Introduction
Breast cancer is one of the most malignant tumors
occurring in females, and approximately 350,000 women worldwide
succumb to the disease annually (1). A number of factors, including
lifestyle, environmental, genetic and biological factors contribute
to the initiation and progression of breast cancer. Transforming
growth factor-β1 (TGF-β1) is secreted from breast cancer cells and
plays an important role in the occurrence and development of breast
cancer (2). TGF-β1 functions as a
suppressor during the early phase of tumor progression and becomes
a cancer-promoting modulator during the late stages of cancer.
Classic TGF-β1 signaling functions through 2 types of
serine/threonine kinase receptors. Upon ligand binding, the type II
receptors activate the type I receptors, which activate and recruit
Smad2 and Smad3 and activated Smad2 and Smad3 combined with Smad4
then translocate to the nucleus to regulate gene expression
(3). Apart from the TGF-β1/Smad
pathway, TGF-β1 can also carry out its role in tumor progression by
activating non-Smad pathways, such as the phosphatidylinositol-3
kinase (PI3K), extracellular signal-regulated kinase [ERK,
mitogen-activated protein kinase (MAPK)], c-Jun NH2-terminal kinase
(JNK) and p38 MAPK pathways and Rho GTPases (4). TGF-β1 is commonly recognized as an
essential promoter of epithelial-mesenchymal transition (EMT) and
TGF-β1-induced EMT is considered to be an important initiator of
the invasive behavior of tumors during cancer progression.
The high-mobility group A (HMGA) family has 3
proteins: HMGA1a, HMGA1b and HMGA2. Although these proteins do not
show direct transcriptional regulation activity, they regulate the
transcriptional activity of several genes by altering the chromatin
structure (5–7). HMGA1 has been confirmed to exert an
oncogenic effect on the initiation and progression of diverse types
of tumors (6,8–14).
The overexpression of HMGA1 has been observed in several malignant
neoplasias, such as thyroid cancer, colon cancer, breast cancer,
lung cancer, ovarian cancer and prostate carcinoma, as well as head
and neck tumors (15–23). HMGA1 has also been reported to
promote the progression of breast tumors by inducing EMT (24).
In this study, we aimed to determine the effects of
TGF-β1 on the expression of HMGA1 in breast cancer cells. To the
best of our knowledge, the present study provides the first link
between TGF-β1 and HMGA1 in breast cancer cells and PI3K signaling
and specificity protein 1 (Sp1) were found to be involved in the
TGF-β1-induced expression of HMGA1. This study also offers further
evidence of the pivotal role of HMGA1 in breast cancer
progression.
Materials and methods
Cell culture, transfection and
antibodies
Human MCF-7 and MDA-MB-231 breast cancer cells
(American Type Culture Collection, Manassas, VA, USA) were cultured
in 37°C in a humidified atmosphere containing 5% CO2.
The cells were maintained in Dulbecco’s modified Eagle’s medium
(DMEM) supplemented with 10% FBS (Gibco Life Technologies Australia
Pty Ltd., Mulgrave, Victoria, Australia). The cells were
transiently transfected with the PGL4/HMGA1 plasmid and normalized
with the use of a co-transfected PTK construct using Lipofectamine
2000 (Invitrogen, Carlsbad, CA, USA). At 24 h after transfection,
the cells were stimulated with the indicated concentrations of
TGF-β1 for 12 h. A luciferase assay kit (Promega, Madison, WI, USA)
was used to measure the reporter activity according to the
manufacturer’s instructions. Luciferase activity was normalized
using a Renilla luciferase internal control. The antibodies
used for immunofluorescence and electrophoretic mobility shift
assay (EMSA) were as follows: anti-HMGA1 (ab129153), anti-Sp1
(ab13370) and anti-p-Sp1 (ab59257) antibodies (Abcam, Cambridge,
MA, USA).
Plasmid construction
The HMGA1 promoter was subcloned into the pGL4.10
basic plasmid (Promega) at the KpnI and HindIII
restriction enzyme sites to drive luciferase expression. The
primers used for amplification were 5′-GACGGTACCT
GCTGGAGGCTGAGGAATCG-3′ and 5′-CAGAAGCTTTA GCAAATGCG GATCTGAAACC-3′
for a 2,164 bp fragment.
