Introduction
Great progress has been made in our understanding of
the development of the anterior pituitary gland and of the
mechanisms that underlie the diagnosis of combined pituitary
hormone deficiency (CPHD). Naturally occurring and transgenic
murine models have demonstrated a role for many of these molecules
in the etiology of CPHD (1,2).
Anatomical abnormalities in the pituitary gland may be associated
with other midline abnormalities and variable endocrine disorders,
ranging from isolated growth hormone deficiency (IGHD) to CPHD
(3,4). CPHD is a severe endocrine disorder
in children. Different types and severities of hormonal
deficiencies with various clinical manifestations are observed in
children with CPHD. The definite diagnosis of CPHD is necessary.
Pituitary magnetic resonance imaging (MRI) and hormones are
essential examinations for the diagnosis of CPHD. Significant
advances in molecular biology and the normal development of the
pituitary gland have led to a greater understanding of the genetic
basis of CPHD and related conditions.
PROP1 has been mapped to chromosome 5q and
encodes a protein of 226 amino acids. The DNA-binding homeodomain
consists of 3 α-helical regions and the majority of mutations
reported to date affect this region. PROP1 is essential for
the differentiation of gonadotrophs in fetal life. The spectrum of
gonadotropin deficiency is again extremely variable, ranging from
hypogonadism and the lack of puberty to spontaneous pubertal
development and infertility (5,6).
However, it is has been suggested that PROP1 is not required
for gonadotroph determination, but is required for differentiation.
A 2-bp deletion (delA301, G302) is now believed to be a mutational
‘hot spot’ within PROP1 (7–9).
To date, mutations in PROP1 are associated with growth
hormone (GH), thyrotropin (TSH), prolactin (PRL) and gonadotropin
deficiencies. Fifteen distinct recessive mutations have been
identified in approximately 147 individuals from 76 to 84 pedigrees
originating in 20 different countries, suggesting that mutations
within PROP1 are the most common genetic cause of CPHD, with
incidence rates quoted between 50 and 100% in familial cases of
CPHD (10–12).
Recently, researchers have found a new class of
short, endogenously non-coding RNAs termed microRNAs (miRNAs or
miRs) in animals and plants (13–15). It is now clear that they play
pivotal roles in a wide array of biological processes, including
differentiation and cell proliferation and apoptosis (16,17). They regulate the expression of
protein-coding genes by degrading or inhibiting the translation of
the targeted mRNAs (18).
Emerging evidence strongly suggests that abnormal miRNA expression
is a common and important characteristic of human diseases
(19,20). To date, a number of studies have
proven that a non-invasive approach for the circulating blood-based
miRNA identification of biomarkers is extremely valuable and useful
in diseases (17,19–21).
miRNA profiling using microarray technology has
recently been developed and applied to the study of a variety of
conditions (22,23). Based on these studies, we can now
perform blood-based miRNA profiling to search for CPHD. In this
study, to ascertain whether circulating miRNA expression signatures
can distinguish children with CPHD from normal (healthy) controls,
we performed genome-wide miRNA expression profiling from serum
samples in children with CPHD and healthy controls. Using
expression profile data and data from reverse
transcription-quantitative PCR (RT-qPCR), our study indicates that
the various levels of specific miRNAs, particularly miR-593 and
miR-511 whose direct target is the PROP1 gene, may serve as
non-invasive diagnostic biomarkers for children with CPHD.
Materials and methods
Blood sample collection
A total of 206 participants at the Department of
Pediatrics of Shandong Provincial Hospital Affiliated to Shandong
University (Jinan, China) between 2009 and 2013 were recruited in
this study. This included 103 children with CPHD (88 boys and 15
girls; age, 11.6±3.5 years; range, 8.2–16.6 years) and 103 normal
(healthy) controls (85 boys and 18 girls; age, 11.2±3.8 years;
range, 7.5–16.0 years). There were no significant differences in
the age and gender between the CPHD group and the control group
(P>0.05). All children had at least one anterior pituitary
hormone deficiency in addition to GHD, and were therefore diagnosed
as having CPHD. Whole blood samples (4 ml) were collected from the
children with CPHD and the normal controls into K2-EDTA-coated
tubes. Subsequently, 3 ml of the whole blood was centrifuged at
1,000 × g for 10 min, and then the serum (approximately 1 ml) was
aliquoted into an RNase-free tube. All the serum samples were
stored at -80°C prior to RNA extraction. The study was approved by
the institutional review board of the hospital. Written informed
consent was obtained from the parents of all the subjects.
miRNA microarray
Total RNA from 7 children with CPHD and 7 normal
controls was isolated using TRIzol reagent (Invitrogen, Carlsbad,
CA, USA) and the miRNeasy Mini kit (Qiagen, Hilden, Germany)
according to manufacturer’s instructions. For each sample that
passed RNA quantity measurement using the NanoDrop 1000
spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) 1
μg of total RNA was 3′-end-labeled with Hy3™ fluorescent
label using the miRCURY™ Hy3™/Hy5™ Power Labeling kit (Exiqon,
Vedbaek, Denmark), and hybridized to miRCURY™ LNA Arrays
(version 18.0), according to the manufacturer’s instructions. The
seventh generation of miRCURY™ LNA Arrays (version 18.0)
(Exiqon) contains 3,100 capture probes, covering all human, mouse
and rat miRNAs annotated in miRBase 18.0. In addition, this array
contains capture probes for 25 miRPlus™ human miRNAs. Following
hybridization, the slides were washed several times using the Wash
buffer kit (Exiqon), and dried by centrifugation for 5 min at 400
rpm. The slides were scanned using the Axon GenePix 4000B
microarray scanner (Axon Instruments, Foster City, CA, USA).
Scanned images were imported into GenePix Pro 6.0 software (Axon
Instruments) for grid alignment and data extraction. The data were
normalized using median normalization. Following normalization,
differentially expressed miRNAs were identified through fold change
filtering. Hierarchical clustering was performed using MEV software
(version 4.6, TIGR).
Bioinformatics: sequence analysis
We researched microRNAs that were associated with
children with CPHD through the miRBase (http://microrna.sanger.ac.uk/). Putative targets were
identified using the microrna.org (http://www.microrna.org/microrna/home.do), TargetScan
(http://www.targetscan.org/vert_40/)
and RNAhybrid databases (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html).
Cell lines, culture and transfection
The human embryonic kidney (HEK)293T cells were
preserved at our institute and cultured in Dulbecco’s modified
Eagle’s medium (DMEM; Sigma, St. Louis, MO, USA) supplemented with
10% fetal calf serum (FCS). Exponentially growing cells were used
for the experiments.
