Introduction
Alzheimer’s disease (AD) is a prominent
neurodegenerative disorder characterized by the progressive loss of
memory and other cognitive functions. Despite considerable progress
in genetics and cell biology, there are still a number of
unresolved issues regarding the mechanisms responsible for
neurodegeneration, as well as the molecular and pathological
components. Extracellular amyloid-β (Aβ), which is derived from a
larger protein known as amyloid precursor protein (APP), is
believed to be responsible for the death of neurons and dementia in
AD. The increased APP expression may increase the risk of
developing AD (1,2). APP levels can be regulated at the
genomic, transcriptional or translational level and participate in
the degradation of neurons. Genetic variants in the APP promoter
increase APP transcription by 2 to 3-fold and have been reported to
increase the risk of developing AD. APP can be processed by a group
of secretases, where α-secretase produces soluble fragments, and β-
and γ-secretase generate Aβ from APP (2). Several lines of evidence have
suggested that Aβ regulates neuronal and synaptic activities and
that the accumulation of Aβ in the brain causes an intriguing
combination of aberrant network activity and synaptic depression
(3). The β-secretase, β-site APP
cleaving enzyme (BACE-1), is a rate-limiting enzyme of Aβ
generation and a key target of drugs for AD (1,4,5).
MicroRNAs (miRNAs or miRs) are endogenous, short,
non-coding RNAs, which act as important post-transcriptional
regulators of gene expression by binding with their target mRNAs
and are essential for neuronal function and survival (5,6).
Several miRs have been shown to be important in neuropathology by
downregulating AD-related proteins, such as APP and BACE-1. It has
been demonstrated that miR-16, -101, -106a/b, -147 and -160a
function as APP suppressors; another study using an APP/presenilin
1 (PS1) mutant mouse model of AD revealed a negative correlation
between BACE-1 protein levels and 2 miRs (miR-298 and -328);
miR-298 and -328 downregulated BACE-1 expression through direct
interaction (5–7).
Several cerebral spinal fluid (CSF)- or blood-based
markers, such as Aβ, soluble APP (sAPP)α/sAPPβ, tau, phosphorylated
tau (p-tau) and BACE-1, have been proposed as biomarkers for
predicting future cognitive decline in healthy individuals and the
progression to dementia in patients who are cognitively impaired
(8,9). However, there is still an urgent
need for the availability of more biomarkers that can detect AD in
the predementia phase (1,6,8,9).
The expression of AD-related proteins, such as APP and BACE-1, is
controlled by a number of regulatory factors. The levels of these
regulatory factors may be altered before changes occur in the
levels of AD-related proteins. Thus, regulatory factors, such as
miRs, may serve as potential biomarkers for AD, particulary in the
diagnosis of mild cognitive impairment (MCI). In the present study,
miRs that potentially target the 3′ untranslated region (3′-UTR) of
APP or BACE-1 were predicted using retrieve engines, as previously
described (10). Bioinformatics
analysis revealed that miR-384 was a miR that can potentially
target both the 3′-UTRs of APP and BACE-1. The effects of miR-384
on APP and BACE-1 expression were examined, and the levels of
miR-384 in samples from mice with AD, as well as in patients with
MCI and dementia of Alzheimer’s type (DAT) were measured.
Materials and methods
Study population
This study was approved by the Ethics Committee of
Xuanwu Hospital of Capital Medical University, Beijing, China and
the written informed consent was obtained from all participants.
Thirty-two patients with MCI (13 females, 19 males; mean age,
63.2±6.1 years), 45 patients with DAT (18 females, 27 males; mean
age, 64.2±5.8 years) were included in this study. A total of 50
control subjects, including 28 females and 22 males were also
included (mean age, 63.9±5.7 years). Matched CSF and blood samples
were drawn from 7 patients with DAT (2 females, 5 males; mean age,
65.1±3.5 years) and 7 control subjects (2 females, 5 males; mean
age, 66.2±4.1 years). Venous blood was collected using a vacuum
tube within 2 h after CSF collection. Serum was isolated within 1 h
after blood collection. The samples were stored at −80°C until
analysis. Homocysteine (HCY) and apolipoprotein E (ApoE) levels
were determined using the Hitachi 7600 biochemical analyzer
(Hitachi, Ltd., Tokyo, Japan). The Aβ, tau and p-tau levels were
determined using an ELISA kit (Cusabio Biotech Co., Ltd., Suffolk,
UK).
