Introduction
Hepatitis C virus (HCV) is the cause of chronic
infection in approximately 180 million individuals worldwide
(1). However, the elucidation of
the pathogenesis of HCV-associated liver disease is hampered by the
absence of an appropriate small animal model. HCV genomic RNA can
be used as either mRNA for translation or a template for RNA
replication. The translation initiation of the HCV genome is
controlled by an internal ribosome-entry site (IRES) within the 5′
untranslated region (UTR) (2).
The 3′UTR has been shown to serve as a replication onset site for
recognition and nucleotide incorporation by non-structural protein
5B (NS5B), an RNA-dependent RNA polymerase (3). The NS5B polymerase is a 65-kDa
protein responsible for HCV RNA replication (4) that recognizes a specific RNA
sequence in the 3′UTR of the genomic RNA and replicates the primary
negative strand RNA first, and then the positive-strand RNA
according to the negative strand RNA template (5). It has been shown that NS5B alone can
copy an RNA template containing the HCV UTR in vitro or
in vivo, without the need for any other viral/host factors
(6). Based on the HCV elements
mentioned above (2–6), a HCV subgenomic replicon
(sub-replicon) was designed in our laboratory. Zebrafish is a good
model organism for human diseases, and a number of disease models
have emerged from studies using this organism (7,8).
There are several advantages to using zebrafish embryos to develop
models of liver diseases (8–10);
the zebrafish exhibits high homology to humans genetically
(11) and is small and
easy-to-handle experimentally. We therefore explored the potential
of using zebrafish as a host for HCV replication. In our previous
study, we demonstrated that zebrafish may be a model organism to
host HCV (12). The HCV
sub-replicon was created with 2 vectors, one with HCV ns5b and the
other containing the minus strand of the HCV 5′UTR, core and 3′UTR
by co-injecting into zebrafish zygotes. The sub-replicon amplified
in the liver showed a significant expression of HCV core RNA and
protein. As HCV is a plus strand RNA virus, we then intended to
generate a plus strand HCV sub-replicon zebrafish model for
anti-HCV drug screening. In this study, we describe the
construction of a HCV subgenomic expression vector using the mouse
hepatocyte nuclear factor 4 (mHNF4) promoter, the HCV 5′UTR-core
protein, the enhanced green fluorescent protein (EGFP) reporter
gene and the NS5B 3′UTR. This study describes the use of a
zebrafish model for further investigations into the mechanisms of
HCV replication and the pathology of HCV infection in the liver
in vivo. This model may also aid drug evaluation studies and
may thus aid in the discovery of new anti-HCV drugs.
Materials and methods
Plasmids, reagents and antibodies
HCV strain 1b (J4L6s; Accession no. AF054247) was
provided by Dr H.S. Chen (Institute of Medicinal Biotechnology,
Beijing, China). The sub-replicon construct (pH5B) was constructed
by the insertion of the HCV 5′UTR-core downstream of the mHNF4a
promoter (the promoter sequence was cloned and identified by
sequencing in our preliminary experiment) into the pIRES2-EGFP
plasmid, followed by the insertion of the IRES-NS5B-3′UTR after the
EGFP gene using the NotI and XbaI restriction sites
(Fig. 1A). All the constructs
were confirmed by DNA sequencing. The antibodies against NS5B
(ab35586), the core protein (sc-8334) and GFP (ab2740) were
purchased from Abcam (Cambridge, UK) and Santa Cruz Biotechnology,
Inc. (Santa Cruz, CA, USA), respectively.
 | Figure 1Replication of the hepatitis C virus
(HCV) sub-replicon in zebrafish larvae. (A) Schematic diagram of
the HCV sub-replicon vector, pH5B. The mouse hepatocyte nuclear
factor 4 (mHNF4a) promoter was included for transcription. The HCV
5′ untranslated region (5′UTR)-core sequence was inserted
downstream of the HNF4a promoter, followed by internal
ribosome-entry site (IRES)-enhanced green fluorescent protein
(EGFP), and IRES-non-structural protein 5B (NS5B)-3′UTR. (B)
Fluorescence microscopic observation of the HCV sub-replicon in the
liver of pH5B-injected zebrafish larvae at 10 days
post-fertilization (dpf) using a green fluorescent protein (GFP)
filter (480 nm excitation, 505 nm emission; image, x100, original).