Stability-enhanced miRNAs, stability-inhibited
miRNAs and negative control RNA-oligonucleotides were obtained from
Ambion Inc. (Austin, TX, USA). The day prior to transfection, the
HEK293T cells were seeded in antibiotic-free medium. The
transfection of miRNAs was carried out using Lipofectamine 2000 in
accordance with the manufacturer’s instructions (Invitrogen).
Stability-enhanced miRNAs, stability-inhibited miRNAs and negative
control RNA-oligonucleotides were transfected at a final
concentration of 50 nM unless otherwise indicated. The level of
miR-593 and miR-511 expression in the transfected HEK293T cells was
assayed by RT-qPCR 48 h after transfection as described below.
RT-qPCR
Total RNA from was extracted from the serum samples
and cultured cells using a miRNeasy Mini kit (Qiagen) designed to
isolate small molecular weight nucleic acids. The concentration and
purity of the total RNA samples were measured using the SmartSpec
Plus spectrophotometer (Bio-Rad, Hercules, CA, USA). The ratio of
A260:A280 was used to indicate the purity of total RNA.
cDNA was generated using the miScript Reverse
Transcription (RT) kit (Qiagen). According to the manufacturer’s
instructions, 1 μg total RNA, 1 μl miScript reverse
transcriptase mix, 4 μl 5X miScript RT buffer and
appropriate volume RNase-free water were mixed well and incubated
for 60 min at 37°C, and then incubated for 5 min at 95°C to
inactivate miScript reverse transcriptase mix. All reverse
transcription procedures and no-template controls were run at the
same time.
A miScript SYBR-Green PCR kit (Qiagen) was used to
measure the expression of mature miR-593 and miR-511 in the samples
and cells following reverse transcription. Quantitative (real-time)
PCR was performed using on an Mx3005P qPCR System (Stratagene, La
Jolla, CA, USA). Following the manufacturer’s instructions, the 20
μl PCR mixture included 2 μl reverse transcription
product, 10 μl 2X QuantiTect SYBR-Green PCR Master Mix, 2
μl 10X miScript Universal Primer, 2 μl 10X miScript
Primer Assay (for miR-593 and miR-511; Qiagen) and 4 μl
RNase-free water. The reaction mixtures were incubated at 95°C for
15 min, followed by 40 amplification cycles of 94°C for 15 sec,
55°C for 30 sec and 70°C for 30 sec. We also quantified transcripts
of U6 small nuclear RNA using the Hs_RNU6B_2 miScript Primer Assay
(Qiagen) for normalizing the levels of miR-593 and miR-511.
Hs_RNU6B_2 was used as an endogenous control. Each sample was
analyzed 2 times. The threshold cycle (Ct) was defined as the
fractional cycle number at which the fluorescence exceeds the given
threshold. The obtained data were translated into the log2 scale,
as previously described (29).
The 2-ΔΔCt method was used to analyze the relative
expression of the miRNAs.
Western blot analysis
The HEK293T cells were transfected with miR-593
precursor, miR-511 precursor, miR-593 inhibitor, miR-511 inhibitor,
or the negative control in 6-well plates. Following transfection,
the cells were cultured for 48 h, protein was extracted using
mammalian protein extraction reagent (Pierce, Rockford, IL, USA)
supplemented with protease inhibitors cocktail (Sigma). Protein
samples (50 μg) were resolved by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then
transferred onto PVDF membranes. The membranes were blocked with
TBST buffer (TBS plus 0.1% Tween-20) containing 5% w/v non-fat milk
and hybridized with primary antibody (rabbit polyclonal antibody,
ab94500), followed by incubation with specific HRP-conjugated
secondary antibody (anti-rabbit IgG antibody, ab191866). Protein
bands were visualized using the ECL detecting system (Amersham
Biosciences, Uppsala, Sweden). Rabbit polyclonal anti-PROP1
(1:1,000; ab94500; Abcam, Cambridge, MA, USA) and anti-rabbit IgG
antibody (1:1,000; ab191866; Abcam) were used. Monoclonal
anti-GAPDH (1:5,000; ab181602; Abcam) was used as a loading
control.
Luciferase activity assay
A 230-bp fragment of the wild-type PROP1
3′-untranslated region (3′-UTR) containing the putative miR-593 or
miR-511 binding site was amplified by PCR using the following
primers: forward, 5′-ACCAAGCTTGTACCA CCAAGGTGATCCC-3′ and reverse,
5′-ACCACTAGTGCA GGCAGCTCCACCGAGGCATC-3′. A mutant 3′-UTR of
PROP1 was synthesized by PCR, whose sequence contained
5′-TCGTGAATATACAAGAAAATG-3′ or
5′-GCTACTGG AAGAGACAG
GGCAAG-3′ (the letters in italic and bold font indicate nucleotides
which are mutated) and cloned downstream of the luciferase gene in
the pMIR-report luciferase vector (Ambion, Inc.). This construct,
named PROP1-3′-UTR or PROP1-3′-UTR-Mut was used for
the transfection of HEK293T cell lines. HEK293T cells were cultured
in 24-well plates. In each well, 10 ng of Renilla
luciferase, phRL-TK vector (Promega, Madison, WI, USA) were
co-transfected to normalize for transfection effciency. A total of
500 ng of REPORT, PROP1-3′-UTR or PROP1-3′-UTR-Mut
together with 10 nM miR-593 or miR-511 or the negative control was
also co-transfected into the cells in 24-well plates. Transfection
was carried out using Lipofectamine 2000 and Opti-MEM I reduced
serum medium (Life Technologies, Carlsbad, CA, USA) in a final
volume of 0.5 ml. The transfection of the same combinations of
plasmid and RNAs as repeated 3 times. After 48 h, the cells were
harvested with 100 μl PLB reagent (Promega) and 20 μl
cell lysates prepared in Reporter Lysis Buffer (Promega), Firefly
luciferase activity was measured for each well using the Dual
luciferase assay kit (Promega) with an analytical luminometer
(TD-20/20; Turner Designs, Sunnyvale, CA, USA) according to the
manufacturer’s instructions. Briefly, a 10% volume of cell lysate
(20 μl) was added to 100 μl of LAR II, and then the
reaction was terminated by the addition of 100 μl Stop and
Glo® Reagent. Normalized relative luciferase activity
(RLA) was calculated using the following formula: RLA = (Firefly
luciferase)/(Renilla luciferase).
Statistical analysis
The differences between groups were estimated using
the Pearson χ2 test, Student’s t-test and ANOVA test.
Receiver operating characteristic (ROC) curves and the area under
the curve (AUC) calculations were performed to determine the
threshold value of the children with CPHD and the normal controls.
SPSS 15.0 software (SPSS Inc., Chicago, IL, USA) was used for all
statistical analyses, and a value of P<0.05 was considered to
indicate a statistically significant difference. Data are expressed
as the means ± standard deviation from at least 3 separate
experiments.