RNA isolation and reverse transcription
quantitative (real-time) PCR (RT-qPCR)
The MCF-7 cells and MDA-MB-231 cells were treated
with or without LY294002 and wortmannin (Sigma, St. Louis, MO,
USA), two selective inhibitors of PI3K for 2 h, and maintained in
culture medium with TGF-β1 for 9 h. Total RNA was extracted from
the MCF-7 cells or MDA-MB-231 cells using TRIzol reagent (Sigma)
and the total RNA was reverse transcribed into cDNA using the
first-strand synthesis kit (Gibco-BRL, Carlsbad, CA, USA). HMGA1
mRNA was amplified using the primers forward,
5′-AGGGAAGATGAGTGAGTCG-3′ and reverse, 5′-AAGCTG CTCCTCCAGTGAG-3′
and β-actin was utilized as a control using the primers forward,
5′-ATCTGGCACCACACCT-3′ and reverse, 5′-CGTCATACTCCTGCTT-3′.
Quantitative PCR was performed using iQ SYBR-Green Supermix and
regulated using a spectrofluorimetric thermal iCycler iQ5 (Bio-Rad,
Hercules, CA, USA). The gene-specific primers were amplified with a
denaturation step (95°C for 3 min), followed by 39 cycles of
denaturation (95°C for 10 sec), annealing (55°C for 10 sec) and
extension (72°C for 30 sec). Quantitative values were obtained from
the cycle threshold (Ct) value. Samples from 3 separate experiments
were analyzed in duplicate. The results from RT-qPCR were expressed
as 2-ΔΔCT using β-actin as a reference.
Immunofluorescence
The cells were seeded on a round glass cover placed
into a 6 well microtiter plate (Corning Life Sciences, Oneonta, NY,
USA). The cells treated with 10 ng/ml TGF-β1 (Sigma) were fixed
with 4% paraformal-dehyde-PBS for 15 min at room temperature,
washed with PBS twice and and then permeabilized by incubation with
0.1% Triton X-100 for 5 min. The cells were washed twice with PBS
again and stained with anti-HMGA1 antibody for 2 h at room
temperature. After washing with PBS, each sample was incubated with
Alexa Fluor-conjugated secondary antibodies (Boster, Wuhan, China)
for 1 h. The samples were analyzed under a fluorescence microscope
(Olympus, Tokyo, Japan).
Luciferase report gene assay
The cells were cultured in serum-free medium for 12
h in 12-well plates. The cells were then transfected with 0.5
μg of the HMGA1 vector containing luciferase or with the
empty control vector, PGL4.1, using Lipofectamine 2000
(Invitrogen). After 6 h of transfection, the cells were treated
with or without TGF-β1 (Sigma) at the indicated concentrations for
addition 12 h. Luciferase activity was assessed using the
Luciferase assay system (Promega) and was normalized using a
Renilla luciferase internal control.
EMSA
EMSA was performed using the LightShift
Chemiluminescent EMSA kit (Pierce Biotechnology, Rockford, IL, USA)
with slight modifications. Briefly, 5 μg of nuclear extracts
were pre-incubated with 50 ng-μl
poly(deoxyinosinic-deoxycytidylic acid), 10 mmol/l Tris-HCl (pH
7.5), 50 mmol/l KCl, 1 mmol/l dithiothreitol (DTT), 5 mmol/l
MgCl2 and 0.05% NP-40 for 10 min at room temperature.
Following pre- incubation, the samples were incubated for 10 min at
room temperature with a biotin-labeled probe (5′-ATTCGATCGG
GGCGGGGCGAGC-3′) or an unlabeled competitor probe (200-fold molar
excess), or incubated for 60 min at 4°C with the appropriate
antibody. Electrophoresis, electrophoretic transfer and the
detection of the biotin-labeled DNA were carried out according to
the manufacturer’s instructions.
Cell proliferation, migration and
invasion assays
For proliferation assays, cells were seeded (5,000
cells/well) and counted using an automated cell counter (Nexcelom
Bioscience, Lawrence, MA, USA). For colony formation assay, cells
were seeded (500 cells/well) and maintained for 7 days. Each
experiment was carried out in triplicate and performed at least
twice. For the invasion assays, 10,000 cells were resuspended in
serum-free medium and placed in the upper chamber of a 24-well
Matrigel™ Invasion Chamber (BD Biosciences, San Diego, CA, USA)
coated with Matrigel. Cell invasion was calculated as the
percentage of total cells that had invaded the bottom chamber
containing complete medium with serum. Cell migration determined in
a similar manner, except that Matrigel was omitted.
Tissue microarray and immunohistochemical
analysis
Tissue microarrays (BR1921a, BR10010a; US Biomax,
Inc., Rockville, MD, USA), consisting 159 breast cancer cases and
32 normal cases and 50 paired cases of ductal breast cancer and
corresponding lymph node metastasis with breast cancer, were
utilized. The tissues were histologically interpretable and
analyzed for the correlation with clinicopathological parameters.
