Introduction
MicroRNAs (miRNAs or miRs) are short non-coding RNAs
(~22 nt long) that can repress translation by binding imperfectly
to their target mRNA. The miRNAs are transcribed and processed by
Drosha and Dicer and are then loaded into an RNA-induced silencing
complex (RISC) that leads to the regulation of translation
(1).
Available evidence suggests that miRNAs are involved
in the regulation of a wide range of biological processes,
including cell proliferation, apoptosis, cell differentiation and
embryonic development (2–4). The important regulatory roles of
miRNAs during chondrogenesis were recently identified. miR-92a has
been reported to be highly enriched in chondrogenic progenitors,
and it has been found that its inactivation stabilizes the mRNA
expression of the bone morphogenetic protein (BMP) antagonist gene,
noggin3, leading to the repression of BMP signaling and the
abnormal function of chondrogenic progenitors, which results in the
unsustainable survival of chondrogenic progenitors (5). The downregulation of miR-181b was
identified during the chondrogenic differentiation of transforming
growth factor (TGF)-β3-stimulated limb mesenchymal cells to
negatively regulate chondrocyte differentiation by reducing matrix
metalloproteinase (MMP)-13 expression and inducing the expression
of type Ⅱ collagen (6). In a
comparison of the changes occurring in miRNA expression levels
during the chondrogenesis of mesenchymal stem cells (MSCs) in
vitro, it was discovered that the expression of miR-140 was
significantly altered (7).
Furthermore, it was also discovered that miR-140 stimulated
chondrogenesis in vitro by targeting Ras-related small
GTPases (RALA) and thereby affecting SRY (sex determining region
Y)-box (SOX)9 at the protein level (7). Another study on miR-140 demonstrated
that equine cord blood-derived mesenchymal stromal cells expressed
significantly higher levels of miR-140 after 14 days of
chondrogenic differentiation (8).
Furthermore, chemokine ligand 12 and disintegrin and
metalloproteinase with thrombosponin motifs were verified as direct
targets of miR-140 (8). The
functional role of miR-23b was also found to be the induction of
chondrogenic differentiation through the negative inhibition of
protein kinase A (PKA) signaling (9).
However, the majority of previous studies have
focused on miRNA expression profiles in bone marrow-derived MSCs or
stromal cells (10–12). Human adipose-derived stem cells
(hADSCs) display a differentiation capacity that shows their
potential for use in regenerative medical and tissue engineering
applications due to their easy accessibility, isolation and
expandability to clinical scales in a comparatively short period of
time (13,14). It has been demonstrated that the
differentiation potential of hADSCs resembles that of MSCs. The
similarities between these adult stem cells extend to the
biochemical levels, including multiple surface proteins, such as
CD29 and CD44 (10,15).
In this study, we examined the expression profiles
of miRNAs during the chondrogenic differentiation of hADSCs using a
miRNA microarray. The differentially expressed miRNAs between the
undifferentiated hADSCs and chondrogenically differentiated hADSCs
were verified by northern blot analysis. We then predicted their
putative target genes through bioinformatics analysis and confirmed
1 miRNA target. The results of the present study provide new
insight into the function of miRNAs during the chondrogenic
differentiation of hADSCs.
Materials and methods
Isolation of hADSCs and induction of
chondrogenic differentiation
For the isolation hADSCs, 3 samples of adipose
tissue were obtained from donors who underwent elective liposuction
or other abdominal surgery at the First Affiliated Hospital of
Chongqing Medical University (Chongqing, China). All the donors
provided written informed consent and signed approval forms, and
the study was approved by the relevant ethics committee. Healthy
hADSCs (passage 3) were harvested, resuspended in incomplete
chondrogenic medium (basal medium) at a density of
2.5×107 cells/ml, placed in a 24-well plate, and allowed
to adhere at 37°C for 90 min. Subsequently, 500 ml of complete
chondrogenic medium were added to each well. The complete
chondrogenic medium (Cyagen Biosciences, Inc., Santa Clara, CA,
USA) contained Dulbecco’s modified Eagle medium/mutrient mixture
F-12 (DMEM/F-12), 5 ng/ml fibroblast growth factor (FGF)-2, 10
ng/ml TGF-β1, 50 μg/ml Vc and 10−7 M
dexamethasone. After 24 h of incubation, the cell droplets
coalesced and became spherical. The complete medium was changed
every 3 days.