Results
Global serum miRNA profiling and data
analysis in children with CPHD
miRNA expression profiles from the serum samples of
7 children with CPHD and 7 normal controls were analyzed by
microarray analysis. Hierarchical clustering analyses of the
overall expression profile divided the samples into 2 groups:
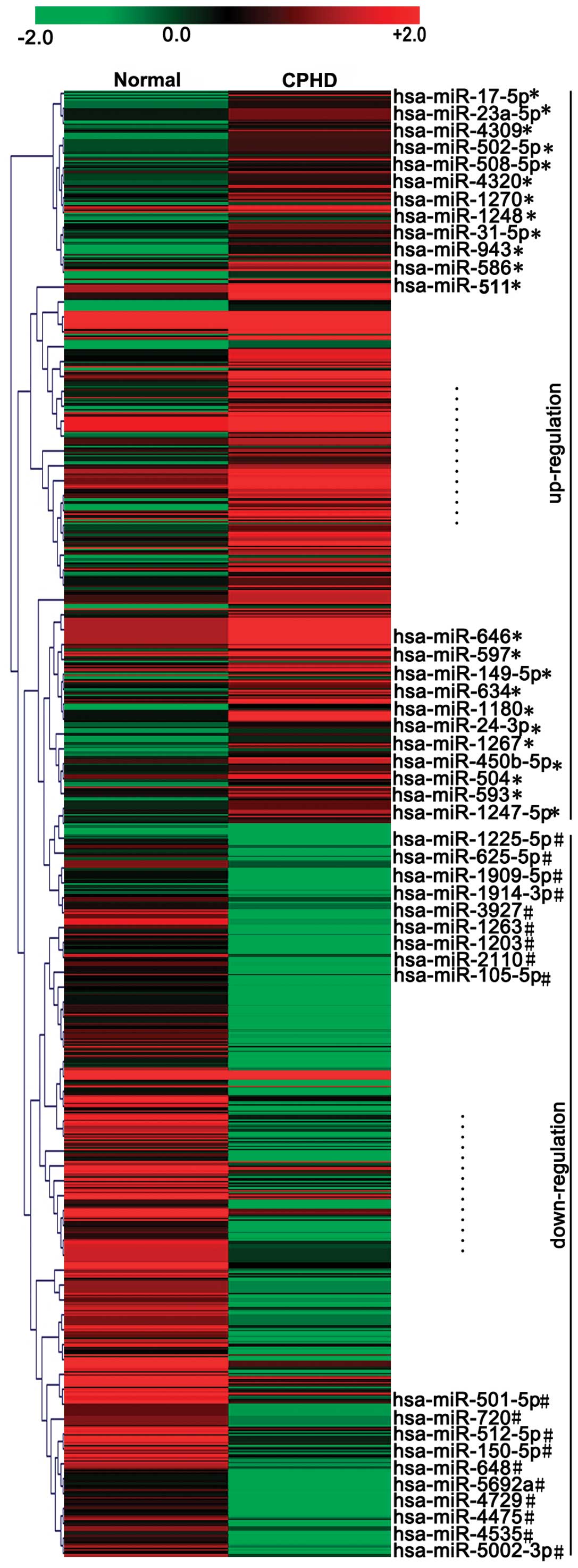
children with CPHD and normal (healthy) controls (Fig. 1). The threshold set for up- and
downregulated miRNAs was a fold change ≥6.0 and a P-value ≤0.05.
Our results revealed 23 upregulated and 19 downregulated miRNAs
with abnormal expression levels in children with CPHD compared with
the normal controls (Fig. 1 and
Table I). The expression levels
of miR-17-5p, miR-593, miR-23a-5p, miR-586, miR-1180, miR-508-5p,
miR-511, miR-646, miR-634, miR-149-5p, miR-24-3p, miR-1267, miR-504
and miR-1270 were upregulated (Fig.
2A) (P<0.05). However, the expression levels of miR-1225-5p,
miR-1909-5p, miR-512-5p, miR-3927, miR-1203, miR-2110, miR-501-5p,
miR-648, miR-4729, miR-4475, miR-1914-3p and miR-5002-5p were
downregulated (Fig. 2B)
(P<0.05) in the children with CPHD compared with the normal
controls.
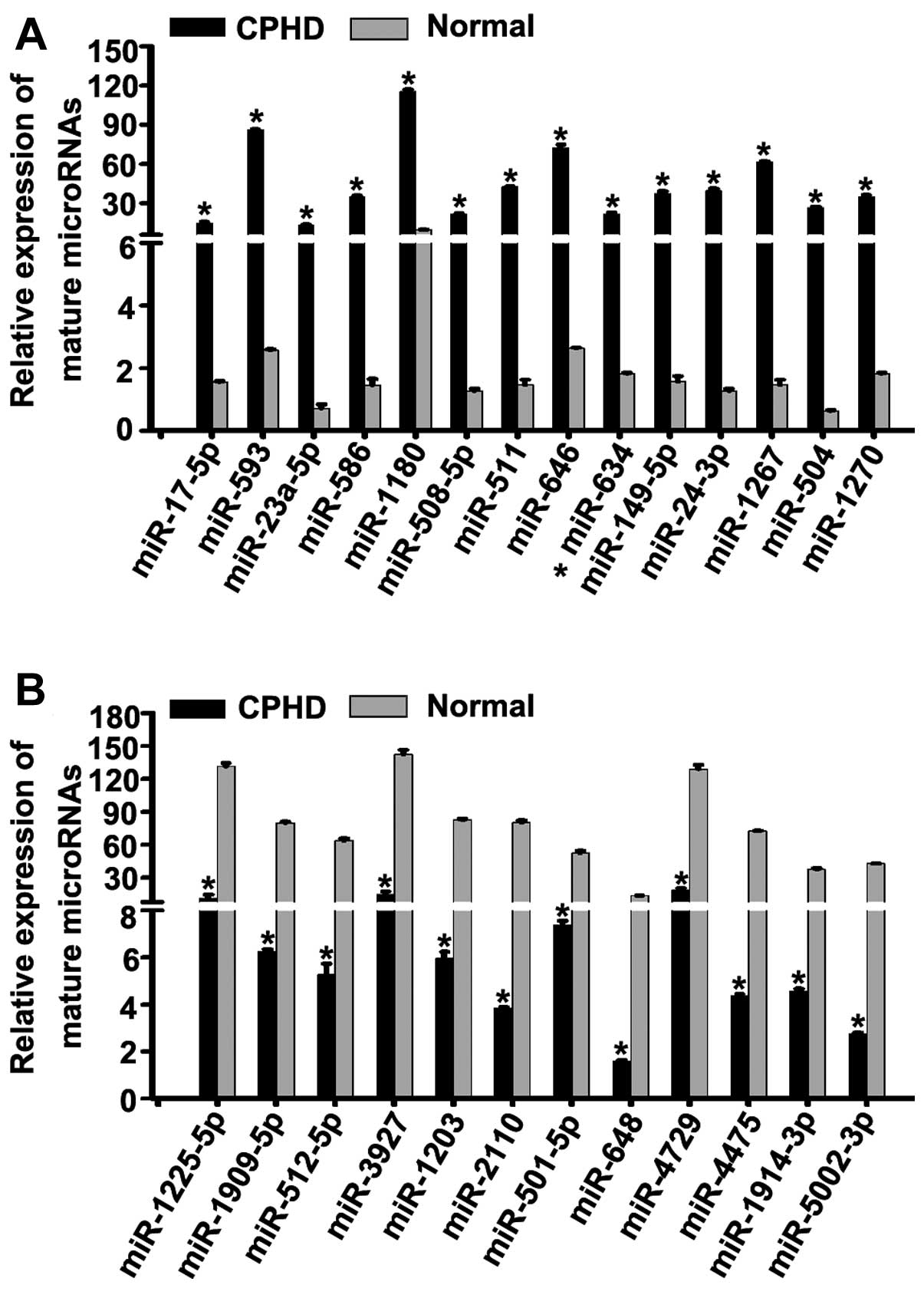 | Figure 2Identification of candidate miRNAs.