APP/PS1 double-transgenic and wild-type
(WT) mice
For our study purposes, 3-, 6- and 9-month-old
APP/PS1 double-transgenic mice with a C57BL/6J genetic background
were purchased from the Institute of Laboratory Animal Science,
Chinese Academy of Medical Sciences and Comparative Medical Center,
Beijing, China. All the animal protocols were approved by the
Ethics Committee of Xuanwu Hospital of Capital Medical University.
Non-transgenic mice were used as the WT controls. The mice were
anesthetized with ether and blood was taken by removing the
eyeballs, and then CSF-like fluid was collected as previously
described (11). Briefly, the
mice were sacrificed and their brains were removed into a 35-mm
dish. The cranial cavity and cerebral ventricles (lateral, third
and fourth ventricles) were rinsed with 1 ml PBS, and CSF was thus
harvested with PBS, the washing solution being CSF-like fluid. The
hippocampi were then isolated for miR-384 qPCR detection. The
samples were placed in liquid nitrogen until analysis. There were 5
mice examined in each group.
Cell culture
The SH-SY5Y and HEK293 cell lines were purchased
from the Shanghai Institute of Cell Biology, Shanghai, China. The
cells were grown in antibiotic-free DMEM supplemented with 10% FBS
at 37°C with 5% CO2. Primary mouse hippocampal neurons
were isolated from mouse fetuses. Sixteen-day pregnant WT mice were
sacrificed by CO2 inhalation, and the primary mouse
hippocampal neurons were then isolated as previously described
(12).
Transfection
The cells were transfected with 100 nM (final
concentration) miR-384 mimic oligonucleotide, miR-384 inhibitor
oligonucleotide or a non-specific control siRNA (GenePharma,
Shanghai, China) using Lipofectamine™ 2000 reagent (Invitrogen Life
Technologies, Carsbad, CA, USA) following the manufacturer’s
instructions. Cells treated with Lipofectamine 2000 reagent only
served as the negative control There were 5 samples tested in each
group.
Aβ42 treatment
Synthetic Aβ42 peptides (Bachem, Heidelberg,
Germany) dissolved in PBS were aged by incubation at 37°C for 24 h
with shaking at 1,000 rpm to allow fibril formation, as previously
described (13). The cells were
treated for 0, 12, 24, 36, 48 and 72 h with either a mock treatment
containing PBS or 5 μM aged Aβ42, followed by miR-384 detection.
There were 5 samples tested in each group.
5-aza-deoxycytidine (5-Aza-dC)
treatment
The SH-SY5Y cells were exposed to 0.2 μM 5-Aza-dC
(Sigma, St. Louis, MO, USA) dissolved in dimethyl sulfoxide for 96
h. The medium and the 5-Aza-dC were replaced every day, as
previously described (14). The
levels of miR-384 in the harvested cells were detected by qPCR as
follows: The 0 μM 5-Aza-dC group served as the control. The CpG
islands upstream of miR-384 were analyzed by the CpG Island
Searcher. There were 5 samples tested in each group.
Isolation of mRNA and qPCR analysis
Total RNA from the harvested cells was isolated
using TRIzol Reagent (Invitrogen Life Technologies). The isolated
RNA was reverse transcribed using PrimeScript™ RT reagent (Takara
Bio, Inc., Shiga, Japan). The mRNA expression of APP and BACE-1 was
determined using SYBR®-Green qPCR (Takara Bio, Inc.) in
the Light Cycler 480 System (Roche Diagnostics GmbH, Mannheim,
Germany). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used
to normalize the target genes. The primers used for PCR were as
follows: APP forward, 5′-TTGCGAAACTCATC TTCACTGG-3′ and reverse,
5′-CAGTGGGCAACACACAAA CTCTAC-3′; BACE-1 forward,
5′-AGGCAGTCTCTGGTAT ACACCCATC-3′ and reverse, 5′-TGCCACTGTCCACAATG
CTC-3′; and GAPDH forward, 5′-GCACCGTCAAGGCTGAG AAC-3′ and reverse,
5′-TGGTGAAGACG CCAGTGGA-3′.
Isolation of miRs and qPCR analysis
Total RNA from the CSF, plasma and serum samples was
extracted by a spin column method using the miRNeasy Serum/Plasma
kit (Qiagen, Hilden, Germany) according to the manufacturer’s
instructions. Total RNA in the hippocampal tissues from the animals
and cultured cells was extracted by a spin column method using the
miRNeasy kit (Qiagen). The miRs were reverse transcribed into cDNA
using the miScript II RT kit (Qiagen) in 10-μl reaction system.
miR-384 was detected by the TaqMan qPCR method (Qiagen), using U6
snRNA as an endogenous control.