The cell surface markers, CD29, CD44, CD49 and CD45,
were detected using a flow cytometer (Beckman Coulter, Inc., Brea,
CA, USA). The corresponding fluorescein isothiocyanate
(FITC)-conjugated monoclonal mouse anti-human antibodies were as
follows: CD29 (1:100, Cat. no. 119-15141; Raybiotech, Inc.,
Norcross, GA, USA), CD44 (1:100, Cat. no. 119-15548; Raybiotech,
Inc.), CD49 (1:100, Cat. no. ABIN118708, antibodies-online.com,
Aachen, Germany) and CD45 (1:100, Cat. no. 119-15144; Raybiotech,
Inc.). The cells were incubated with the relevant antibodies at 4°C
for 20 min, washed with phosphate-buffered saline (PBS), 0.1%
NaN3 and 5% FBS, and then analyzed using a
fluorescence-activated cell sorting (FACS) Calibur (Becton
Dickinson, Franklin Lakes, NJ, USA). The data were analyzed using
CellQuest3.1f software (Becton Dickinson).
Immunohistochemical examination
hADSCs (passage 3) before and after the induction of
chondrogenesis were subjected to immunohistochemical examination.
The cells were washed 3 times with PBS and fixed with 95% alcohol
for 10 min at room temperature. The cells were permeabilized with
permeable solution for 10 min. After washing, endogenous peroxidase
was quenched in the cells with 3% H2O2 in
ethanol for 10 min, washed 3 times with Tris-buffered saline (TBS,
pH 7.6), incubated with blocking solution for 10 min and then
incubated with monoclonal mouse anti-human antibodies against
collagen type II (ColII; ab3092; Abcam, Cambridge, UK) for 2 h.
Subsequently, the cells were washed 3 times with TBS and incubated
with a biotin-conjugated second antibody for 10 min. After washing
with TBS, the cells were added to an enzyme (HRP
streptavidin)-conjugated anti-biotin solution for 10 min and
washed. The proteins were displayed using AEC mounting solution
(Fuzhou Maixin Biotechnology Development Co., Ltd., Fuzhou, China)
and then visualized and photographed.
Total RNA extraction
Total RNA from the hADSCs prior to (controls) and
after chondrogenic induction was isolated using the mirVana
kit® (Applied Biosystems, Foster City, CA, USA)
according to the manufacturer’s instructions, and the concentration
was determined by the ratio of the absorbance at 260 to that at 280
nm using a Nanodrop® ND-1000 spectrophotometer (Thermo
Scientific, Waltham, MA, USA).
Western blot analysis
Western blot analysis was performed to determine the
protein expression of BMPR2, SOX2 and GAPDH. Total protein from the
cells were lysed by RIPA buffer. GAPDH was used as an endogenous
normalizer. Polyclonal rabbit anti-human BMPR2 (1:1000, sc-20737;
Santa Cruz Biotechnology, Santa Cruz, CA, USA), SOX2 (1:1000;
sc-17319; Santa Cruz Biotechnology) and GAPDH (1:1000; sc-367714,
Santa Cruz Biotechnology) antibodies were used.
miRNA microarray analysis
After the RNA was isolated from the hADSCs prior to
and after chondrogenic induction were compared, miRNA labeling was
then performed using the miRCURY™ Hy3™/Hy5™ Power labeling kit
(Exiqon, Vedbaek, Denmark). The Hy3TM -labeled samples
were then hybridized at 56°C overnight on a miRCURY™ LNA Array
(v.16.0) (Exiqon), which contains probes for 1,223 human miRNAs, in
a 12-Bay Hybridization System (Nimblegen Systems, Inc., Madison,
WI, USA). Following hybridization, the slides were washed 5 times
using a wash buffer kit (Exiqon) and dried. The slides were then
scanned using a Axon GenePix 4000B microarray scanner (Axon
Instruments, Foster City, CA, USA). We performed a fold change
filtering to determine the differential expression of the miRNAs
before and after the induction of the differentiation of the hADSCs
in all 3 sets of hADSCs. The threshold used to screen the up- or
down-regulated miRNAs was a fold change ≥2.0. Average linkage
hierarchical clustering was performed to obtain clusters of data
sets and generate a heatmap, using Gene Cluster and Treeview
software (http://www.eisenlab.org/eisen/).
