 3D) was found in the 3′UTR
of OTX2 by miRanda, TargetScan and PicTar. Furthermore, RT-PCR
revealed that the mRNA levels of CD164 and OTX2 were not
significantly affected by miR-219 in the D283-MED cells (Fig. 3E and F).
Overexpression of CD164 promotes the
proliferation, migration and invasion of D283-MED cells
We confirmed that miR-219 inhibited proliferation,
migration and invasion, and reduced CD164 expression in D283-MED
cells. We hypothesized that CD164 plays an opposite role to miR-219
in D283-MED cells. Firstly, the D283-MED cells were transfected
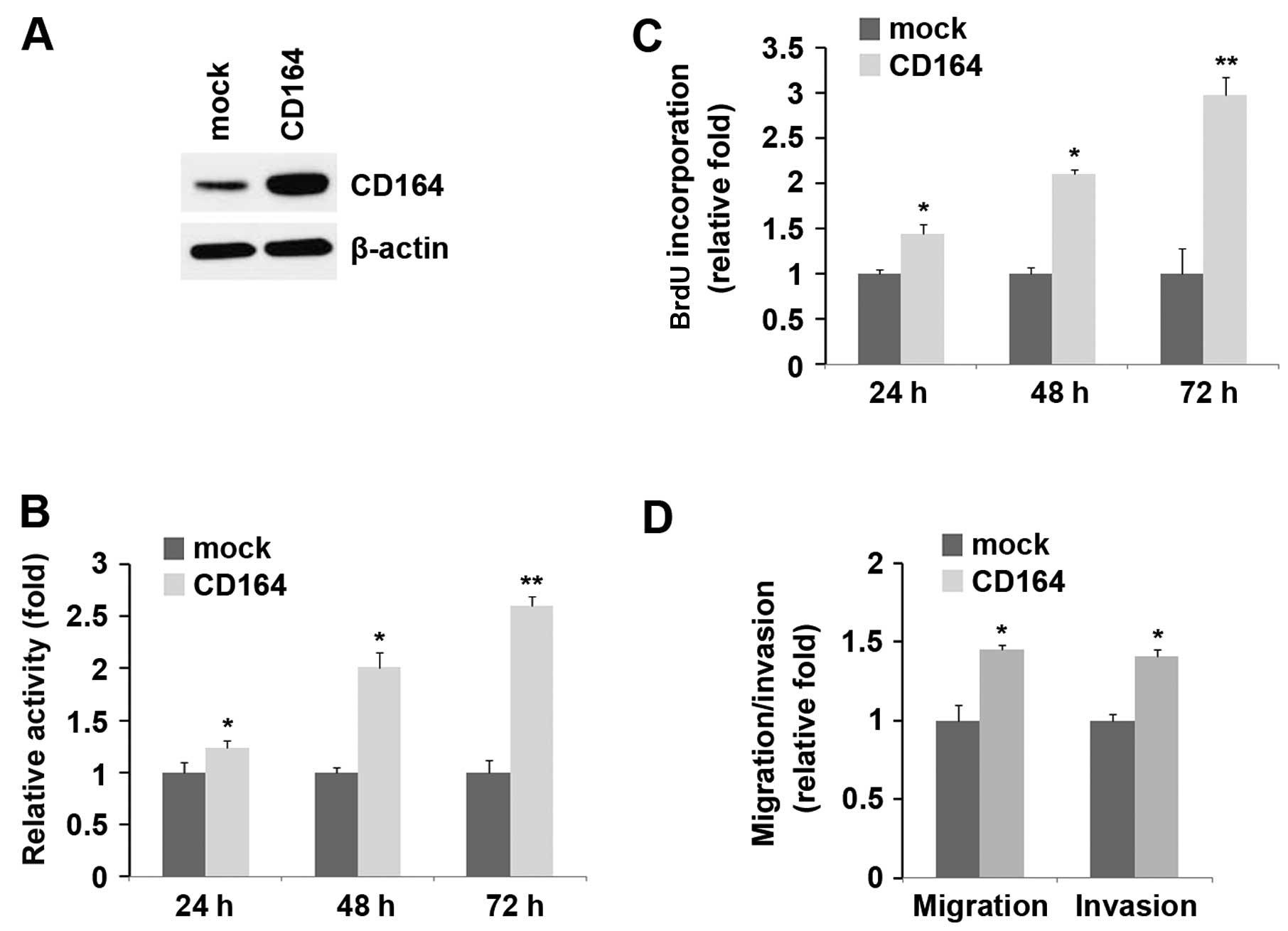
with CD164 expression plasmids. Following transfection, CD164
expression was detected by western blot analysis and the results
revealed that the CD164 expression plasmids markedly increased
CD164 protein expression in the D283-MED cells (Fig. 4A). After 24, 48 and 72 h of stable
transfection, the proliferation rates of the D283-MED cells were
determined by MTT assay. The results revealed that the
overexpression of CD164 markedly increased the proliferation rate
of the D283-MED cells by approximately 25–150% and the promotion of
cell growth was time-dependent (Fig.