Red arrows indicate positive signals of GFP in the liver. (C)
Confocal DIC images of pH5B-injected zebrafish larvae at 10 dpf.
The left, the middle and the right images are at fluorescence
field, bright field and the overlay, respectively. White arrows
indicate the positive signal of GFP in the liver, but are different
parts from the fluorescent images of the liver shown in (B). (D)
Upper panel shows templates and primers for the reverse
transcription reactions; lower panel shows reverse
transcription-polymerase chain reaction (RT-PCR) results of the
target genes, core, NS5B and EGFP, as well as
β-actin at 10 dpf in the zebrafish larvae injected with the HCV
sub-replicon vector. β-actin was used as a loading control. (E)
Whole mount in situ hybridizations were performed on larvae
at 10 dpf using antisense or sense RNA probes. Red indicate show
the positive signals (including NS5B-positive or -negative,
core-positive or -negative, and transferrin-positive) in the larvae
livers (x80 magnification, original). (F) Western blots of the GFP,
HCV core and NS5B proteins from the larvae at 10 dpf were detected
using anti-EGFP, anti-core and anti-NS5B antibodies, respectively.
β-actin was used as a loading control. |
Zebrafish microinjection and fluorescence
microscopy
The adult zebrafish strain AB (Danio rerio)
was obtained from Dr Anming Meng (Tsinghua University, Beijing,
China). The zebrafish were maintained in a controlled environment
under a 14-h light/10-h dark cycle at 28±1°C.
The pH5B construct was linearized and injected into
the blastomere of 1-8-cell stage embryos at 1 ng/μl. Larvae
positive for GFP were examined at 8 and 12 days post-fertilization
(dpf), using a fluorescence microscope (IX51; Olympus, Tokyo,
Japan) with GFP filters (480 nm excitation, 505 nm emission).
Reverse transcription-polymerase chain
reaction (RT-PCR)
Total RNA was isolated from the larvae using TRIzol
reagent (Invitrogen, Carlsbad, CA, USA). The cDNA was synthesized
from 1 μg of the total RNA using AMV reverse transcriptase
(Promega, Madison, WI, USA) with different reverse transcription
primers: NS5B-R and d(T)18 for transcript resulting from
mHNF4a promoter, and core-F for HCV sub-replicon replication
products. The target cDNA templates were then amplified by PCR
using Taq polymerase (Takara Bio Inc., Shiga, Japan) and the primer
pairs for the core protein, NS5B and β-actin are listed in Table I. The cDNA template of
non-transgenic wild-type zebrafish larvae was used as a negative
control. PCR was performed with 0.5 μl of cDNA under the following
cycling conditions: 94°C for 5 min, then 30 cycles of 94°C for 60
sec, 55°C for 30 sec and 72 °C for 1 min, followed by an additional
extension step at 72°C for 10 min to allow for complete synthesis.
The RT-PCR products were subjected to 1.5% agarose gel
electrophoresis. β-actin was used as a control.
 | Table IRT-PCR primer sequences for HCV. |
Table I
RT-PCR primer sequences for HCV.