Northern blot analysis of miRNA
expression
Total RNA was isolated using TRIzol reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA) according to the
manufacturer’s instructions. For the miRNA northern blot analyses,
15 μg of the total RNA was separated on 15% denaturing
polyacrylamide gels, electrotransferred to GeneScreen Plus
membranes and hybridized using Ultra-Hyb-Oligo buffer (Ambion,
Carlsbad, CA, USA) overnight at 42°C. Oligonucleotides
complementary to the mature miRNAs were end-labeled with T4 kinase
(Roche Applied Science, Indianapolis, IN, USA) and used as probes.
The probe sequences were as follows: miR-196a antisense,
5′-CCCAACAACATGAAACTACCTA-3′; miR-193b antisense,
5′-AGCGGGACTTTGAGGGCCAGTT-3′; miR-383 antisense,
5′-AGCCACAATCACCTTCTGATCT-3′; miR-490-5p antisense,
5′-ACCCACCTGGAGATCCATGG-3′; miR-1307 antisense,
5′-AGCCGGTCGAGGTCCGGTCGA-3′; and U6 antisense,
5′-GCCATGCTAATCTTCTCTGTATC-3′.
Bioinformatics analysis
The miRNA targets were predicted using the
algorithms, TargetScan (www.targetscan.org), MiRanda (www.microrna.org) and miRBase Targets
(microrna.sanger.ac.uk). The predicted targets were intersected
using MatchMiner to identify the genes that were commonly predicted
by the 3 different algorithms.
Fluorescent reporter assays
The human BMP receptor type 2 (BMPR2) 3′
untranslated region (3′UTR) harboring the miR-490-5p target
sequence and the seed sequence mutated version (BMPR2-3′UTR-mut)
were synthesized by GenePharma (Shanghai, China) and then ligated
after the luc ORF into the pMIR-REPORT luciferase vector (Ambion).
For the fluorescent reporter assay, the cells were seeded in a
48-well plate and co-transfected with miR-490-5p mimics and BMPR2
3′UTR or BMPR2-3′UTR-mut. The cells were lysed 48 h after
transfection, and the luciferase intensity was measured.
Knockdown of PREX2a by siRNA
To knockdown the expression of BMPR2, a BMPR2 siRNA
lentivirus was purchased from Applied Biological Materials, Inc.
(Cat. no. i002001d; Richmond, BC, Canada). A scrambled siRNA GFP
Lentivector was used as a control (Cat. no. LV015-G, Applied
Biological Materials, Inc.).
Enzyme-linked immunosorbent assay
(ELISA)
The culture medium was collected at 12, 15, and 18
days following the induction of chondrogenic differentiation. ELISA
was performed to detect the secreted concentrations of the
chondrogenic markers, collagen, type II, alpha 1 (Col2A1),
collagen, type X, alpha 1 (Col10A1) and aggrecan using specific
ELISA assay kits (the ELISA kits were obtained from www.antibodies-online.com, and the catalog numbers
were ABIN415072, ABIN1114237, ABIN1113295, respectively) according
to the manufacturer’s instructions.
Statistical analysis
The paired t-test was applied to examine the
differences between the basal cell cultures and the cultures of
hADSCs subjected to chondrogenic differentiation. A value of
p<0.05 was considered to indicate a statistically significant
difference. All the statistical analyses were two-sided and were
carried out using the SPSS statistical software version 13.0 (SPSS,
Inc., Chicago, IL, USA).
Results
Confirmation of chondrogenic
differentiation of hADSCs
hADSCs at passage 3, which included small, round
individual or several cells clumped together suspended in medium,
were cultured in basal or chondrogenic induction medium for 18
days. In the basal medium, the cells were tightly packed (Fig. 1A, left panel), whereas the
morphology of the cells cultured in the chondrogenic induction
medium was altered and consisted of polygon-shaped cells, which is
a typical characteristic of chondrocyte morphology, as shown in
Fig. 1A (right panel). The cell
surface markers in the cultured cells were detected by flow
cytometry at passage 3 following incubation with FITC-labeled
antibodies. In accordance with the antigenic profiles of hADSCs
reported previously (15,16,20), the cells were positive for CD29,
CD44 and CD49 and negative for CD45 (Fig. 1B). To ascertain whether the hADSCs
had undergone chondrogenic differentiation in the chondrogenic
induction medium, the expression of proteins associated with
chondrogenesis was detected by immunohistochemical examination
(Fig. 1C). Following culture in
chondrogenic induction medium for 18 days, the level of ColII in
the cells subjected to chondrogenic differentiation was
significantly higher than that in the cells cultured in basal
medium. These results provide evidence of chondrogenic
differentiation in the cells exposed to the chondrogenic induction
medium.