Immunohistochemical staining was performed as described in our
previous study (25). Rabbit
polyclonal HMGA1 antibody (1:50; Abcam) was used. Ethical approval
for this study was obtained from the Human Research Ethics Advisory
Committee of the University of South China, Hengyang, China.
Statistical analysis
All experiments were performed with 3 replicates and
the results were expressed as the means ± SEM. Statistical analysis
was performed using SPSS software, version 13.0. χ2
tests were applied to assess the statistical significance. P-values
<0.05 were considered to indicate statistically significant
differences.
Results
Expression of HMGA1 is enhanced by TGF-β1
in breast cancer cells
TGF-β1 is an essential cytokine which has high
expression level in the majority of mammary tumors, particularly in
estrogen receptor (ER)-negative (ER-) tumors,
stimulating mammary carcinoma cell invasion and metastatic
potential (26). In this study,
to investigate the effects of TGF-β1 on the expression of HMGA1,
the 2 breast cancer cell lines, MCF-7 and MDA-MB-231, with or
without ER expression, respectively were utilized. As shown in
Fig. 1A, the HMGA1 mRNA level was
increased by stimulation with TGF-β1 in both the MCF-7 and
MDA-MB-231 cells in a dose- and time-dependent manner (Fig. 1A and B). Immunofluorescence assay
revealed that the protein expression of HMGA1 was also elevated by
TGF-β1 in both the MCF-7 and MDA-MB-231 cells (Fig. 1C). These data indicate that TGF-β1
functions as a positive modulator of HMGA1 expression in breast
cancer cells independently of the existence of ER.
TGF-β1 upregulates HMGA1 expression
through the PI3K/Akt pathway
Several pathways and molecules, such as PI3K/Akt,
MAPK and Smad3, have been shown to be involved in TGF-β1-induced
cellular molecular events (27,28). LY294002 and wortmannin, 2
inhibitors of the PI3K/Akt pathway, were utilized to elucidate the
underlying mechanisms of the TGF-β1-induced expression of HMGA1. As
shown in Fig. 2A, the
TGF-β1-induced mRNA expression of HMGA1 was abrogated by treatment
with LY294002 and wortmannin in both the MCF-7 and MDA-MB-231
cells. Immunofluorescence staining also revealed that LY294002 and
wortmannin reversed the TGF-β1-induced protein expression of HMGA1
in the MCF-7 and MDA-MB-231 cells (Fig. 2B). These results indicate the
potential involvement of the PI3K/Akt pathway in the TGF-β1-induced
expression of HMGA1 in breast cancer cells.
TGF-β1 upregulates HMGA1 expression by
enhancing the promoter activity of HMGA1 in breast cancer
cells
To further unravel the mechanisms through which
TGF-β1 enhances the expression of HMGA1, the HMGA1 promoter
sequence containing the GC box was obtained from the MCF-7 cells.
As shown in Fig. 3A and B, TGF-β1
enhanced the promoter activity of HMGA1 in a dose-dependent manner
in both the MCF-7 and MDA-MB-231 cells. Sp1 has been identified to
be an essential transcription factor for the modulation of HMGA1
promoter activity by binding to the GC box located in the promoter
sequence of HMGA1 (29). To
further understand the role of Sp1 in the TGF-β1-induced expression
of HMGA1, EMSA was performed on the breast cancer cells. It was
found that TGF-β1 markedly increased the binding affinity of Sp1 to
the HMGA1 promoter and the p-Sp1 binding to the HMGA1 promoter was
also increased in response to TGF-β1 in the MCF-7 cells (Fig. 3C). These data suggest that Sp1
plays an essential role in the TGF-β1-induced expression of HMGA1
in breast cancer cells.