(A) We detected the expression levels of 23 upregulated miNAs in
children with combined pituitary hormone deficiency (CPHD) and
normal (healthy) controls by RT-qPCR. The expression levels of
miR-17-5p, miR-593, miR-23a-5p, miR-586, miR-1180, miR-508-5p,
miR-511, miR-646, miR-634, miR-149-5p, miR-24-3p, miR-1267, miR-504
and miR-1270 were upregulated compared to the normal controls. (B)
We examined the expression levels of 19 downregulated miRNAs in
children with CPHD and normal controls by RT-qPCR. The expression
levels of miR-1225-5p, miR-1909-5p, miR-512-5p, miR-3927, miR-1203,
miR-2110, miR-501-5p, miR-648, miR-4729, miR-4475, miR-1914-3p and
miR-5002-5p were downregulated compared to the normal controls.
*P<0.05. Each bar represents the mean value ±
standard deviation from 3 independent experiments. |
 | Table IDifferentially expressed miRNAs in
children with CPHD with at least a 6-fold change in epxression
selected by microarray data analysis. |
Table I
Differentially expressed miRNAs in
children with CPHD with at least a 6-fold change in epxression
selected by microarray data analysis.
| No. | Name | Corrected
P-value | Fold change | Regulation |
|---|
| 1 | hsa-miR-17-5p | 0.00038 | 9.737702 | Up |
| 2 | hsa-miR-23a-5p | 0.00004531 | 7.1713095 | Up |
| 3 | hsa-miR-4309 | 0.000814 | 13.075409 | Up |
| 4 | hsa-miR-502-5p | 0.0000028 | 12.420118 | Up |
| 5 | hsa-miR-508-5p | 0.000018 | 8.289489 | Up |
| 6 | hsa-miR-4320 | 0.000029 | 7.698423 | Up |
| 7 | hsa-miR-1270 | 0.00000478 | 7.7110386 | Up |
| 8 | hsa-miR-1248 | 0.001356 | 16.126984 | Up |
| 9 | hsa-miR-31-5p | 0.000029 | 8.6424675 | Up |
| 10 | hsa-miR-943 | 0.00645 | 7.1091013 | Up |
| 11 | hsa-miR-586 | 0.000781 | 9.900925 | Up |
| 12 | hsa-miR-511 | 0.00000569 | 13.058901 | Up |
| 13 | hsa-miR-646 | 0.0000056 | 6.3428288 | Up |
| 14 | hsa-miR-597 | 0.0000848 | 8.018958 | Up |
| 15 | hsa-miR-149-5p | 0.00264 | 8.6524935 | Up |
| 16 | hsa-miR-634 | 0.000157 | 10.592684 | Up |
| 17 | hsa-miR-1180 | 0.000457 | 11.971244 | Up |
| 18 | hsa-miR-24-3p | 0.0000063 | 12.153815 | Up |
| 19 | hsa-miR-1267 | 0.000487 | 6.773687 | Up |
| 20 |
hsa-miR-450b-5p | 0.000158 | 8.018958 | Up |
| 21 | hsa-miR-504 | 0.0000056 | 6.3428288 | Up |
| 22 | hsa-miR-593 | 0.000158 | 8.018958 | Up |
| 23 |
hsa-miR-1247-5p | 0.0000651 | 6.3428288 | Up |
| 1 |
hsa-miR-1225-5p | 0.000024 | −6.7412977 | Down |
| 2 | hsa-miR-625-5p | 0.000467 | −12.856266 | Down |
| 3 |
hsa-miR-1909-5p | 0.000267 | −11.81031 | Down |
| 4 |
hsa-miR-1914-3p | 0.000984 | −8.549995 | Down |
| 5 | hsa-miR-3927 | 0.000289 | −14.380777 | Down |