| Core | F:
5′-AGCGGTCGCAACCTCGTGGAA-3′ |
| Core | R:
5′-GCGGAAGCTGGGATGGTCAAAC-3′ |
| NS5B | F:
5′-GCTCGCCTTATCGTATTCC-3′ |
| NS5B | R:
5′-AGTCGTCAGCACGCCAC-3′ |
| GFP | F:
5′-ACGGCGTGCAGTGCTT-3′ |
| GFP | R:
5′-TGGGTGCTCAGGTAGTGG-3′ |
| β-actin | F:
5′-AGGGAAATCGTGGGTGACATCAAA-3′ |
| β-actin | R:
5′-ACTCATCGTACTCCTGCTTGCTGA-3′ |
| Chemokine20 | F:
5′-TCTCTTCTCACCTGCCCTAA-3′ |
| Chemokine20 | R:
5′-ATTGCTTGCACCTTCTCCCTC-3′ |
| AHSG | F:
5′-GGAAGGCAGCGGTGAAA-3′ |
| AHSG | R:
5′-ATGGTCTGGCCCGAGTG-3′ |
| Hsp70 | F:
5′-GCGACACCTCTGGAAAC-3′ |
| Hsp70 | R:
5′-TGCTCAGCCTGCCCTTG-3′ |
| ScarF2 | F:
5′-CTCTTGCGTCTACAGGG-3′ |
| ScarF2 | R:
5′-GCTCAGCGGTTTCTATT-3′ |
| Leugpcr | F:
5′-GGTGTTTGTCTGGGTTG-3′ |
| Leugpcr | R:
5′-GGTCTGAGTGAAGAGGGA-3′ |
| ACC | F:
5′-TTAGACCTGGATCAACGGCG-3′ |
| ACC | R:
5′-CATGATCTGTCCTGTACGGG-3′ |
| Heparanase | F:
5′-CAAGCGTTTAGTCACTCGGC-3′ |
| Heparanase | R:
5′-GGTTGCATTCCACGAGTTGTC-3′ |
| Leptin
receptor | F:
5′-GTCACACTGATGATGTCACAGAACCAGATG-3′ |
| Leptin
receptor | R:
5′-GCTAAAGACCTCTATTACCTCGAGATGACC-3′ |
| C-myc | F:
5′-CCCAGCCGGAGACAGTCGCTCTCCACCGCG-3′ |
| C-myc | R:
5′-CCACAGTCACCACATCAATTTCTTCCTCC-3′ |
Western blot analysis
Zebrafish larvae total protein was extracted using
lysis buffer and separated by 12% SDS-PAGE. The protein bands were
transferred onto nitrocellulose membranes followed by blocking of
the membranes in TBS containing 10% skim milk. The membranes were
incubated with anti-core or anti-NS5B antibodies (Abcam) at 1:2,000
dilutions in TBS containing 1% skim milk, and the membranes were
then washed and incubated with HRP-conjugated goat anti-mouse or
goat anti-rabbit IgGs (1:2,000 dilutions; Zhongshan Jinqiao
Biotechnology Co., Ltd., Beijing, China) for 1 h at room
temperature. Proteins were detected using the
SuperSignal® West Pico Chemiluminescent Substrate
(Thermo Scientific, Rockford, IL, USA) with the
AlphaEase® FC Imaging System (Alpha Innotech Corp., San
Leandro, CA, USA).
Whole mount in situ hybridization
The core (nt 430–702) and NS5B (nt 8067–8459) gene
sequences of the J4L6 strain were used as templates for
hybridization probe synthesis, using the DIG RNA Labeling kit
(Roche Diagnostics Scandinavia AB, Bromma, Sweden). Whole mount
in situ hybridization was performed as previously described
(13). Briefly, larvae at 10 dpf
were fixed with 4% paraformaldehyde for 10 h at 4°C and washed with
1X phosphate-buffered saline containing Tween-20 (PBST). The larvae
were treated with proteinase K and DNase I separately,
pre-hybridized at 65°C for 4 h, and hybridized with the RNA probes
such as core and NS5B respectively, at 65°C
overnight. The residual probe was washed with 0.2X saline sodium
citrate, followed by incubation with anti-Dig-AP (Roche Diagnostics
Scandinavia AB) at 4°C overnight. After washing with 1X PBST, the
samples were developed with BCIP/NBT solution for 30 min and this
reaction was terminated by washing with 1X PBST. Hybridized signals
of the core and NS5B genes were detected using the
purple colorigenic substrate. The larvae were observed under a
light microscope (SZ61TRC; Olympus).