Analysis of miRNA expression before and
after the induction of chondrogenic differentiation
The miRNA expression levels of hADSCs before and
after the induction of chondrogenic differentiation were detected
using miRNA microarray chips. The miRNAs that exhibited a
differential expression between the undifferentiated hADSCs and the
hADSCs subjected to chondrogenic differentiation were identified
(Fig. 2). The miRNAs that
exhibited at least a 2-fold change in expression in the hADSCs
before and after the induction of chondrogenic differentiation are
listed in Table I, and these
include 12 upregulated miRNAs (miR-196a, miR-143, miR-383,
miR-193b, let-7i, miR-26a, miR-539, miR-199a-3p, miR-337-5p,
miR-146a-5p, miR-646, and miR-381) and 8 downregulated miRNAs
(miR-490-5p, miR-1307, miR-125b, miR-96-3p, miR-302-3p, miR-23a-3p,
miR-590, and miR-510).
 | Table IMicroarray analysis of average
expression of miRNAs in hADSCs isolated from 3 sets of samples
subjected to chondrogenic differentiation. |
Table I
Microarray analysis of average
expression of miRNAs in hADSCs isolated from 3 sets of samples
subjected to chondrogenic differentiation.
| miRNA | Gene ID | Average fold
change | p-value |
|---|
| miR-196a | 406972 | 6.87 | 0.0021 |
| miR-143 | 406935 | 4.52 | 0.0038 |
| miR-383 | 494332 | 5.16 | 0.0149 |
| miR-193b | 574455 | 5.62 | 0.0057 |
| let-7i | 406891 | 3.41 | 0.0025 |
| miR-26a | 407015 | 3.72 | 0.0031 |
| miR-539 | 664612 | 2.39 | 0.0041 |
| miR-199a-3p | 406976 | 3.19 | 0.0131 |
| miR-337-5p | 442905 | 2.81 | 0.0093 |
| miR-146a-5p | 406938 | 2.16 | 0.0071 |
| miR-646 | 693231 | 3.05 | 0.0049 |
| miR-381 | 494330 | 2.47 | 0.0051 |
| miR-490-5p | 574443 | 0.13 | 0.0017 |
| miR-1307 | 100302174 | 0.38 | 0.0081 |
| miR-125b | 406911 | 0.29 | 0.0038 |
| miR-96-3p | 407053 | 0.41 | 0.0217 |
| miR-302-3p | 407028 | 0.47 | 0.0029 |
| miR-23a-3p | 407018 | 0.35 | 0.0024 |
| miR-590 | 693175 | 0.23 | 0.0036 |
| miR-510 | 574515 | 0.35 | 0.0077 |
Validation of the microarray results by
northern blot analysis
In order to confirm the results of the microarray
analysis, we conducted a northern blot analysis to detect the
expression levels of 8 representative differentially expressed
miRNAs identified using the microarray, including 4 upregulated
miRNAs (miR-196a, miR-193b, miR-383 and miR-143) and 4
downregulated miRNAs (miR-490-5p, miR-1307, miR-125b and miR-590).