| 6 | hsa-miR-1263 | 0.000451 | −9.568466 | Down |
| 7 | hsa-miR-1203 | 0.000167 | −10.962637 | Down |
| 8 | hsa-miR-2110 | 0.000287 | −11.463881 | Down |
| 9 | hsa-miR-105-5p | 0.0000671 | −6.78255 | Down |
| 10 | hsa-miR-501-5p | 0.0000205 | −8.172279 | Down |
| 11 | hsa-miR-720 | 0.000091 | −10.1659565 | Down |
| 12 | hsa-miR-512-5p | 0.00036 | −17.136076 | Down |
| 13 | hsa-miR-150-5p | 0.000497 | −8.718422 | Down |
| 14 | hsa-miR-648 | −7.745552 | −6.179926 | Down |
| 15 | hsa-miR-5692 | −10.1659565 | −7.8253 | Down |
| 16 | hsa-miR-4729 | −7.745552 | −13.084776 | Down |
| 17 | hsa-miR-4475 | −6.1597967 | −9.571002 | Down |
| 18 | hsa-miR-4535 | −7.383092 | −7.945036 | Down |
| 19 |
hsa-miR-5002-3p | −5.559328 | −10.245269 | Down |
Prediction and identification of the
candidate miRNAs, miR-593 and miR-511, which directly target the
PROP1 gene
Among the targets predicted by the microrna.org,
TargetScan databases and RNAhybrid database online search programs,
we identified the PROP1 gene as a possible target of miR-593
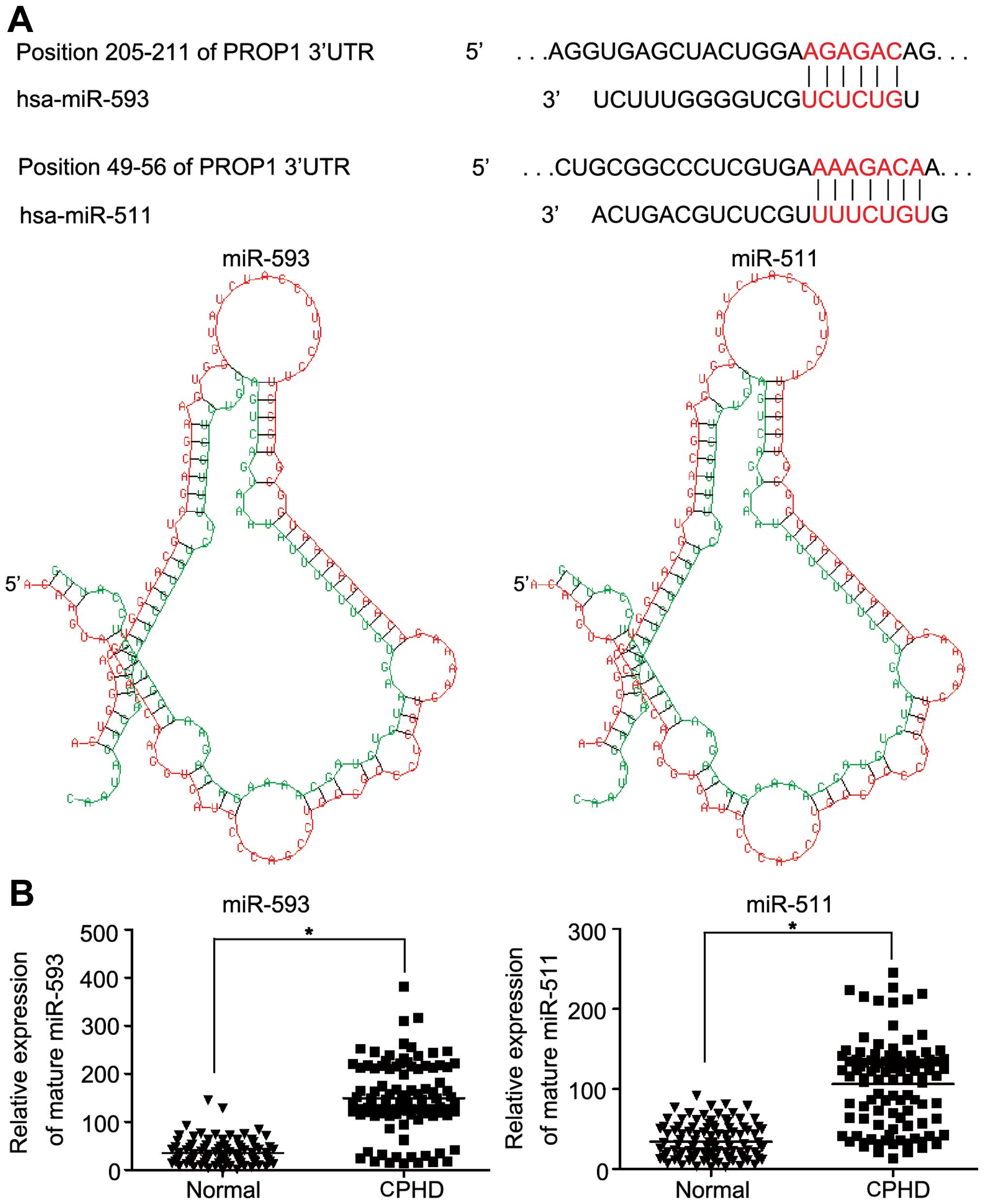
and miR-511 (Fig. 3A). The
results of the database search suggested the association of miR-593
and miR-511 with the PROP1 gene. The expression levels of
miR-593 and miR-511 in the 103 children with CPHD and the normal
controls were examined. The mean expression level of miR-593 was
149.06±72.34 and that of miR-511 was 106.18±54.08. The mean
expression levels of miR-593 (149.06±72.34 vs. 34.89±24.61) and
miR-511 (106.18±54.08 vs. 34.21±21.53) were upregulated compared to
the normal controls (Fig. 3B;
P<0.05).
To verify the direct interaction between miR-593 and
miR-511 and the 3′-UTR of the PROP1 gene, we cloned the
3′-UTR region that was predicted to interact with miR-593 and
miR-511 into a luciferase reporter vector (Fig. 4A). The HEK293T cells were
transfected with the miR-593 precursor, miR-511 precursor, miR-593
inhibitor, miR-511 inhibitor or control oligonucleotides. As is
shown in Fig. 4B, the
upregulation of miR-593 and miR-511 inhibited PROP1 protein
expression by approximately 87%. The luciferase activity of the
reporter plasmid with the wild-type 3′-UTR of the PROP1 gene
was markedly decreased in the cells transfected with the miR-593
precursor and miR-511 precursor compared to the luciferase activity
of the reporter plasmid with the mutant 3′-UTR of the PROP1
gene (Fig. 4C; P<0.05).
Conversely, the luciferase activity of the reporter plasmid was not
affected following transfection with miR-593 inhibitor and miR-511
inhibitor compared to the anti-sense (AS)-miR-control (Fig. 4C) (P<0.05).