4B). This was further revealed by BrdU incorporation assay,
showing that transfection with CD164 resulted in increased DNA
synthesis activity per viable cell and the promotion of cell growth
was time-dependent in the D283-MED cells (Fig. 4C).
Given that CD164 markedly increased the
proliferation of D283-MED cells, we then sought to determine
whether CD164 has any effect on the migration and invasion of
D283-MED cells. Of note, the results of cell migration and invasion
assay revealed that the overexpression of CD164 not only increased
the migration of D283-MED cells, but also promoted their invasion
(Fig. 4D).
CD164 regulates cell cycle-associated
gene expression in D283-MED cells
To identify the cell cycle-associated genes that may
be involved in the CD164-mediated regulation of the proliferation
of D283-MED cells, we used Affymetrix GeneChip DNA microarrays to
compare the gene expression profiles in the D283-MED cells
transfected with CD164 expression plasmids or the empty vector.
RNA was isolated from the D283-MED cells transfected
with CD164 expression plasmids or the empty vector (mock). Total
RNA was used to generate cRNA, which was labeled with biotin as
recommended by Affymetrix. cRNA was then hybridized to Affymetrix
Hu95A GeneChips, which contain approximately 12,000 human
oligonucleotide probe sets. Analysis of the significantly regulated
genes, as described in Materials and methods, identified 99 genes
which were regulated by >2-fold (i.e.,either upregulated or
downregulated) by CD164 in the D283-MED cells. We identified 6
genes, c-myc, PCNA, CDK6, CK19, Ki67 and CDK1, that were commonly
associated with cell cycle regulation (Fig. 5).
Discussion
The role of miRNAs in medulloblastoma has only
recently been addressed and more recently, studies have implicated
miRNAs as important regulators of medulloblastoma cell survival,
proliferation and migration. For example, miR-128a increases the
intracellular reactive oxygen species (ROS) level by targeting
Bmi-1 and inhibits medulloblastoma cell growth by promoting
senescence (12); the pleiotropic
effects of miR-183-96-182 converge to regulate the cell survival,
proliferation and migration of medulloblastoma (13); miR-199b-5p can impair cancer stem
cell growth through the negative regulation of HES1 in
medulloblastoma (14); miR-182
promotes the leptomeningeal spread of non-sonic
hedgehog-medulloblastoma (15);
miR-218 acts as a tumor suppressor by targeting multiple cancer
phenotype-associated genes in medulloblastoma (16); miR-21 suppression impedes
medulloblastoma cell migration (17); the ectopic expression of miR-383
in medulloblastoma cells leads to the suppression of cell growth
(18); miR-124 inhibits cell
proliferation in medulloblastoma cells (19). Recent studies have reported that
miR-219 is required for normal oligodendrocyte
differentiation/myelination and inhibits hepatocellular carcinoma
cell proliferation (20,21). Although the downregulation of
miR-219 expression is known to be involved in the development of
medulloblastoma, the functional role of miR-219 in medulloblastoma
has not been previously described (22).