However, only 5 miRNAs [miR-196a, miR-193b, miR-383 (upregulated),
miR-490-5p and miR-1307 (downregulated)] produced northern blot
analysis signals. The results from northern blot analysis also
revealed that miR-196a was overexpressed in all the 3 samples,
whereas miR-193b and miR-383 were overexpressed in only samples 1
and 3 between the undifferentiated hADSCs and the hADSCs which were
subjected to chondrogenic differentiation. Of the 2 downregulated
miRNAs, miR-490-5p was significantly downregulated in all 3
samples, and the expression of miR-1307 was down-regulated only in
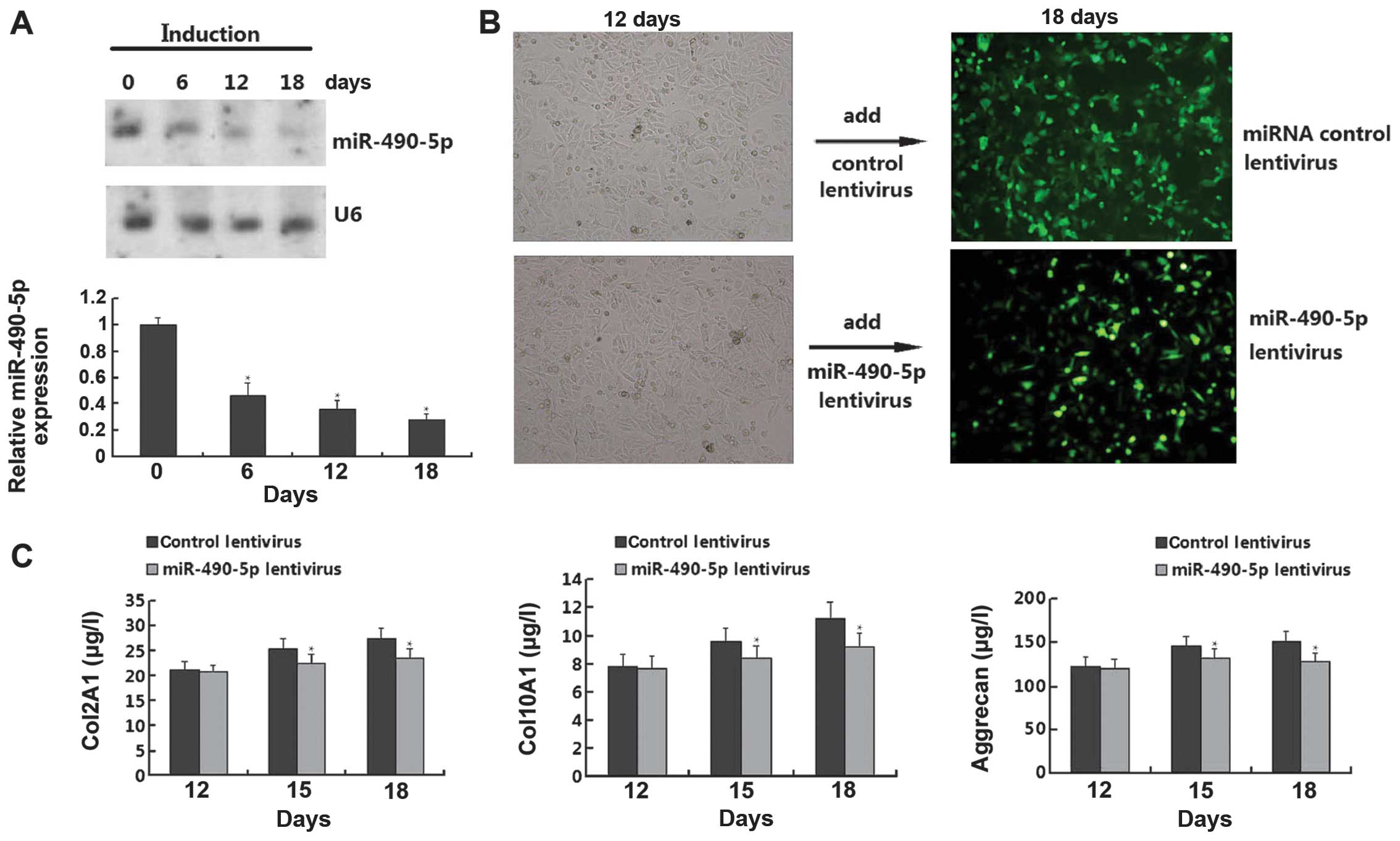
sample 2 (Fig. 3).