Reporter vectors and DNA constructs
Reporter vectors containing the putative miRNA
target sites from the APP and BACE-1 3′-UTRs were synthesized with
double-stranded oligonucleotides perfectly complementary to
putative miRNA target sites and oligonucleotides in which the seed
regions were mutated. The APP oligonucleotides had following the
sequence (seed region shown in bold): 5′-CCCAAGCTTTATATTTGT
GATATAGGAATAAGCTTGGG-3′ and 3′-GGGTTCGAAA
TATAAACACTATATCCTTATTCGAACCC-5′. The mutant APP target
oligonucleotides had nucleotides 3 through 6 of the seed region
mutated (italicized): 5′-CCCAAGCTTTATATTT
GTGATATCGTAGTAAGCTTGGG-3′
and 3′-GGGTTCG
AAATATAAACACTATAGCATCATTCGAACCC-5′.
The BACE-1 oligonucleotides had the sequence (seed region shown in
bold): 5′-CCC AAGCTTTGGGATCAAAGCTAGGAAA AAGCTTGGG- 3′
and 3′-GGGTTCGAAACCCTAGTTTC GATCCTTTTTCGAACCC-5′. The
mutant BACE-1 target oligonucleotides had nucleotides 7 through 13
of the seed region mutated (italicized):
5′-CCCAAGCTTTGGTACCCACGCC
ATGCGAAAGCTTGGG-3′ and
3′-GGGTTCGAAACCAT
GGGTGCGGTACGCTTTCGAACCC-5′.
We utilized established methods to clone these synthetic versions
of the putative miRNA target sites into a luciferase reporter gene
(pMIR-REPORT; Ambion Inc., Austin, TX, USA) (11). A total of 10,000 HEK-293 cells
were plated in 24 well plates. The following day, the cells were
transfected with a miRNA mimic oligonucleotide, reporter vectors
bearing either the miRNA target sequence or the miRNA seed region
mutant target sequence, and one tenth of the molar volume of
pRL-SV40, a Renilla luciferase control vector. We utilized
Arrest-In transfection reagent (Open Biosystems Inc., Lafayette,
CO, USA); any differences in transfection efficiency were accounted
for by measuring Renilla luciferase activity. At 48 h
post-transfection, the cells were lysed using 100 μl of GLB (Glo
Lysis Buffer; Promega, Madison, WI, USA). Firefly and Renilla
luciferase activities were measured using a dual luciferase
reporter assay kit (Promega), as per the manufacturer’s
instructions. Firefly luciferase activity was normalized to Renilla
luciferase activity. There were 5 samples examined in each
group.
Western blot analysis
Western blot analysis was performed as previously
described (15). Briefly,
proteins (30 μg/well) were separated by 10% SDS-polyacrylamide gel
electrophoresis (SDS-PAGE; Bio-Rad, Hercules, CA, USA). Proteins in
the gel were transferred to nitrocellulose membranes (Pall Life
Sciences, Port Washington, NY, USA). In the sequence, the membranes
were incubated with anti-APP antibody (diluted 1:400; Abcam,
London, UK), anti-BACE-1 antibody (diluted 1:1,000, Abcam), or
anti-GAPDH antibody (diluted 1:400, Abcam) at room temperature for
1.5 h. The membranes were washed and incubated with anti-IgG
antibody conjugated to horseradish peroxidase at room temperature
for 1 h. Subsequently, the membranes were incubated with substrate
for peroxidase and chemiluminescence enhancer (KPL Inc.,
Gaithersburg, MD, USA) for 1 min and exposed immediately to X-ray
film for 1–5 min. The films were then revealed in the conventional
manner. The amount of each protein was measured by densitometric
analysis and standard relative to the GAPDH. There were 5 samples
examined in each group.
In vitro BACE-1 activity assay
BACE-1 activity was determined by a fluorescence
resonance energy transfer method using a kit from Sigma. The BACE-1
activity in [fluorescence units (FU)] was quantified through the
standard curve. Negative control (no enzyme) reaction and standard
curve blank were contained in the assay. There were 5 samples
examined in each group. All reactions were performed in
duplicate.
Statistical analysis and ROC curve
analysis
Statistical analyses were performed using SPSS 13.0
software for Windows. For normally distributed data, the results
are expressed as the means ± standard deviation (SD); differences
between 2 groups were assessed by t-tests. Differences between
multiple groups were analyzed using the Mann-Whitney U test, while
correlations were determined by computing Spearman rank correlation
coefficients. P-values <0.05 were considered to indicate
significantly different differences.
Results
Bioinformatics analysis
A total of 62 miRs were found to be putatively
target on the 3′-UTR of BACE-1, and 34 miRs were found to be
putatively target on the 3′-UTR of APP. miR-384 was an miR that may
target the 3′-UTRs of BACE-1 and APP (Fig. 1).