Treatment with drugs
Ribavirin and oxymatrine were obtained from the
National Institute for the Control of Pharmaceutical and Biological
Products (Beijing, China). Ribavirin at final concentrations 1000,
100 and 10 μg/ml, and oxymatrine at concentrations of 200, 20 and 2
μg/ml were added to the zebrafish cultivation water, and were
incubated with the zebrafish larvae injected or not with pH5B at 5
dpf for another 5 days. The larvae were then collected for
analysis.
Results
Replication of the HCV sub-replicon in
zebrafish larvae
To ensure that the HCV sub-replicon could be
expressed in the larva liver, a liver specific promoter, mHNF4a
(14–16), was introduced upstream of the gene
expression cassette, followed by the HCV 5′UTR-core,
encephalomyocarditis virus (EMCV) IRES, EGFP, EMCV IRES and HCV
NS5B 3′UTR genes. The inclusion of the reporter gene encoding EGFP
enabled the easy detection of the HCV sub-replicon construct by
fluorescence examination (Fig. 1B and
C), indicating HCV protein expression. EGFP fluorescence was
observed mainly in the liver of the larvae, and to a lesser extent
in the intestines and pancreas in the injected zebrafish
larvae.
We further examined the transcription of the HCV
core and NS5B genes in zebrafish using the specific
primer, NS5B-R, that only binds to the NS5B mRNA 3′end to test the
transcript of the sub-replicon, and an universal primer
d(T)18 by RT-PCR. The results revealed thatthe HCV
core and NS5B genes were expressed in the
microinjected larvae, whereas no expression was detected in the
wild-type (WT) larva control (Fig.
1D). The HCV sub-replicon transcript contained genetic
information for the 5′UTR, core, EGFP,
NS5B and 3′UTR, which are the elements which respond
to HCV RNA replicatiion apart from EGFP. When NS5B
polymerase is translated, the sub-replicon transcript can be used
as a template for the replication of the HCV sub-replicon negative
strand that harbors antisense RNA for the core, EGFP and NS5B.
Thus, the production of the HCV negative strand RNA is likely to be
performed by NS5B since no endogenous RNA-dependent RNA polymerase
is known to exist in zebrafish. The negative strand of HCV RNA was
detected by reverse transcription in vitro using total RNA
as a template and the specific forward primer core-F that only
binds to the negative strand sequence of the core 3′end to produce
the positive strand core cDNA, followed by PCR with the core primer
pair and NS5B primer pair to amplify the negative strand sequence.
Thus, the products of core, NS5B and EGFP are a sign of the
successful replication of the HCV subgenomic replicon in zebrafish
(Fig. 1D). No HCV product was
detected in the WT zebrafish by RT-PCR (Fig. 1D).
Hybridization in situ was carried out for
exploring the localization of HCV gene expression in zebrafish.
With the antisense RNA probes or the sense RNA probes of the
core and NS5B genes, the hybridized signals were
mainly detected in the zebrafish liver, with minor signals detected
in the intestines and pancreas of the larvae at 10 dpf (Fig. 1E). By contrast, no HCV signals
were detected in the WT generation. By control, the liver-specific
gene, transferrin, was detected only by the antisense
transferrin RNA probe, and was shown as negative by the
sense transferrin RNA probe. These results indicate that the
existence of the HCV antisense RNA sequence demonstrates the
potential of the HCV sub-replicon to replicate in zebrafish liver
(the HCV sense probe results in Fig.
1E).
Furthermore, the HCV core, EGFP and NS5B proteins
were examined by western blot analysis of the zebrafish larvae
injected with pH5B the WT larva control. Bands of the core, EGFP
and NS5B proteins were detected at the predicted sizes with
anti-core, anti-EGFP or anti-N5B antibodies, respectively, but no
signals were detected in the WT control (Fig. 1F). These result indicates that the
HCV sub-replicon was indeed expressed and that its transcript
became a RNA template for the successful HCV sub-genome replication
in the pH5B-injected zebrafish.