Effect of miR-490-5p on chondrogenic
differentiation of hADSCs
Since we confirmed that miR-196a and miR-490-5p were
overexpressed and downregualted, respectively, in all 3 samples, we
conducted functional analysis to detect the role of the 2 miRNAs in
the chondrogenic differentiation of hADSCs. However, we only found
that miR-490-5p, but not miR-196a (data not shown), had an obvious
effect on chondrogenic differentiation. As shown in Fig. 4A, the chondrogenic differentiation
of the hADSCs resulted in the downregulation of miR-490-5p in a
time-dependent manner. However, when the cells were transfected
with miR-490-5p lentivirus on day 12, the cell morphology was
reversed on day 18 compared with the control lentivirus group. In
order to further confirm the results that miR-490-5p regulates the
chondrogenic differentiation of hADSCs, we performed an ELISA for
the quantitative measurement of chondrogenic differentiation
markers (Col2A1, Col10A1 and aggrecan); the levels of these markers
were gradually upregulated on days 12, 15, and 18; however, the
levels of these markers were downregulated in the cells transfected
with the miR-490-5p lentivirus (Fig.
4C). Thus, these results demonstrate that miR-490-5p inhibits
the chondrogenic differentiation of hADSCs.
BMPR2 as a direct target gene of
miR-490-5p
Bioinformatics analysis identified SOX2 and BMPR2 as
putative targets of miR-490-5p (Fig.
5A), which are related to chondrogenic differentiation. A
western blot analysis was then performed to examine the protein
expression of the predicted targets SOX-2 and BMPR2. The results
revealed that the expression levels of SOX2 and BMPR2 in the hADSCs
subjected to chondrogenic differentiation were upregulated, and the
transfection of the cells with miR-490-5p lentivirus resulted in
the downregulation of BMPR2 after the induction of hADSC
differentiation, whereas the expression of SOX2 was not obviously
affected (Fig. 5B). Furthermore,
luciferase reporter assays confirmed that miR-490-5p suppressed the
BMPR2 3′UTR luciferase activity by 40%, and the luciferase activity
was completely reversed following transfection with mutated BMPR2
3′UTR (Fig. 5C). Therefore, we
confirmed that BMPR2 is a direct target of miR-490-5p.
Knockdown of BMPR2 inhibits the hADSC
chondrogenic differentiation
To investigate the functional role of BMPR2 in the
chondrogenic differentiation of hADSCs, BMPR2 siRNA lentivirus was
transfected into the hADSCs and this resulted in a significant
decrease in BMPR2 protein expression in a time-dependent manner
following the induction of chondrogenic differentiation (Fig. 6A). The immunohistochemical
examination illustrated that ColII was downregulated in the cells
transfected with BMPR2 siRNA lentivirus compared with the cells
transfected with the control lentivirus after the induction of
chondrogenic differentiation for 18 days (Fig. 6B). Consistent with the effect of
miR-490-5p, we used ELISA analysis and found that the levels of
Col2A1, Col10A1 and aggrecan were downregulated on days 12, 15 and
18 in the cells transfected with BMPR2 siRNA lentivirus (Fig. 6C).
Discussion
The use of stem cells for cartilage regeneration is
hampered by a lack of knowledge of the molecular mechanisms of
chondrogenesis, which require the stringent control of a program
for gene activation and suppression in response to biological cues
(17,18). Due to the complexity of the
biological network associated with cellular differentiation, the
identification of regulatory miRNAs and their direct targets is
valuable (19). Although the
functions of some individual miRNAs in chondrogenesis are known,
the mechanisms through which a set of miRNAs regulate the onset of
a tissue-specific phenotype remain ambiguous (20). The analysis of miRNA expression
profiles may substantially help to determine the mechanisms
responsible for chondrocyte development and may eventually lead to
the development of therapeutic interventions (21).
In support of the hypothesis that miRNAs play a key
role in chondrogenesis, in this study, we investigated
chondro-miRNAs to provide insight into the specific involvement of
miRNAs in the chondrogenesis of hADSCs. We used the miRNA
microarray technique to determine the miRNA expression profiles and
screen miRNAs with a significant change in expression (>2-fold)
before and after the induction of chondrogenic differentiation. A
northern blot analysis was conducted to confirm that miR-196a and
miR-490-5p were indeed differentially expressed in all 3 samples,
which is consistent with the microarray results, whereas miR-193b
and miR-383 were found to be upregulated in only 2 samples, and
miR-1307 was downregulated in only 1 sample. Our findings on the
reduction in the expression of miR-490-5p were in line with those
of a previous study (22).