MiR-384 suppresses the expression of
BACE-1 and APP
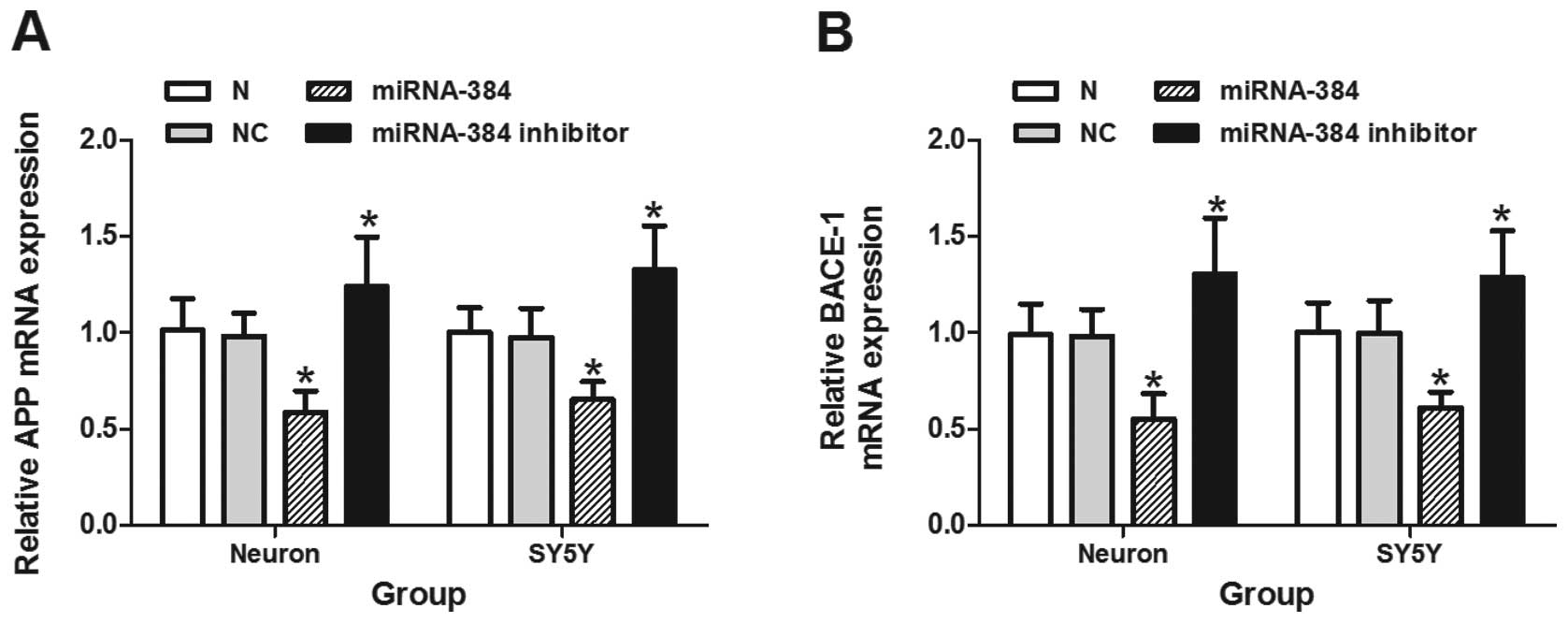
As illustrated in Figs. 2 and 3A–D, both the mRNA and protein
expression of BACE-1 and APP was markedly decreased following
transfection with miRNA-384 mimic oligonucleotide in primary mouse
hippocampal neurons and SH-SY5Y cells (P<0.05). The miR-384
inhibitor oligonucleotide induced a significant upregulation in the
mRNA and protein expression of APP and BACE-1 compared with the
groups transfected with the non-specific control siRNA or the
negative control (P<0.05). We observed a 57% downregulation in
endogenous miR-384 under this condition (Fig. 3E). The activity of BACE-1 in the
primary mouse hippocampal neurons and SH-SY5Y cells was also
significantly suppressed by miR-384 and was significantly increased
by the miR-384 inhibitor oligonucleotide (P<0.05) (Fig. 3F).