 | Figure 3specificity protein 1 (Sp1) is
involved in transforming growth factor-β1 (TGF-β1)-induced high
mobility group A1 (HMGA1) promoter activity in breast cancer. (A
and B) Effects of TGF-β1 on HMGA1 promoter activity. MCF-7 cells
and MDA-MB-231 cells were transfected with the HMGA1
promoter/vector containing luciferase or with the empty control
vector, PGL4.1, using Lipofectamine 2000 for 24 h, and the cells
were treated with the indicated concentrations of TGF-β1. Twelve
hours later, the cells were subjected to luciferase activity assay
(n=3). (C) Binding of Sp1 and p-Sp1 (Thr453) to HMGA1 promoter was
enriched following 12 h of TGF-β1 treatment in electrophoretic
mobility shift assay (EMSA) (Materials and methods). Lane 1, probe;
lane 2, probe + nucleic protein complex; lane 3, probe + nucleic
protein complex + cold probe; lane 4, probe + nucleic protein
complex + Sp1 antibody(−TGF-β1); lane 5, probe + nucleic protein
complex + Sp1 antibody(+TGF-β1); lane 6, probe + nucleic protein
complex +p-Sp1 antibody(-TGF-β1); lane 7, probe + nucleic protein
complex + p-Sp1 antibody (+TGF-β1). *P<0.01, compared
with the group not treated with TGF-β1. |
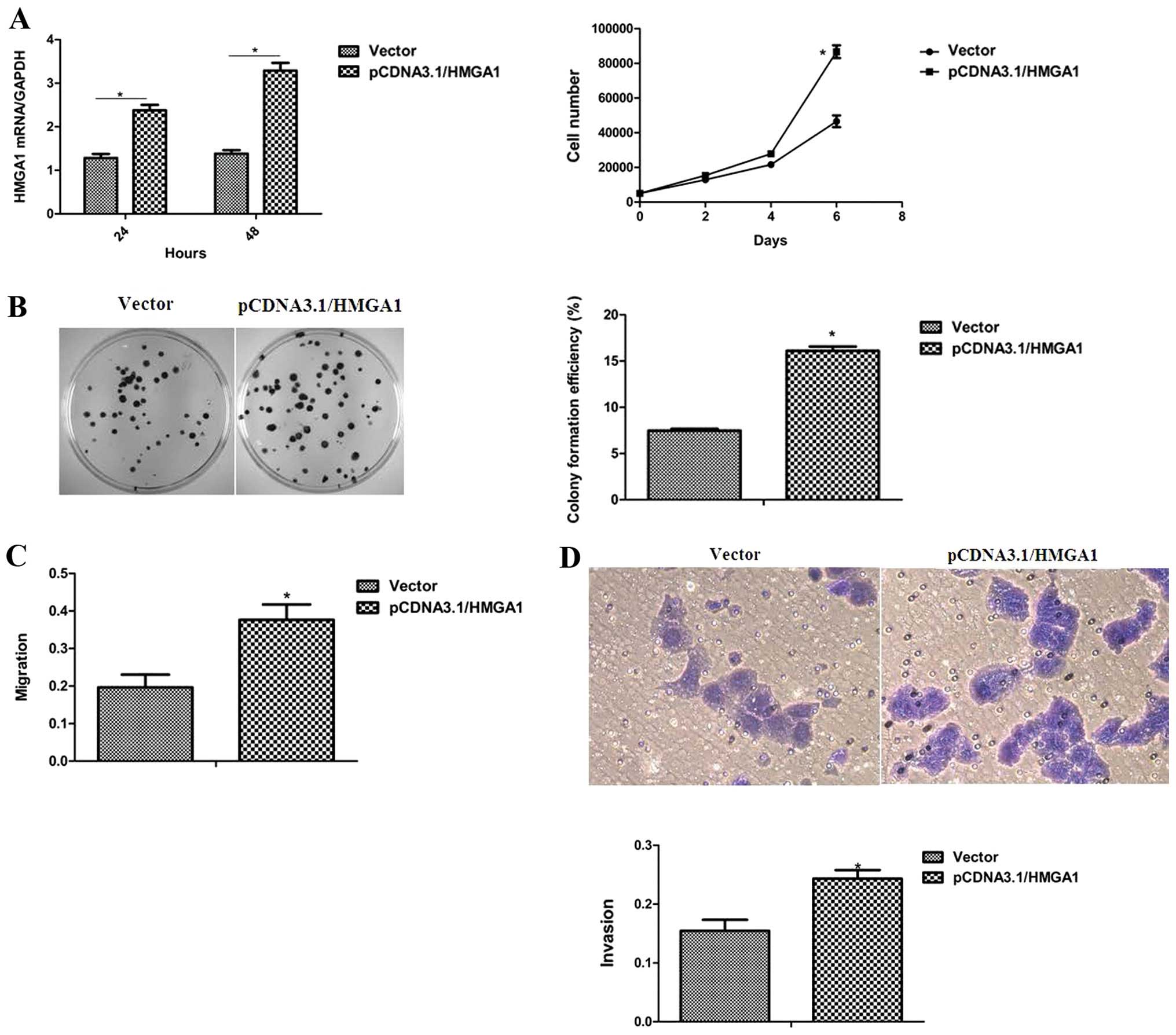
HMGA1 enhances the proliferation and
migration ability of breast cancer cells
To determine the role of HMGA1 in the oncogenic
characteristics of breast cancer cells, the MCF-7 cells with an
ectopic HMGA1 expression were utilized. As shown in Fig. 4A and B, the elevated expression of
HMGA1 in the MCF-7 cells led to an obvious enhancement of the
proliferation and colony-forming ability. The effects of HMGA1 on
cell migration and invasion, properties of breast cancer
progression, were further assessed and it was observed that the
ectopic expression of HMGA1 in the MCF-7 cells markedly promoted
cell migration and invasion (Fig. 4C
and D). These results define an essential role of HMGA1 in the
determination of the cellular oncogenic properties of breast
cancer.