Since miR-196a and miR-490-5p were significantly
differentially expressed in all 3 samples, we conducted a
functional analysis. However, we found that miR-490-5p, but not
miR-196a (data not shown), had an obvious effect on chondrogenic
differentiation. Accompanying hADSC differentiation, the expression
of miR-490-5p was gradually downregulated and transfection with
miR-490-5p lentivirus reversed the differentiation ability of the
hADSCs. Using a luciferase reporter system, miR-490-5p was proven
to target the 3′UTR of BMPR2. Thus, miR-490-5p inhibits hADSC
differentiation by suppressing BMPR2 expression.
The confirmation of proper chondrogenesis is usually
determined by an analysis of ‘end-stage’ differentiation markers,
such as Col2A1, Col10A1 and aggrecan (23–25). The strongest upregulation of the
proteins was observed in the hADSCs subjected to chondrogenic
differentiation, which confirmed that the hADSCs had differentiated
into chondrocytes. Moreover, we identified that miR-490-5p
lentivirus partly rescued the expression of these markers, and the
downregulation of BMPR2 by siRNA lentivirus had similar effects as
the overexpression of miR-490-5p, which further indicates that
miR-490-5p inhibits the process of chondrogenesis by targeting
BMPR2.
In conclusion, we discovered a set of miRNAs that
are differentially expressed during the process of hADSC
chondrogenic differentiation and confirmed that miR-490-5p has the
potential to inhibit the chondrogenesis of hADSCs. To enhance our
understanding of the regulatory mechanisms of these differentially
expressed miRNAs during chondrogenic differentiation, further
studies are required. Our results shed new light for further
investigation into the molecular mechanisms of the chondrogenic
differentiation of hADSCs.
Acknowledgments
The present study was sponsored by the Science and
Technology Agency of Guizhou Province, China [Contract number: LS
(2011) no. 032].
References
|
1
|
Dong S, Yang B, Guo H and Kang F:
MicroRNAs regulate osteogenesis and chondrogenesis. Biochem Biophys
Res Commun. 418:587–591. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huang J, Zhao L, Xing L and Chen D:
MicroRNA-204 regulates Runx2 protein expression and mesenchymal
progenitor cell differentiation. Stem Cells. 28:357–364. 2010.
|
|
3
|
Tomé M, López-Romero P, Albo C, Sepúlveda
JC, Fernández-Gutiérrez B, Dopazo A, Bernad A and González MA:
miR-335 orchestrates cell proliferation, migration and
differentiation in human mesenchymal stem cells. Cell Death Differ.
18:985–995. 2011. View Article : Google Scholar :
|
|
4
|
Wagner W, Horn P, Castoldi M, Diehlmann A,
Bork S, Saffrich R, Benes V, Blake J, Pfister S, Eckstein V and Ho
AD: Replicative senescence of mesenchymal stem cells: a continuous
and organized process. PloS One. 3:e22132008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ning G, Liu X, Dai M, Meng A and Wang Q:
MicroRNA-92a upholds Bmp signaling by targeting noggin3 during
pharyngeal cartilage formation. Dev Cell. 24:283–295. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Song J, Lee M, Kim D, Han J, Chun CH and
Jin EJ: MicroRNA-181b regulates articular chondrocytes
differentiation and cartilage integrity. Biochem Biophys Res
Commun. 431:210–214. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Karlsen TA, Jakobsen RB, Mikkelsen TS and
Brinchmann JE: microRNA-140 targets RALA and regulates chondrogenic
differentiation of human mesenchymal stem cells by translational
enhancement of SOX9 and ACAN. Stem Cells Dev. 23:290–304. 2014.
View Article : Google Scholar
|
|
8
|
Buechli ME, Lamarre J and Koch TG:
MicroRNA-140 expression during chondrogenic differentiation of
equine cord blood-derived mesenchymal stromal cells. Stem Cells
Dev. 22:1288–1296. 2013. View Article : Google Scholar
|
|
9
|
Ham O, Song BW, Lee SY, Choi E, Cha MJ,
Lee CY, Park JH, Kim IK, Chang W, Lim S, Lee CH, Kim S, Jang Y and
Hwang KC: The role of microRNA-23b in the differentiation of MSC
into chondrocyte by targeting protein kinase A signaling.