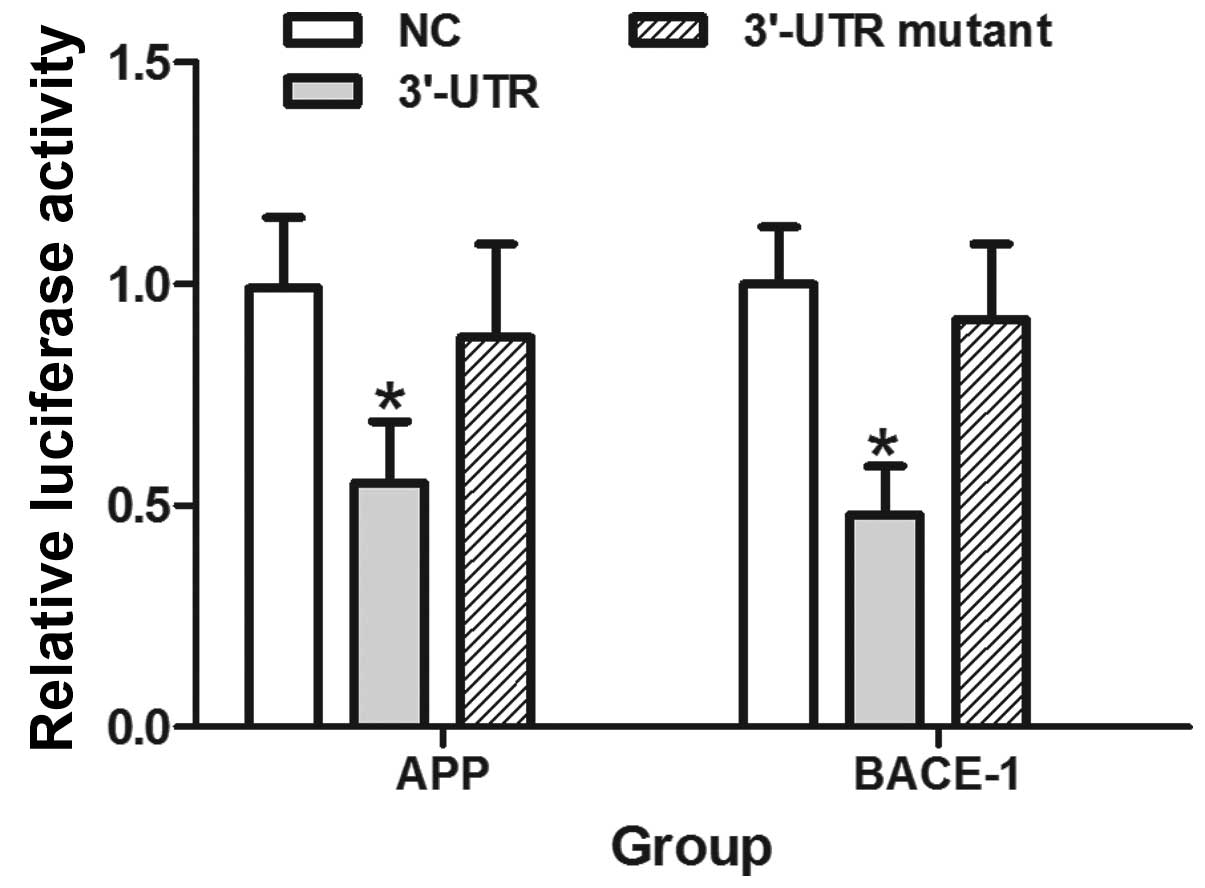
The UTRs of BACE-1 and APP 3′ are both
targets of miR-384
The overexpression of miR-384 significantly reduced
fluorescence from the APP and BACE-1 reporter vectors in the HEK293
cells (P<0.05). These reductions were not observed when the seed
regions mutants of the APP or BACE-1 3′-UTRs were utilized
(Fig. 4).