The expression of HMGA1 is associated
with the type of breast carcinoma
To further indentify the association of HMGA1 with
breast cancer progression, a tissue microarray (BR1921a; US Biomax,
Inc.), consisting of 159 breast cancer cases and 32 breast tumor
adjacent tissues was used. The tissue microarray analysis revealed
that HMGA1 was expressed in 62.3 and 46.9% of the breast tumor
tissues and matched breast cancer adjacent tissues, respectively,
predominantly in the nucleus (Table
I and Fig. 5A). Furthermore,
statistical analysis confirmed that breast tumors with a high
expression of human epidermal growth factor receptor 2 (HER2)
showed a higher expression level of HMGA1 (P=0.007) and a higher
expression level of HMGA1 was also found in the ductal breast cases
compared with the lobular breast cancer cases (P=0.000) (Table II and Fig. 5A). These tissue microarray data
suggest that a differential expression of HMGA1 exists in various
types of breast cancer. To determine the association of HMGA1 with
lymph node metastasis, a tissue microarray (BR10010a; US Biomax,
Inc.), consisting of 50 paired samples of ductal breast cancer
tissues and corresponding lymph node tissues, was utilized. It was
found that 76% of the ductal breast cancer cases showed HMGA1
positive staining and 90% of the paired lymph node samples showed
an expression of HMGA1 (Table
III and Fig. 5B), although
statistical analysis showed no significance (P=0.088).
 | Table IHMGA1 expression in breast carcinoma
and breast cancer adjacent tissue. |
Table I
HMGA1 expression in breast carcinoma
and breast cancer adjacent tissue.
| Tissue type | n | HMGA1 (%)
| P-value |
|---|
| − | + | ++ |
|---|
| Normal | 32 | 17 (53.1) | 13 (40.6) | 2 (6.3) | 0.102 |
| Malignant | 159 | 60 (37.7) | 82 (51.6) | 17 (10.7) | |
 | Table IICorrelation between HMGA1 expression
and clinicopathological parameters. |
Table II
Correlation between HMGA1 expression
and clinicopathological parameters.
| Variables | n | HMGA1
| P-value |
|---|
| − | + | ++ |
|---|
| Age (years), n
(%) | | 0.077 |
| <50 | 87 | 37 (42.5) | 44 (50.6) | 6 (6.9) | |
| ≥50 | 72 | 23 (31.9) | 38 (52.8) | 11 (15.3) | |
| Tumor type, n
(%) | | 0.000 |
| Lobular | 79 | 48 (60.8) | 30 (38.0) | 1 (1.3) | |
| Ductal | 79 | 11 (13.9) | 52 (65.8) | 16 (20.3) | |
| Tumor size, n
(%) | | 0.814 |
| ≤2 cm | 18 | 6 (33.3) | 12 (66.7) | 0 (0.0) | |
| >2 cm | 141 | 54 (38.3) | 70 (49.6) | 17 (12.1) | |
| Node metastasis, n
(%) | | 0.208 |
| Negative | 93 | 39 (41.9) | 45 (48.4) | 9 (9.7) | |
| Positive | 66 | 21 (31.8) | 37 (56.1) | 8 (12.1) | |
| Nuclear grade, n
(%) | | 0.097 |
| 1 | 3 | 0 (0.0) | 2 (66.7) | 1 (33.3) | |
| 2 | 60 | 6 (10.0) | 40 (66.7) | 14 (23.3) | |
| 3 | 12 | 4 (33.3) | 7 (58.3) | 1 (8.3) | |
| ER, n (%) | | 0.090 |
| Negative | 104 | 43 (41.3) | 53 (51.0) | 8 (7.7) | |
| Positive | 55 | 17 (30.9) | 29 (52.7) | 9 (16.4) | |
| PR, n (%) | | 0.105 |
| Negative | 116 | 46 (39.7) | 62 (53.4) | 8 (6.9) | |
| Positive | 43 | 14 (32.6) | 20 (46.5) | 9 (20.9) | |
| HER2, n (%) | | 0.007 |
| Negative | 130 | 54 (41.5) | 66 (50.8) | 10 (7.7) | |
| Positive | 29 | 6 (20.7) | 16 (55.2) | 7 (24.1) | |
 | Table IIIHMGA1 expression in breast carcinoma
and breast cancer lymph node metastastic tissue. |
Table III
HMGA1 expression in breast carcinoma
and breast cancer lymph node metastastic tissue.