In this study, we report the novel regulation of
proliferation, migration and invasion by miR-219 through the
specific inhibition of CD164 in medulloblastoma cells. Our data are
in line with those of previous studies, showing that miR-219 is
downregulated in medulloblastoma samples, suggesting a tumor
growth-inhibitory function (22),
suggesting that the overexpression of miR-219 inhibits
proliferation, migration and invasion and that the knockdown of
miR-219 promotes the proliferation, migration and invasion of
D283-MED cells. This further confirms that miR-219 is a tumor
suppressor gene in medulloblastoma.
Further analysis of the possible mechanisms involved
revealed that CD164 is a putative target of miR-219. We
demonstrated that CD164 protein, but not mRNA expression was
downregulated by miR-219 in the D283-MED cells. Consistent with the
results of previous studies on CD164 in prostate and colon cancer,
our results demonstrated that CD164 promoted the proliferation,
migration and invasion of D283-MED cells (4,6).
In addition, the regulation of CD164 by miR-219 was further
supported by the results that CD164 played an opposite role to
miR-219 in regulating the proliferation, migration and invasion of
medulloblastoma cells. OTX2 is known as an oncogene for
medulloblastoma, and its aberrant overexpression has been shown to
correlate with a poorer survival in patients with medulloblastoma
(8). As prevoiusly demonstrated,
the ectopic OTX2 expression enhanced the proliferation and
tumorigenicity of immortalized primary cells, whereas OTX2
knockdown in medulloblastoma cells prolonged the survival of
animals bearing tumor xenografts (8). In this study, we demonstrated that
miR-219 inhibited OTX2 protein expression in D283-MED cells.
In conclusion, in this study, we describe the
functional roles of miR-219 in D283-MED cells, and demonstrate that
miR-219 decreases D238-MED cell proliferation, migration and
invasion by downregulating CD164. It is more meaningful to
demonstrate whether CD164 expression is downregulated in
medulloblastoma and that miR-219 and CD164 are inversely expressed
in medulloblastoma. Moreover, the modulation of miR-219 in in
vivo studies using medulloblastoma models may shed light on the
utility of the restoration of the expression of the tumor
suppressor, miR-219, as a therapeutic modality for
medulloblastoma.
Acknowledgements
This study was financially supported by grants from
the National Natural Science Foundation of China (nos. 31071126,
31000343, 31171303, 31171297 and 31070694).
References
|
1
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar
|
|
2
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
3
|
Crawford JR, MacDonald TJ and Packer RJ:
Medulloblastoma in childhood: new biological advances. Lancet
Neurol. 6:1073–1085. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zeltzer PM, Boyett JM, Finlay JL, et al:
Metastasis stage, adjuvant treatment, and residual tumor are
prognostic factors for medulloblastoma in children: conclusions
from the Children’s Cancer Group 921 randomized phase III study. J
Clin Oncol. 17:832–845. 1999.PubMed/NCBI
|
|
5
|
Doyonnas R, Yi-Hsin Chan J, Butler LH, et
al: CD164 monoclonal antibodies that block hemopoietic progenitor
cell adhesion and proliferation interact with the first mucin
domain of the CD164 receptor. J Immunol. 165:840–851. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tang J, Zhang L, She X, et al: Inhibiting
CD164 expression in colon cancer cell line HCT116 leads to reduced
cancer cell proliferation, mobility, and metastasis in vitro and in
vivo. Cancer Invest. 30:380–389. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Havens AM, Jung Y, Sun YX, et al: The role
of sialomucin CD164 (MGC-24v or endolyn) in prostate cancer
metastasis. BMC Cancer. 6:1952006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Adamson DC, Shi Q, Wortham M, et al: OTX2
is critical for the maintenance and progression of Shh-independent
medulloblastomas. Cancer Res. 70:181–191. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Luo XG, Zou JN, Wang SZ, Zhang TC and Xi
T: Novobiocin decreases SMYD3 expression and inhibits the migration
of MDA-MB-231 human breast cancer cells. IUBMB Life. 62:194–199.