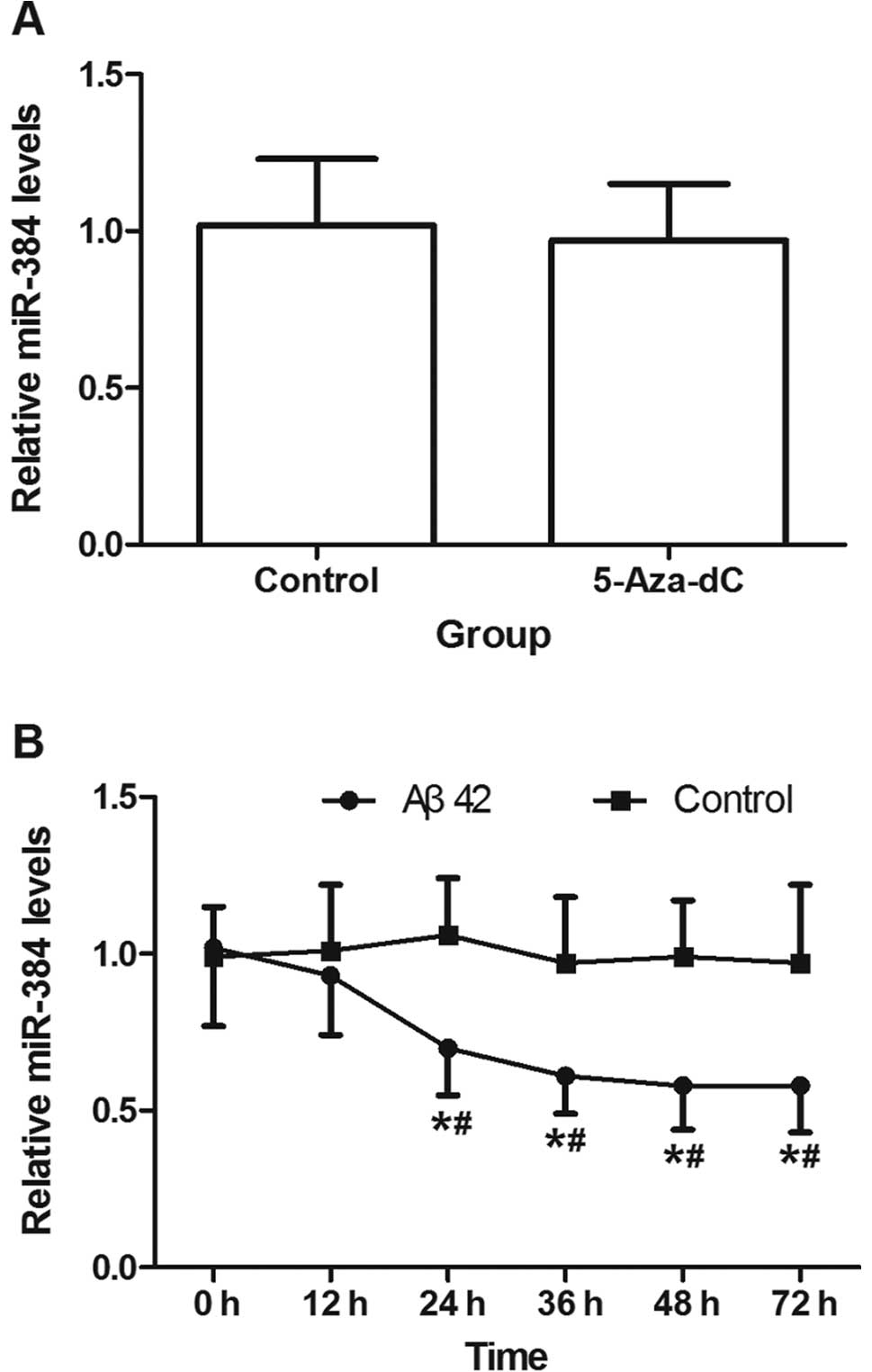
Aβ42 downregulates miR-384 expression in
vitro
5-Aza-dC at a concentration of 0.2 μM did not affect
miR-384 expression in the SH-SY5Y cells (P>0.05) (Fig. 5A). miR-384 expression decreased
after 12 h of incubation with 5 μM aged Aβ42, and continued to
decline until 48 h (Fig. 5B). The
results of bioinformatics analysis suggested that there was no CpG
island upstream of miR-384.
Expression of miR-384 is decreased in the
hippocampus, CSF-like fluid and serum of transgenic mice
The levels of miR-384 were significantly
downregulated in the hippocampi, CSF-like fluid and serum obtained
from the 3-, 6- and 9-month-old APP/PS1 transgenic mice compared
with the WT mice (P<0.05). The levels of miR-384 in the
hippocampi, CSF-like fluid and serum of the 6- and 9-month-old
transgenic mice were significantly lower than those in the
hippocampi, CSF-like fluid and serum from the 3-month-old
transgenic mice (P<0.05) (Fig.
6A–C).
Expression of miR-384 is decreased in the
CSF, serum and plasma of patients with AD
Compared with the control groups, patients with MCI
and DAT had lower levels of miR-384 in the serum and plasma
(P<0.05). The patients with DAT had lower miR-384 levels in
their serum and plasma compared with the MCI groups (P<0.05). We
also found decreased a miR-384 expression in the CSF of patients
with DAT compared with the control group (n=7) (P<0.05)
(Fig. 6D and E).