| Variables | n | HMGA1 (%)
| P-value |
|---|
| − | + | ++ |
|---|
| Tissue type | | 0.088 |
| Breast tumor | 50 | 12 (24.0) | 27 (54.0) | 11 (22.0) | |
| Lymph node | 50 | 5 (10.0) | 33 (66.0) | 12 (24.0) | |
Discussion
HMGA1, as an oncogene, is enriched in adult stem
cells and high-grade/poorly differentiated tumors (6,8–14)
and it is highly expressed during embryogenesis with low or absent
levels in adult tissues. HMGA1 is found to be overexpressed in
almost all aggressive cancers and high levels of HMGA1 point to a
poor prognosis in diverse types of tumors (18,20,21,30). HMGA1 functions in tumor
progression by reprogramming differentiated cells into poorly
differentiated, stem-like cancer cells. HMGA1 has also been
demonstrated to be essential for the reprogramming of somatic cells
to induce pluripotent stem cells (31).
TGF-β1 is a cytokine that modulates many fundamental
aspects of cellular behavior, including growth, differentiation,
migration and apoptosis. In early-stage adenomas, TGF-β1 functions
as a tumor suppressor in normal epithelia by inhibiting cell growth
and it is also one of the key cytokines in promoting EMT during
embryonic development and during the late stages of cancer
progression, leading to tumor cell invasiveness and metastasis. The
present study provides evidence of a link between TGF-β1 and HMGA1
in breast cancer cells. It was found that TGF-β1 induced the
expression of HMGA1 in both triple-positive breast cancer cells and
triple-negative breast cancer cells. Since PI3K signaling has
previously been described as one of the non-Smad signalings
pathways which regulates TGF-β1 signaling pathway (4), we aimed to assess the involvement of
PI3K signaling in the TGF-β1-induced HMGA1 expression. It was found
that PI3K signaling was involved in this process; however, whether
PI3K signaling operates in parallel or in direct coordination with
the Smad proteins in the TGF-β1-induced epxression of HMGA1 remains
to be further elucidated.
We further determined the downstream key molecules
in the PI3K signaling pathway which mediate the TGF-β1- induced
expression of HMGA1 in breast cancer cells. The effects of TGF-β1
on HMGA1 promoter activity were assessed and we found that TGF-β1
markedly increased the promoter activity of HMGA1 in both the MCF-7
and MD-321 cells. Sp1 has previously been confirmed to be an
essential modulator of the regulation of HMGA1 promoter activity
(29) and we determined the
involvement of Sp1 in the TGF-β1-induced HMGA1 expression in breast
cancer cells. It was revealed that TGF-β1 increased the binding
affinity of Sp1 to the HMGA1 promoter in vitro, indicating
that Sp1 takes part in the TGF-β1-induced expression of HMGA1. Sp1
phosporylation has been found to play an essential role in gene
regulation (32). In a previous
study, the phosporylation of Sp1 was shown to be involved in the
gene promoter activity regulation of TRAIL, when vascular smooth
muscle cells (VSMCs) were exposed to fibroblast growth factor
(FGF)-2 (33). Our findings also
suggest that TGF-β1 induces HMGA1 expression by activating Sp1
phosporylation, altering its occupancy at the HMGA1 promoter.