Biomaterials. 33:4500–4507. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bossio C, Mastrangelo R, Morini R, Tonna
N, Coco S, Verderio C, Matteoli M and Bianco F: A simple method to
generate adipose stem cell-derived neurons for screening purposes.
J Mol Neurosci. 51:274–281. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pandey AC, Semon JA, Kaushal D, O’Sullivan
RP, Glowacki J, Gimble JM and Bunnell BA: MicroRNA profiling
reveals age-dependent differential expression of nuclear factor κB
and mitogen-activated protein kinase in adipose and bone
marrow-derived human mesenchymal stem cells. Stem Cell Res Ther.
2(49): 2011
|
|
12
|
Crobu F, Latini V, Marongiu MF, Sogos V,
Scintu F, Porcu S, Casu C, Badiali M, Sanna A, Manchinu MF and
Ristaldi MS: Differentiation of single cell derived human
mesenchymal stem cells into cells with a neuronal phenotype: RNA
and microRNA expression profile. Mol Biol Rep. 39:3995–4007. 2012.
View Article : Google Scholar
|
|
13
|
Guan L, Shaoqing L, Wang Y, Yue H, Liu D,
He L, Bai C, Yan F, Nan X, Shi S and Pei X: In vitro
differentiation of human adipose-derived mesenchymal stem cells
into endothelial-like cells. Chinese Science Bulletin.
51:1863–1868. 2006. View Article : Google Scholar
|
|
14
|
Zhu Y, Liu T, Song K, Fan X, Ma X and Cui
Z: Adipose-derived stem cell: a better stem cell than BMSC. Cell
Biochem Funct. 26:664–675. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peroni D, Scambi I, Pasini A, Lisi V,
Bifari F, Krampera M, Rigotti G, Sbarbati A and Galiè M: Stem
molecular signature of adipose-derived stromal cells. Exp Cell Res.
314:603–615. 2008. View Article : Google Scholar
|
|
16
|
Gronthos S, Franklin DM, Leddy HA, Robey
PG, Storms RW and Gimble JM: Surface protein characterization of
human adipose tissue-derived stromal cells. J Cell Physiol.
189:54–63. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen FH, Rousche KT and Tuan RS:
Technology Insight: adult stem cells in cartilage regeneration and
tissue engineering. Nat Clin Pract Rheumatol. 2:373–382. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vilquin JT and Rosset P: Mesenchymal stem
cells in bone and cartilage repair: current status. Regen Med.
1:589–604. 2006. View Article : Google Scholar
|
|
19
|
Iliopoulos D, Malizos KN, Oikonomou P and
Tsezou A: Integrative microRNA and proteomic approaches identify
novel osteoarthritis genes and their collaborative metabolic and
inflammatory networks. PLoS One. 3:e37402008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Han J, Yang T, Gao J, Wu J, Qiu X, Fan Q
and Ma B: Specific microRNA expression during chondrogenesis of
human mesenchymal stem cells. Int J Mol Med. 25:377–384.
2010.PubMed/NCBI
|
|
21
|
Bakhshandeh B, Soleimani M, Paylakhi SH
and Ghaemi N: A microRNA signature associated with chondrogenic
lineage commitment. J Genet. 91:171–182. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Z, Kang Y, Zhang Z, Zhang H, Duan X,
Liu J, Li X and Liao W: Expression of microRNAs during
chondrogenesis of human adipose-derived stem cells. Osteoarthritis
Cartilage. 20:1638–1646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Indrawattana N, Chen G, Tadokoro M, Shann
LH, Ohgushi H, Tateishi T, Tanaka J and Bunyaratvej A: Growth
factor combination for chondrogenic induction from human
mesenchymal stem cell. Biochem Biophys Res Commun. 320:914–919.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ronzière MC, Perrier E, Mallein-Gerin F
and Freyria AM: Chondrogenic potential of bone marrow- and adipose
tissue-derived adult human mesenchymal stem cells. Biomed Mater
Eng. 20:145–158. 2010.PubMed/NCBI
|
|
25
|
Zhao Q, Eberspaecher H, Lefebvre V and De
Crombrugghe B: Parallel expression of Sox9 and Col2a1 in cells
undergoing chondrogenesis. Dev Dyn. 209:377–386. 1997. View Article : Google Scholar : PubMed/NCBI
|