The levels of miR-384 obtained from plasma or serum
strongly correlated with each other (r=0.957, P<0.05),
indicating that both serum and plasma samples are suitable for
investigations of miR-384 as blood-based biomarkers (Fig. 6F). The levels of miR-384 were
lower in the CSF than those in the serum from a given individual
(P<0.05) (data not shown). There was no correlation observed in
miR-384 expression in the CSF and serum from a given individual
(data not shown). When the cut-off values were set as 0.771 and
0.526 according to the ROC curve analysis, the positive rates of
serum miR-384 were 53.1% (17/32) and 66.7% (30/45) of the MCI and
DAT subjects, respectively.
miR-384 negatively correlates with Aβ42
in serum and CSF
A weak but significant negative correlation was
observed between the levels of miR-384 and Aβ42 in the serum
obtained from patients with MCI (r=−0.372, P<0.05) and DAT
(r=−0.437, P<0.05), as well as the control group (r=−0.463,
P<0.05). A stronger negative correlation was observed between
the levels of miR-384 and Aβ42 in the CSF obtained from patients
with DAT (r=−0.571, P<0.05) and the control group (r=−0.577,
P<0.05). miR-384 expression showed no correlation with HCY,
ApoE, tau and p-tau (data not shown).
Discussion
miRs are integral components of biological networks
with fundamental roles in regulating gene expression (1,2,5).
miR profiles are known to be altered in several regions of the
brain in AD; however, the cause or consequence of the disease
remains unknown. There are no data suggesting a direct genetic link
between miRs or miR recognition elements and neurodegenerative
disease (6–8). The increased expression of APP
correlates with the accelerated accumulation of Aβ in the brain in
AD. As the rate-limiting enzyme of Aβ generation, BACE-1 is a key
drug target for AD. In the present study, we found that miR-384
suppressed the expression of APP and BACE-1 by binding their
3′-UTRs. The activity of BACE-1 was also suppressed by miR-384,
which coincided with the the change in its protein expression. The
suppressive effects of miR-384 on BACE-1 and APP expression may
provide a new direction for targeted therapy for AD. This may allow
researchers to suppress two key proteins using one miR, which may
prove to be more effective with fewer side-effects.
The increase in Aβ42 expression is caused by the
aberrant processing of APP, which is the major constituent of
senile plaques in the brain in AD (16). It has been demonstrated that Aβ42
affects miR profiles (17). miR
downregulation in Aβ-treated hippocampal neurons was observed in
the hippocampus of Aβ-plaque forming APP23 mice at the onset of
plaque formation (13). In this
study, the downregulation of miR-384 expression by Aβ42 in
vitro indicates that excessive Aβ42 generated from transgenic
APP may play a role in the downregulation of miR-384 in APP/PS1
double-transgenic mice, and the decreased expression of miR-384 may
lead to the upregulation of APP. In this manner, a vicious circle
is generated; this vicious circle may play a role in the
progression of AD.
For the majority of diseases, multiple biomarkers
rather than a particular one are required for diagnosis. CSF is in
direct contact with the extracellular space of the brain and can
reflect biochemical changes that occur in the latter (17,18). For these reasons, CSF is the
optimal source of AD biomarkers. However, CSF is not an appropriate
sample for the screening and routine test as requires an invasive
process of sample collection. Previous studies have demonstrated
that miRNAs are stably expressed in animal serum/plasma, and that
their unique expression patterns can serve as ‘fingerprints’ of
various diseases (18,19). Thus, blood-based biomarkers for AD
would be ideal. APP/PS1 double-transgenic mice contain insoluble
amyloid peptides at the age of 6–9 months, concomitant with the
formation of amyloid plaques (19). In this study, we found that the
level of miR-384 was decreased in the hippocampi of 3-month-old
transgenic mice, which suggests that the change in miR-384
expression occurs earlier than the formation of amyloid plaques.
The detection of miR-384 in the CSF-like fluid and serum of 3-, 6-
and 9-month-old transgenic mice demonstrates that miR-384 is a
potential AD biomarker, particularly durin the earlier stages. This
conjecture was further confirmed by clinical detection, which
showed that the miR-384 level in patients with MCI was higher than
the control group and lower than that in patients with DAT in the
CSF and blood samples. This indicates that the level of miR-384 may
change with the progression of AD and may serve as a biomarker for
MCI and DAT.
There is no direct evidence that miRs can move
across the blood-brain barrier (BBB) freely. However, some miRs can
be selectively packaged into microvesicles, exosomes and actively
secreted, and a microvesicle-mediated secretion pathway exists in
BBB (18,20). Although circulating miRs can be
derived from many sources, the decreased secretion of miR-384 may
lead to the decreased level of miR-384 in the blood.
In conclusion, these findings demonstrate that
miR-384 may play a role in the development of AD, and shows great
potential as a novel, non-invasive and easily detected blood-based
biomarker for MCI and DAT.
Acknowledgements
This study was supported by grants from the Natural
Science Foundation of China (no. 81271924) and the Research Fund
for the Doctoral Program of Higher Education of China (no.