Biological impact of pH5B microinjection
into zebrafish larvae
The HCV infection has been reported to cause
alterations in the expression of host genes in human liver cells
(17). Thus, in this study, we
investigated whether the replication of the HCV sub-replicon in the
larvae also causes changes in gene expression using RT-PCR. The
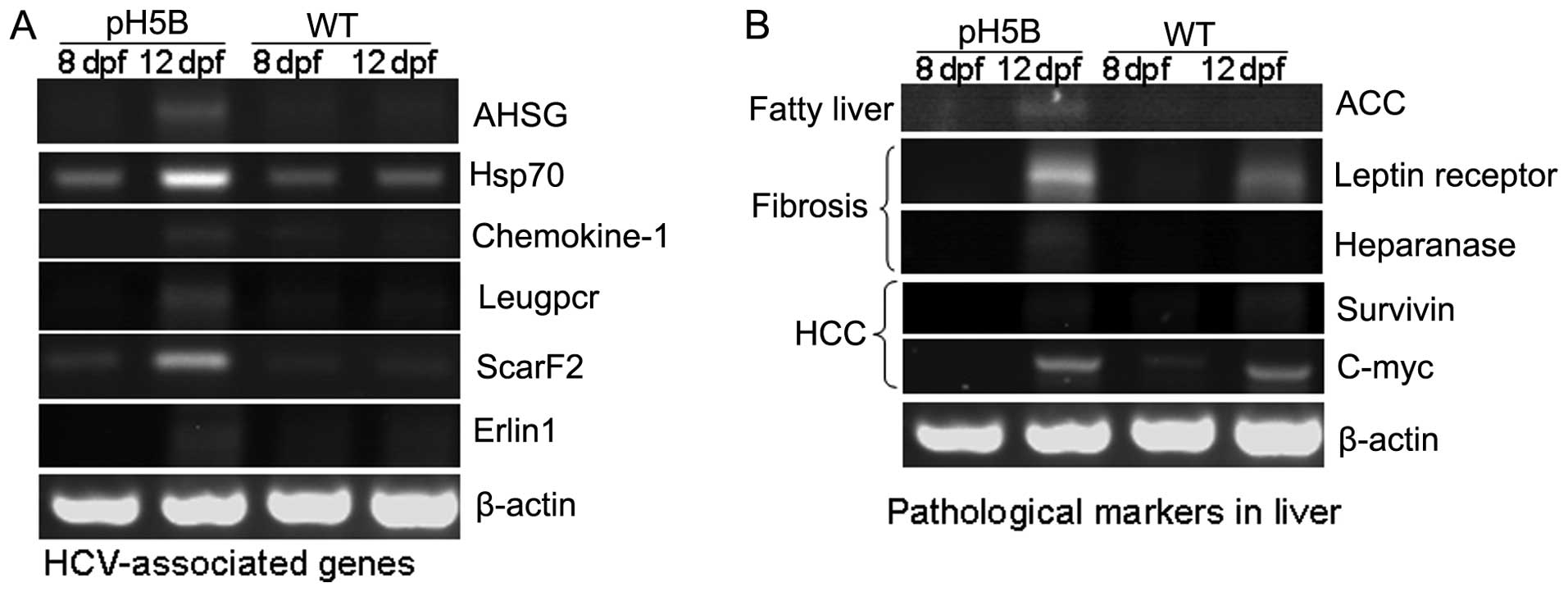
results are presented in Fig. 2A.
The expression of the alpha-2-HS-glycoprotein (AHSG)
(18), Hsp70 (19), chemokine-1 (20), Leucine-rich repeat-containing G
protein-coupled receptor 5 (Leugpcr) (17), scavenger receptor class F, member
2, (ScarF2) (21) and ER
lipid raft associated 1 (Erlin1) (17) genes were increased in the larvae
at 8 and 12 days after the pH5B injection, compared with the
untreated WT controls. The results in the zebrafish are in
accordance with those from previous studies on HCV-infected human
liver cells (17,22), demonstrating the possible
biological impact on the host.