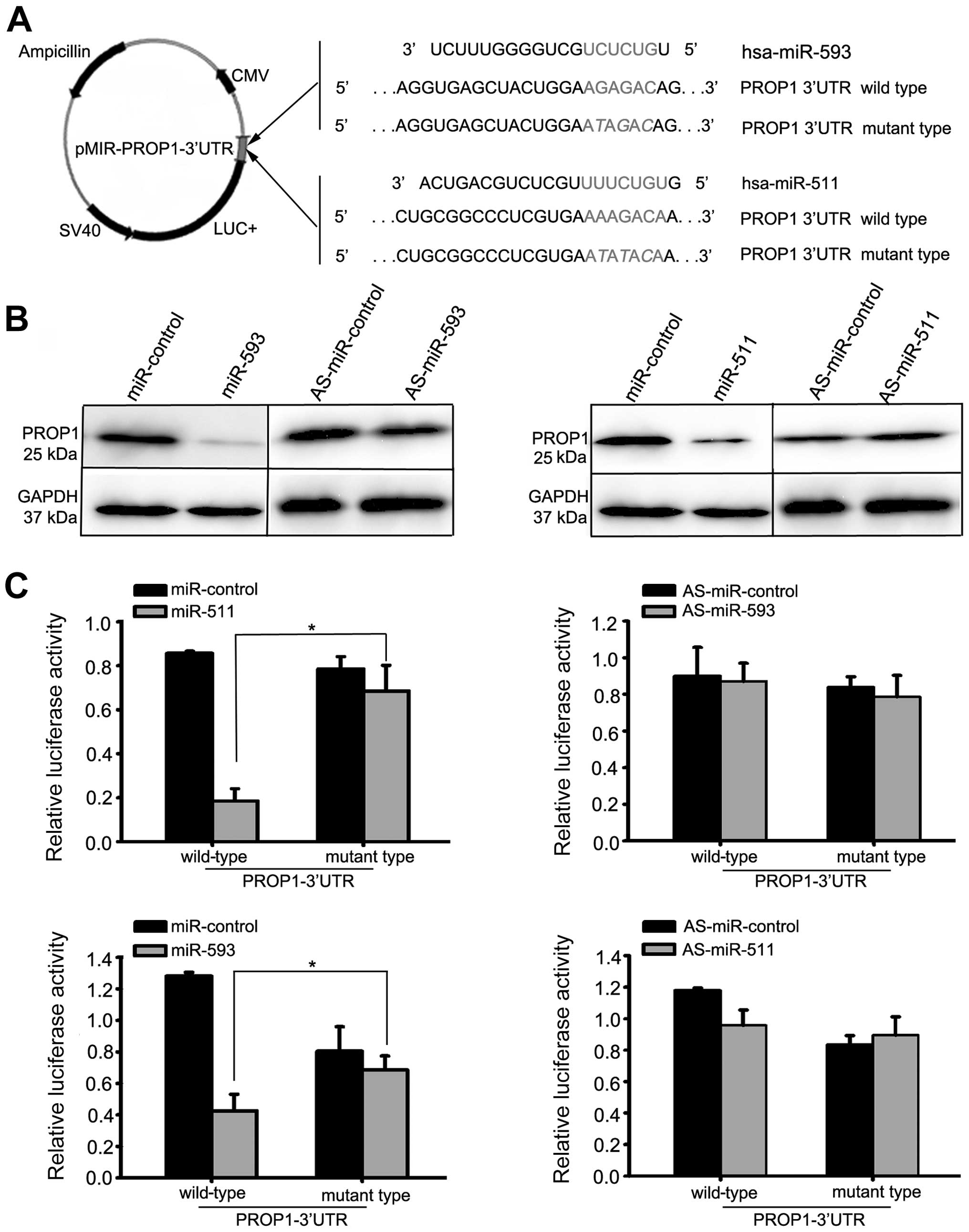 | Figure 4miR-593 and miR-511 target the
3′-untranslated regions (3′-UTR) of the PROP1 gene and
decrease PROP1 expression. (A) Representative nucleotide sequence
matches between possible target sequences and miRNAs. The miR-593
seed sequence (UCUCUG) and miR-511 seed sequence (UUUCUGU) is shown
(gray and italic nucleotides). Schematic graph of the 3′-UTR
binding site for miR-593 or miR-511. PROP1-3′-UTR-wild-type
or PROP1-3′-UT-mutant type was inserted downstream of the
luciferase of pMIR-reporter vector. (B) At 48 h after the
transfection of miR-593 precursor, miR-511 precursor, miR-593
inhibitor, miR-511 inhibitor or control oligonucleotides into the
HEK293T cells, the PROP1 protein level was significantly reduced in
the cells transfected with miR-593 precursor or miR-511 precursor,
as shown by western blot analysis. By contrast, transfection with
miR-593 inhibitor or miR-511 inhibitor did not affect the protein
expression of PROP1. (C) We assessed the luciferase activity by
co-transfecting the luciferase reporter vector bearing the 3′-UTR
of the PROP1 gene with the miR-593 precursor, miR-511
precursor, miR-593 inhibitor, miR-511 inhibitor or control
oligonucleotides. Luciferase activity of reporter plasmid with
wild-type 3′-UTR of the PROP1 gene was markedly decreased in
the cells transfected with miR-593 precursor and miR-511 precursor,
compared with the luciferase activity of the reporter plasmid with
mutant 3′-UTR of PROP1. Each bar represents the mean values
± SD from 3 independent experiments (*P<0.05). |
miR-593 and miR-511 as serum biomarkers
for children with CPHD
The expression of miR-593 and miR-511 in the
children with CPHD was significantly increased compared with the
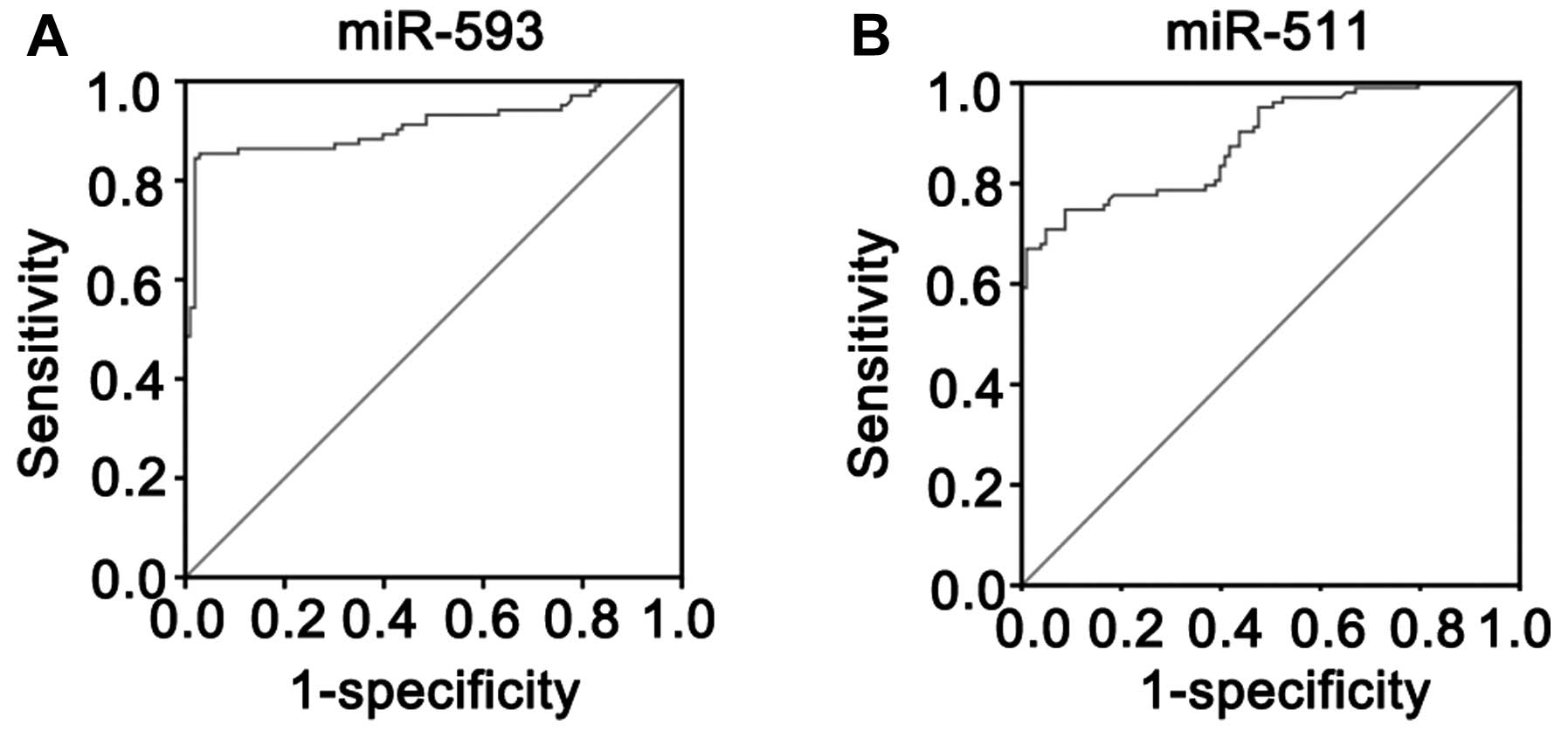
normal controls. The AUC was 0.912±0.020 for miR-593 and was
significantly higher than that of the null hypothesis (true area
was 0.5) (Fig. 5A; P<0.01).