20121107110001). The authors would like to thank Dr Shuang Meng of
the Chinese Center for Disease Control and Prevention, Beijing,
China for the vector construction and fluorescence detection.
References
|
1
|
Weiner MW: Dementia in 2012: further
insights into Alzheimer disease pathogenesis. Nat Rev Neurol.
2:65–66. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Singer O, Marr RA, Rockenstein E, et al:
Targeting BACE1 with siRNAs ameliorates Alzheimer disease
neuropathology in a transgenic model. Nat Neurosci. 8:1343–1349.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tan L, Yu JT, Hu N and Tan L: Non-coding
RNAs in Alzheimer’s disease. Mol Neurobiol. 47:382–393. 2013.
|
|
4
|
Junn E and Mouradian MM: MicroRNAs in
neurodegenerative diseases and their therapeutic potential.
Pharmacol Ther. 133:142–150. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schonrock N, Matamales M, Ittner LM and
Götz J: MicroRNA networks surrounding APP and amyloid-β metabolism
- implications for Alzheimer’s disease. Exp Neurol. 235:447–454.
2012.PubMed/NCBI
|
|
6
|
Boissonneault V, Plante I, Rivest S and
Provost P: MicroRNA-298 and microRNA-328 regulate expression of
mouse beta-amyloid precursor protein-converting enzyme 1. J Biol
Chem. 284:1971–1981. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Faghihi MA, Modarresi F, Khalil AM, et al:
Expression of a noncoding RNA is elevated in Alzheimer’s disease
and drives rapid feed-forward regulation of beta-secretase. Nat
Med. 14:723–730. 2008.
|
|
8
|
Blennow K, Hampel H, Weiner M and
Zetterberg H: Cerebrospinal fluid and plasma biomarkers in
Alzheimer disease. Nat Rev Neurol. 6:131–144. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zetterberg H, Andreasson U, Hansson O, et
al: Elevated cerebrospinal fluid BACE1 activity in incipient
Alzheimer disease. Arch Neurol. 65:1102–1107. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schonrock N, Ke YD, Humphreys D, et al:
Neuronal microRNA deregulation in response to Alzheimer’s disease
amyloid-beta. PLoS One. 5:e110702010.PubMed/NCBI
|
|
11
|
Patel N, Hoang D, Miller N, et al:
MicroRNAs can regulate human APP levels. Mol Neurodegener.
3:102008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wei PC, Tsai CH, Chiu PS and Lai SC:
Matrix metalloproteinase-12 leads to elastin degradation in BALB/c
mice with eosinophilic meningitis caused by Angiostrongylus
cantonensis. Int J Parasitol. 41:1175–1183. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schonrock N, Humphreys DT, Preiss T and
Götz J: Target gene repression mediated by miRNAs miR-181c and
miR-9 both of which are down-regulated by amyloid-β. J Mol
Neurosci. 46:324–335. 2012.PubMed/NCBI
|
|
14
|
Lindner DJ, Wu Y, Haney R, et al:
Thrombospondin-1 expression in melanoma is blocked by methylation
and targeted reversal by 5-Aza-deoxycytidine suppresses
angiogenesis. Matrix Biol. 32:123–132. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu CG, Xu KQ, Xu X, et al:
17Beta-oestradiol regulates the expression of
Na+/K+-ATPase beta1-subunit, sarcoplasmic
reticulum Ca2+-ATPase and carbonic anhydrase iv in H9C2
cells. Clin Exp Pharmacol Physiol. 34:998–1004. 2007.PubMed/NCBI
|
|
16
|
Giedraitis V, Sundelöf J, Irizarry MC, et
al: The normal equilibrium between CSF and plasma amyloid beta
levels is disrupted in Alzheimer’s disease. Neurosci Lett.
427:127–131. 2007.PubMed/NCBI
|
|
17
|
Dassow H and Aigner A: MicroRNAs (miRNAs)
in colorectal cancer: from aberrant expression towards therapy.
Curr Pharm Des. 19:1242–1252. 2013.PubMed/NCBI
|
|
18
|
Zhang Y, Liu D, Chen X, et al: Secreted
monocytic miR-150 enhances targeted endothelial cell migration. Mol
Cell. 39:133–144. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
van Leuven F: Single and multiple
transgenic mice as models for Alzheimer’s disease. Prog Neurobiol.
61:305–312. 2000.
|
|
20
|
Ma R, Jiang T and Kang X: Circulating
microRNAs in cancer: origin, function and application. J Exp Clin
Cancer Res. 31:382012. View Article : Google Scholar : PubMed/NCBI
|