2010.PubMed/NCBI
|
|
10
|
Frasor J, Danes JM, Komm B, Chang KC,
Lyttle CR and Katzenellenbogen BS: Profiling of estrogen up- and
down-regulated gene expression in human breast cancer cells:
insights into gene networks and pathways underlying estrogenic
control of proliferation and cell phenotype. Endocrinology.
144:4562–4574. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sethupathy P, Megraw M and Hatzigeorgiou
AG: A guide through present computational approaches for the
identification of mammalian microRNA targets. Nat Methods.
3:881–886. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Venkataraman S, Alimova I, Fan R, et al:
MicroRNA 128a increases intracellular ROS level by targeting Bmi-1
and inhibits medulloblastoma cancer cell growth by promoting
senescence. PLoS One. 5:e107482010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Weeraratne SD, Amani V, Teider N, et al:
Pleiotropic effects of miR-183~96~182 converge to regulate cell
survival, proliferation and migration in medulloblastoma. Acta
Neuropathol. 123:539–552. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Garzia L1, Andolfo I, Cusanelli E, et al:
MicroRNA-199b-5p impairs cancer stem cells through negative
regulation of HES1 in medulloblastoma. PLoS One. 4:e49982009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bai AH, Milde T, Remke M, Rolli CG,
Hielscher T, Cho YJ, Kool M, Northcott PA, Jugold M, Bazhin AV,
Eichmüller SB, Kulozik AE, Pscherer A, Benner A, Taylor MD, Pomeroy
SL, Kemkemer R, Witt O, Korshunov A, Lichter P and Pfister SM:
MicroRNA-182 promotes leptomeningeal spread of non-sonic
hedgehog-medulloblastoma. Acta Neuropathol. 123:529–538. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Venkataraman S, Birks DK, Balakrishnan I,
Alimova I, Harris PS, Patel PR, Handler MH, Dubuc A, Taylor MD,
Foreman NK and Vibhakar R: MicroRNA 218 acts as a tumor suppressor
by targeting multiple cancer phenotype-associated genes in
medulloblastoma. J Biol Chem. 288:1918–1928. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Grunder E, D’Ambrosio R, Fiaschetti G,
Abela L, Arcaro A, Zuzak T, Ohgaki H, Lv SQ, Shalaby T and Grotzer
M: MicroRNA-21 suppression impedes medulloblastoma cell migration.
Eur J Cancer. 47:2479–2490. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li KK, Pang JC, Lau KM, Zhou L, Mao Y,
Wang Y, Poon WS and Ng HK: MiR-383 is downregulated in
medulloblastoma and targets peroxiredoxin 3 (PRDX3). Brain Pathol.
23:413–425. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li KK, Pang JC, Ching AK, Wong CK, Kong X,
Wang Y, Zhou L, Chen Z and Ng HK: miR-124 is frequently
down-regulated in medulloblastoma and is a negative regulator of
SLC16A1. Hum Pathol. 40:1234–1243. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dugas JC, Cuellar TL, Scholze A, Ason B,
Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT and Barres BA:
Dicer1 and miR-219 are required for normal oligodendrocyte
differentiation and myelination. Neuron. 65:597–611. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang N, Lin J, Ruan J, Su N, Qing R, Liu
F, He B, Lv C, Zheng D and Luo R: MiR-219-5p inhibits
hepatocellular carcinoma cell proliferation by targeting
glypican-3. FEBS Lett. 586:884–891. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ferretti E, De Smaele E, Po A, Di
Marcotullio L, Tosi E, Espinola MS, Di Rocco C, Riccardi R,
Giangaspero F, Farcomeni A, Nofroni I, Laneve P, Gioia U,
Caffarelli E, Bozzoni I, Screpanti I and Gulino A: MicroRNA
profiling in human MB. Int J Cancer. 124:568–577. 2009. View Article : Google Scholar
|