The AUC was 0.785±0.023 for miR-511 and was significantly higher
than that of the null hypothesis (true area was 0.5) (Fig. 5B; P<0.01). According to
Youden’s index, the optimal operating point of the expression level
of miR-593 and miR-511 in the serum was 108.27 and 63.43,
respectively. At this cut-off point, the sensitivity was 82.54 and
84.86%, and the specificity was 98.15 and 91.36% for miR-593 and
miR-511, respectively.
Discussion
The pituitary gland produces hormones that play
important roles in the development and the homeostasis of the body.
A deficiency of two or more of these pituitary hormones, known as
CPHD, may present in infants or children due to an unknown etiology
and is considered congenital or idiopathic. To date, to the best of
our knowledge, there is no study available on miRNAs in CPHD.
miRNAs are a class of small non-coding RNAs and have been
discovered in animals and plants. miRNAs are 19–22 nucleotide
non-coding RNAs and are able to bind complementary sequences in the
3′-UTR of target mRNAs to induce their degradation or translational
repression (24–26). They are encoded by genes that are
presumably transcribed into single or clustered primary
transcripts, which are processed and produce mature miRNAs. They
function as regulators of disease initiation, progression and
metastasis. miRNAs are a novel class of regulatory molecules with
the ability to control gene expression at the post-transcriptional
level. They appear decrease protein expression by blocking the
translation of mRNAs into proteins. Therefore, the identification
of CPHD-specific miRNAs is critical for understanding their role in
the pathophysiological basis of CPHD and may prove useful for
finding novel therapeutic methods. Intriguingly, it has been
suggested that miRNAs are present in human peripheral blood in a
consistent, reproducible and stable manner (19). More importantly, a series of
studies have demonstrated that at least in some pathological
conditions, such as cancer, heart failure and liver damage,
circulating miRNAs may in part reflect tissue damage (13,14,27,28). This discovery opens up the
possibility of using miRNAs as non-invasive biomarkers for
CPHD.
Hitherto, there data on the association between
miRNAs and CPHD are limited. Microarray analysis is a powerful
technology that is able to perform genome-wide analysis in one
experiment. miRNA expression profiles can characterize miRNAs that
are differentially regulated under different experimental
conditions. Thus, in this study, we used a global miRNA microarray
to identify specific miRNAs in CPHD. Our results revealed 23
upregulated and 19 downregulated miRNAs that were abnormally
expressed in children with CPHD compared with the normal controls.
The expression levels of miR-17-5p, miR-593, miR-23a-5p, miR-586,
miR-1180, miR-508-5p, miR-511, miR-646, miR-634, miR-149-5p,
miR-24-3p, miR-1267, miR-504 and miR-1270 were upregulated. The
expression levels of miR-1225-5p, miR-1909-5p, miR-512-5p,
miR-3927, miR-1203, miR-2110, miR-501-5p, miR-648, miR-4729,
miR-4475, miR-1914-3p and miR-5002-5p were downregulated in the
children with CPHD. These data indicate that the characteristics of
serum miRNA expression are associated with children with CPHD and
provide a valuable repertoire that can be used to discover
circulating miRNA-based biomarkers for the diagnosis of CPHD. To
better understand the role of specific miRNAs in the diagnosis of
CPHD, the microrna.org, TargetScan and RNAhybrid database online
searching programs were used to predict putative targets. We
identified that the PROP1 gene was a possible target of
miR-593 and miR-511; further experimental procedures were carried
out to validate these findings. Our results revealed that the
expression levels of miR-593 or miR-511 in the serum of children
with CPHD were upregulated compared with the normal controls.
miR-593 and miR-511 directly targeted the PROP1 gene and
this was confirmed by western blot analysis and luciferase activity
assay following the transfection of HEK293T cells with miR-593
precursor, miR-511 precursor, miR-593 inhibitor, miR-511 inhibitor
or control oligonucleotides. The above findings support the
hypothesis that miRNAs may be involved in the development of CPHD.
In clinical practice, the tissue in children with CPHD is not
readily accessible. The ROC curve and the AUC for the expression
levels of the serum of the 103 children with CPHD and the 103
normal controls were calculated. The AUC was 0.912±0.020 for
miR-593 and the AUC was 0.785±0.023 for miR-511. Thus, the
expression levels of miR-593 and miR-511 in serum may serve as a
molecular marker for children with CPHD. To identify an optimal
cut-off point to detect children with CPHD, Youden’s index was used
in this study. According to Youden’s index, the optimal operating
point of the blood expression level of miR-593 and miR-511 was
108.27 and 63.43, respectively. At this cut-off point, the
sensitivity was 82.54 and 84.86%, specificity was 98.15 and 91.36%
for miR-593 and miR-511, respectively. Taken together, these data
suggest that miR-593 and miR-511 directly target the PROP1
gene and may serve as serum biomarkers for children with CPHD.
In conclusion, in the present study, we found that
the levels of miR-593 and miR-511 in the serum of children with
CPHD were significantly increased and that these miRNAs directly
targeted the PROP1 gene. This suggests that circulating
levels of miR-593 or miR-511 may serve as novel biomarkers for the
clinical diagnosis of CPHD. Thus, miRNAs cannot be overlooked as a
class of molecules that regulate biological functions and CPHD. Our
data broaden the understanding of the functions of miRNAs in
children with CPHD. Considering the small sample size used in the
present study, investigations including a larger scale of patients
are warranted. Additionally, further studies are required to reveal
the exact time course of miR-593 and miR-511 in the serum of
children with CPHD. Although circulating miRNA levels can be
detected by real-time PCR, the underlying mechanisms responsible
for the increased circulating miRNA levels and whether they have
pathophysiological functions in CPHD require further
investigation.
Acknowledgments
We would like to thank Professor Qiaoming Zhi for
providing technical support with the in vitro experiments.