We also found that the ectopic expression of HMGA1
had a profound effect on oncogenic properties, including
proliferation, migration and invasion and these results are
consistent with those reported in the study by Shah et al
(14) showing HMGA1 silencing in
triple-negative breast cancer cells. To further unravel the
significance of HMGA1 in clinical prognosis, we detected the
expression of HMGA1 in a tissue microarray containing 159 breast
cancer cases and 32 breast tumor adjacent tissues. We discovered
that breast tumors with HER2 expression showed a higher expression
level of HMGA1 and a higher expression level of HMGA1 was found in
the ductal breast cancer cases compared with the lobular breast
cancer cases. The associations of nuclear grade, tumor size and
node metastasis with HMGA1 expression in breast tumor tissues was
not found. Increasing evidence indicates that HMGA1 is of
importance in maintaining a de-differentiated, pluripotent
stem-like state (31) and HMGA1
has been demonstrated to reprogram somatic cells to induce
pluripotent stem cells (iPSCs) (34). EMT is of importance for stem cells
and metastatic tumors. The link between HMGA1 and EMT was revealed
in MCF-7 cells, in which the enforced expression of HMGA1 resulted
in metastatic progression and histological changes consistent with
EMT (24). Although a growing
body of evidences suggests the essential role of HMGA1 in tumor
metastatic progression, we did not obtain the data for the
correlation between HMGA1 expression and node metastasis in the
tissue microarray. The disparity of HMGA1 between the cellular
behaviors and clinical prognosis may be partly explained by the
sample number used in this tissue microarray may not be sufficient
and further investigations are required to clarify the importance
of HMGA1 in the diagnosis and prognosis of breast cancer
patients.
In conclusion, to the best of our knowledge, the
present study provides the first evidence of the role of TGF-β1 in
the regulation of HMGA1 expression in breast cancer cells and PI3K
signaling and Sp1 were found to be involved in the TGF-β1-induced
expression of HMGA1. Through Smads signaling, TGF-β1 is a well
known inducer of EMT leading to tumor metastasis progression, and
our data suggest that TGF-β1 promotes EMT by increasing the
expression of HMGA1, offering a novel pathway for TGF-β1-induced
EMT in breast cancer. Our study, together with data from previous
studies, provides compelling evidence of the crucial role of HMGA1
in breast cancer progression.
Acknowledgments
This study was supported by projects from the
National Natural Science Foundation of P.R. China (Grant No.
81272355, 31200573, 81170807 and 81372824), the Hunan Provincial
Natural Science Foundation of China (12JJ3116) and the Education
Department of Hunan Province Youth Fund (12B108).
References
|
1
|
Porter PL: Global trends in breast cancer
incidence and mortality. Salud Publica Mex. 51(Suppl 2): s141–s146.
2009. View Article : Google Scholar
|
|
2
|
Yang L, Pang Y and Moses HL: TGF-beta and
immune cells: an important regulatory axis in the tumor
microenvironment and progression. Trends Immunol. 31:220–227. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Smith AL, Robin TP and Ford HL: Molecular
pathways: targeting the TGF-β pathway for cancer therapy. Clin
Cancer Res. 18:4514–4521. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Derynck R, Akhurst RJ and Balmain A:
TGF-beta signaling in tumor suppression and cancer progression. Nat
Genet. 29:117–129. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Reeves R: Nuclear functions of the HMG
proteins. Biochim Biophys Acta. 1799:3–14. 2010. View Article : Google Scholar
|
|
6
|
Resar LM: The high mobility group A1 gene:
transforming inflammatory signals into cancer? Cancer Res.
70:436–439. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fusco A and Fedele M: Roles of HMGA
proteins in cancer. Nat Rev Cancer. 7:899–910. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ben-Porath I, Thomson MW, Carey VJ, et al:
An embryonic stem cell-like gene expression signature in poorly
differentiated aggressive human tumors. Nat Genet. 40:499–507.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chou BK, Mali P, Huang X, et al: Efficient
human iPS cell derivation by a non-integrating plasmid from blood
cells with unique epigenetic and gene expression signatures. Cell
Res. 21:518–529. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Karp JE, Smith BD, Resar LS, et al: Phase
1 and pharmacokinetic study of bolus-infusion flavopiridol followed
by cytosine arabinoside and mitoxantrone for acute leukemias.
Blood. 117:3302–3310. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nelson DM, Joseph B, Hillion J, Segal J,
Karp JE and Resar LM: Flavopiridol induces BCL-2 expression and
represses oncogenic transcription factors in leukemic blasts from
adults with refractory acute myeloid leukemia. Leuk Lymphoma.
52:1999–2006. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schuldenfrei A, Belton A, Kowalski J, et
al: HMGA1 drives stem cell, inflammatory pathway, and cell cycle
progression genes during lymphoid tumorigenesis. BMC Genomics.
12(549): 2011
|
|
13
|
Belton A, Gabrovsky A, Bae YK, et al:
HMGA1 induces intestinal polyposis in transgenic mice and drives
tumor progression and stem cell properties in colon cancer cells.
PLoS One. 7:e300342012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shah SN and Resar LM: High mobility group
A1 and cancer: potential biomarker and therapeutic target. Histol
Histopathol. 27:567–579. 2012.PubMed/NCBI
|
|
15
|
Abe N, Watanabe T, Masaki T, et al:
Pancreatic duct cell carcinomas express high levels of high
mobility group I(Y) proteins. Cancer Res. 60:3117–3122.