To determine the pathological changes in the
microinjected zebrafish larvae, we used RT-PCR to detect the
expression of the marker genes for liver pathology (8) (Fig.
2B). In the first stage of liver disease, fatty liver,
acetyl-CoA carboxylase (ACC) (23) is the limiting enzyme in the
biosynthesis of fatty acids, and its high expression can promote
the synthesis of fat. Our results revealed that the expression of
ACC was almost undetectable in the WT larvae at 8 and 12 dpf, but
was significantly increased in the pH5B-injected larvae at 12 dpf
compared to the injected larvae at 8 dpf (Fig. 2B). In the second stage of liver
disease, liver fibrosis, the leptin receptor (24) and heparanase (25) are known as marker genes and these
were selected for examination in our study. Our results revealed
that the expression of leptin receptor and heparanase was
significantly increased in the pH5B-injected larvae compared with
the WT larvae (Fig. 2B). In the
third stage of liver disease, hepatocellular carcinoma, the level
of c-myc and survivin (26) in
the injected larvae was similar to that in the WT larvae at 12 dpf.
These findings indicate that the zebrafish HCV sub-replicon model
can partly mimic HCV activity in the human liver.
Evaluation of the zebrafish HCV
sub-replication model with anti-HCV drugs
The larvae displaying EGFP fluorescence were treated
with various concentrations of ribavirin or oxymatrine from 5 to 10
dpf, and the core gene was then tested as the sign of the
evaluation of the HCV sub-replicon model by RT-PCR. Incubation of
the larvae in the ribavirin- or oxymatrine-containing water for 5
days markedly inhibited the amplification of the negative strand
core RNA of the HCV sub-replicon (Fig. 3). The inhibition occurred in a
dose-dependent manner both in the ribavirin- and oxymatrine-treated
groups. Ribavirin at 100 μg/ml, or oxymatrine at concentrations ≥20
μg/ml, significantly suppressed the replication of the HCV
sub-replicon. The positive strand core RNA is included in the
figure as a reference to show the successful microinjection and
transcription of the pH5B vector into the zebrafish larvae
(Fig. 3). As the positive strand
HCV core RNA maintained a steady level during the course of
treatment, the anti-HCV mechanisms of the 2 drugs apperared to be
associated with HCV sub-genomic replication-related events. None of
the larvae died or grew abnormally during the course of treatment,
indicating a good level of safety of the drugs at the doses used.
These results indicate that the zebrafish-hosted HCV sub-replicon
amplification system is a suitable animal model with which to
evaluate anti-HCV drugs or drug candidates, particularly in the
case of water-soluble agents.
Discussion
In this study, we developed a zebrafish HCV
subgenomic replicon model that can be transcribed by the mHNF4a
promoter. All the proteins encoded by the HCV sub-replicon
construct were detected in the microinjected zebrafish larvae by
western blot analysis, which indicates that the mHNF4a promoter
functioned properly in zebrafish liver and that the key factors for
HCV RNA repliction, core and HCV RNA-dependent polymerase (NS5B)
were successfully translated. As the negative strand RNA was an
intermediate product of replication, its existence demonstrates
that the HCV sub-genome can be replicated in zebrafish. These
results indicated the successful development of a zebrafish HCV
subgenomic replication model. Moreover, the changes observed in the
transcription levels of HCV-associated genes and the liver
pathological marker genes in the pH5B-injected zebrafish larvae are
in agreement with those observed in HCV-infected human liver cells
(17), which further confirm the
successful creation of the zebrafish model of HCV.