We are grateful to our patients and the families who consented to
be a part of this study. This study was supported in part by grants
from the National Youthful Science Foundation of China (no.
81101858), and the Shandong Science and Technology Commission of
China (no. 2013GSF11817).
References
|
1
|
Kandemir N, Vurallı D, Taşkıran E, Gönç N,
Özön A, Alikaşifoğlu A and Yılmaz E: Frequency of mutations in
PROP-1 gene in Turkish children with combined pituitary hormone
deficiency. Turk J Pediatr. 54:570–575. 2012.
|
|
2
|
Kelberman D, Turton JP, Woods KS, et al:
Molecular analysis of novel PROP1 mutations associated with
combined pituitary hormone deficiency (CPHD). Clin Endocrinol
(Oxf). 70:96–103. 2009. View Article : Google Scholar
|
|
3
|
Reynaud R, Albarel F, Saveanu A, et al:
Pituitary stalk interruption syndrome in 83 patients: novel HESX1
mutation and severe hormonal prognosis in malformative forms. Eur J
Endocrinol. 164:457–465. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tauber M, Chevrel J, Diene G, et al:
Long-term evolution of endocrine disorders and effect of GH therapy
in 35 patients with pituitary stalk interruption syndrome. Horm
Res. 64:266–273. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cruz JB, Nunes VS, Clara SA, Perone D,
Kopp P and Nogueira CR: Molecular analysis of the PROP1 and HESX1
genes in patients with septo-optic dysplasia and/or pituitary
hormone deficiency. Arq Bras Endocrinol Metabol. 54:482–487. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ward RD, Raetzman LT, Suh H, Stone BM,
Nasonkin IO and Camper SA: Role of PROP1 in pituitary gland growth.
Mol Endocrinol. 19:698–710. 2005. View Article : Google Scholar
|
|
7
|
Cogan JD, Wu W, Phillips JA III, et al:
The PROP1 2-base pair deletion is a common cause of combined
pituitary hormone deficiency. J Clin Endocrinol Metab.
83:3346–3349. 1998.PubMed/NCBI
|
|
8
|
Cohen LE, Wondisford FE, Salvatoni A,
Maghnie M, Brucker-Davis F, Weintraub BD and Radovick S: A ‘hot
spot’ in the Pit-1 gene responsible for combined pituitary hormone
deficiency: clinical and molecular correlates. J Clin Endocrinol
Metab. 80:679–684. 1995.PubMed/NCBI
|
|
9
|
Wu W, Cogan JD, Pfaffle RW, et al:
Mutations in PROP1 cause familial combined pituitary hormone
deficiency. Nat Genet. 18:147–149. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Arroyo A, Pernasetti F, Vasilyev VV, Amato
P, Yen SS and Mellon PL: A unique case of combined pituitary
hormone deficiency caused by a PROP1 gene mutation (R120C)
associated with normal height and absent puberty. Clin Endocrinol
(Oxf). 57:283–291. 2002. View Article : Google Scholar
|
|
11
|
Deladoëy J1, Flück C, Büyükgebiz A, et al:
‘Hot spot’ in the PROP1 gene responsible for combined pituitary
hormone deficiency. J Clin Endocrinol Metab. 84:1645–1650.
1999.
|
|
12
|
Parks JS, Brown MR, Hurley DL, Phelps CJ
and Wajnrajch MP: Heritable disorders of pituitary development. J
Clin Endocrinol Metab. 84:4362–4370. 1999.PubMed/NCBI
|
|
13
|
Heneghan HM, Miller N, Lowery AJ, Sweeney
KJ, Newell J and Kerin MJ: Circulating microRNAs as novel minimally
invasive biomarkers for breast cancer. Ann Surg. 251:499–505. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tijsen AJ, Creemers EE, Moerland PD, de
Windt LJ, van der Wal AC, Kok WE and Pinto YM: MiR423-5p as a
circulating biomarker for heart failure. Circ Res. 106:1035–1039.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vlassov VV, Laktionov PP and Rykova EY:
Circulating nucleic acids as a potential source for cancer
biomarkers. Curr Mol Med. 10:142–165. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cortez MA and Calin GA: MicroRNA
identification in plasma and serum: a new tool to diagnose and
monitor diseases. Expert Opin Biol Ther. 9:703–711. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hoekstra M, van der Lans CA, Halvorsen B,
et al: The peripheral blood mononuclear cell microRNA signature of
coronary artery disease. Biochem Biophys Res Commun. 394:792–797.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tan KS, Armugam A, Sepramaniam S, Lim KY,
Setyowati KD, Wang CW and Jeyaseelan K: Expression profile of
MicroRNAs in young stroke patients. PLoS One. 4:e76892009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen X, Ba Y, Ma L, et al:
Characterization of microRNAs in serum: a novel class of biomarkers
for diagnosis of cancer and other diseases. Cell Res. 18:997–1006.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zampetaki A, Kiechl S, Drozdov I, et al:
Plasma microRNA profiling reveals loss of endothelial miR-126 and
other microRNAs in type 2 diabetes. Circ Res. 107:810–817. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo X, Guo L, Ji J, et al: miRNA-331-3p
directly targets E2F1 and induces growth arrest in human gastric
cancer. Biochem Biophys Res Commun. 398:1–6. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ratert N, Meyer HA, Jung M, et al: miRNA
profiling identifies candidate mirnas for bladder cancer diagnosis
and clinical outcome. J Mol Diagn. 15:695–705. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sethi S, Kong D, Land S, Dyson G, Sakr WA
and Sarkar FH: Comprehensive molecular oncogenomic profiling and
miRNA analysis of prostate cancer. Am J Transl Res. 5:200–211.
2013.PubMed/NCBI
|
|
24
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lau NC, Lim LP, Weinstein EG and Bartel
DP: An abundant class of tiny RNAs with probable regulatory roles
in Caenorhabditis elegans. Science. 294:858–862. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee RC and Ambros V: An extensive class of
small RNAs in Caenorhabditis elegans. Science. 294:862–864. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Si H, Sun X, Chen Y, Cao Y, Chen S, Wang H
and Hu C: Circulating microRNA-92a and microRNA-21 as novel
minimally invasive biomarkers for primary breast cancer. J Cancer
Res Clin Oncol. 139:223–229. 2013. View Article : Google Scholar :
|
|
28
|
Wang K, Zhang S, Marzolf B, et al:
Circulating microRNAs, potential biomarkers for drug-induced liver
injury. Proc Natl Acad Sci USA. 106:4402–4407. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang GK, Zhu JQ, Zhang JT, et al:
Circulating microRNA: a novel potential biomarker for early
diagnosis of acute myocardial infarction in humans. Eur Heart J.
31:659–666. 2010. View Article : Google Scholar : PubMed/NCBI
|