2000.PubMed/NCBI
|
|
16
|
Chiappetta G, Tallini G, De Biasio MC, et
al: Detection of high mobility group I HMGI(Y) protein in the
diagnosis of thyroid tumors: HMGI(Y) expression represents a
potential diagnostic indicator of carcinoma. Cancer Res.
58:4193–4198. 1998.PubMed/NCBI
|
|
17
|
Chiappetta G, Manfioletti G, Pentimalli F,
et al: High mobility group HMGI(Y) protein expression in human
colorectal hyperplastic and neoplastic diseases. Int J Cancer.
91:147–151. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chiappetta G, Botti G, Monaco M, et al:
HMGA1 protein over-expression in human breast carcinomas:
correlation with ErbB2 expression. Clin Cancer Res. 10:7637–7644.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Meyer B, Loeschke S, Schultze A, et al:
HMGA2 overexpression in non-small cell lung cancer. Mol Carcinog.
46:503–511. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Masciullo V, Baldassarre G, Pentimalli F,
et al: HMGA1 protein over-expression is a frequent feature of
epithelial ovarian carcinomas. Carcinogenesis. 24:1191–1198. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tamimi Y, van der Poel HG, Denyn MM, et
al: Increased expression of high mobility group protein I(Y) in
high grade prostatic cancer determined by in situ hybridization.
Cancer Res. 53:5512–5516. 1993.PubMed/NCBI
|
|
22
|
Miyazawa J, Mitoro A, Kawashiri S, Chada
KK and Imai K: Expression of mesenchyme-specific gene HMGA2 in
squamous cell carcinomas of the oral cavity. Cancer Res.
64:2024–2029. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rho YS, Lim YC, Park IS, et al: High
mobility group HMGI(Y) protein expression in head and neck squamous
cell carcinoma. Acta Otolaryngol. 127:76–81. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dolde CE, Mukherjee M, Cho C and Resar LM:
HMG-I/Y in human breast cancer cell lines. Breast Cancer Res Treat.
71:181–191. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhong J, Cao RX, Zu XY, et al:
Identification and characterization of novel spliced variants of
PRMT2 in breast carcinoma. FEBS J. 279:316–335. 2012. View Article : Google Scholar
|
|
26
|
Welch DR, Fabra A and Nakajima M:
Transforming growth factor beta stimulates mammary adenocarcinoma
cell invasion and metastatic potential. Proc Natl Acad Sci USA.
87:7678–7682. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Derynck R and Zhang YE: Smad-dependent and
Smad-independent pathways in TGF-β family signalling. Nature.
425:577–584. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Moustakas A and Heldin CH: Non-Smad TGF-β
signals. J Cell Sci. 118:3573–3584. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Massimi I, Guerrieri F, Petroni M, Veschi
V, Truffa S, Screpanti I, Frati L, Levrero M, Gulino A and Giannini
G: The HMGA1 protoncogene frequently deregulated in cancer is a
transcriptional target of E2F1. Mol Carcinog. 52:526–534. 2013.
View Article : Google Scholar
|
|
30
|
Abe N, Watanabe T, Izumisato Y, et al:
High mobility group A1 is expressed in metastatic adenocarcinoma to
the liver and intrahepatic cholangiocarcinoma, but not in
hepatocellular carcinoma: its potential use in the diagnosis of
liver neoplasms. J Gastroenterol. 38:1144–1149. 2003. View Article : Google Scholar
|
|
31
|
Shah SN, Kerr C, Cope L, et al: HMGA1
reprograms somatic cells into pluripotent stem cells by inducing
stem cell transcriptional networks. PLoS One. 7:e485332012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Q, Ji Y, Wang X and Evers BM:
Isolation and molecular characterization of the 5′-upstream region
of the human TRAIL gene. Biochem Biophys Res Commun. 276:466–471.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chan J, Prado-Lourenco L, Khachigian LM,
Bennett MR, Di Bartolo BA and Kavurma MM: TRAIL promotes VSMC
proliferation and neointima formation in a FGF-2-, Sp1
phosphorylation-, and NFkappaB-dependent manner. Circ Res.
106:1061–1071. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Flohr AM, Rogalla P, Bonk U, et al: High
mobility group protein HMGA1 expression in breast cancer reveals a
positive correlation with tumour grade. Histol Histopathol.
18:999–1004. 2003.PubMed/NCBI
|