The inhibitory effects of ribavirin and oxymatrine,
two anti-virus drugs used in clinical practice, against the HCV
sub-genomic replication indicate that the zebrafish HCV
sub-replicon model may serve as a valuable platform with which to
study the molecular events in HCV genomic replication and to
evaluate preventive and therapeutic strategies to combat HCV
infection. Furthermore, the results of this study confirm the
suitability of zebrafish as a HCV small animal model. Compared with
mouse models of HCV (27–30), this model system has several
advantages for drug screening and evaluation. First, the HCV
sub-replicon replicates actively and steadily in zebrafish tissue;
second, the procedure for creating the sub-replicon-positive larva
is straight forward and simple; and third, this easy-to-handle
small biological model seems to be suitable for drug screening. The
major disadvantages of this model are that the HCV sub-replicon is
possibly more suitable for water-soluble compounds and the dose
calculation is complicated.
Our results also provide evidence that zebrafish
liver cells may contain biological circumstances compatible to
human HCV replication that usually occur in human hepatocytes.
Although there may be other viral or host factors that contribute
to HCV RNA replication, the HCV core and NS5B proteins together
with the HCV 5′UTR and 3′UTR RNA sequence are capable of copying
the subgenomic RNA in the zebrafish model, and similar results were
reported in the study by Lee et al (6).
In conclusion, in this study, we confirm the use of
zebrafish as an in vivo biological model system for HCV
replication, with potential applications in the evaluation of
anti-HCV drugs.
Acknowledgments
This study was supported by the ‘Innovative Group
Grant’ from the Ministry of Education (China), the 11th 5-year ‘New
Drug R&D Program’ of the Ministry of Science and Technology
(China) (J.-D.J.), a National S&T Major Special Project on
Major New Drug Innovation grant (China) (item no.
2009ZX09301-003-6-2) (J.-P.Z.) and the National Natural Science
Foundation of China (no. 30772681) (J.-P.Z.). We would also like to
thank Wei-Xian Wang and Jie Meng for their assistance with the
microinjection of the zebrafish.
Abbreviations:
|
HCV
|
hepatitis C virus
|
|
UTR
|
untranslated region
|
|
NS5B
|
non-structural protein 5B
|
|
mHNF4
|
mouse hepatocyte nuclear factor 4
|
|
EGFP
|
enhanced green fluorescent protein
|
|
IRES
|
internal ribosome entry site
|
|
dpf
|
days post-fertilization
|
|
EMCV
|
encephalomyocarditis virus
|
|
WT
|
wild-type
|
|
ACC
|
acetyl-CoA carboxylase
|
|
PBST
|
phosphate-buffered saline containing
Tween-20
|
References
|
1
|
Gottwein JM and Bukh J: Hepatitis C virus
host cell interactions uncovered. Proc Natl Acad Sci USA.
104:13215–13216. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Friebe P, Lohmann V, Krieger N and
Bartenschlager R: Sequences in the 5′ nontranslated region of
hepatitis C virus required for RNA replication. J Virol.
75:12047–12057. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee KJ, Choi J, Ou JH and Lai MM: The
C-terminal transmembrane domain of hepatitis C virus (HCV) RNA
polymerase is essential for HCV replication in vivo. J Virol.
78:3797–3802. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Behrens SE, Tomei L and De Francesco R:
Identification and properties of the RNA-dependent RNA polymerase
of hepatitis C virus. EMBO J. 15:12–22. 1996.PubMed/NCBI
|
|
5
|
Lohmann V, Körner F, Herian U and
Bartenschlager R: Biochemical properties of hepatitis C virus NS5B
RNA-dependent RNA polymerase and identification of amino acid
sequence motifs essential for enzymatic activity. J Virol.
71:8416–8428. 1997.PubMed/NCBI
|
|
6
|
Lee S, Lee JH, Kee YH, Park MY and Myung
H: Partial reconstitution of hepatitis C virus RNA polymerization
by heterologous expression of NS5B polymerase and template RNA in
bacterial cell. Virus Res. 114:158–163. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liang JO and Rubinstein AL: Patterning of
the zebrafish embryo by nodal signals. Curr Top Dev Biol.
55:143–171. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rekha RD, Amali AA, Her GM, et al:
Thioacetamide accelerates steatohepatitis, cirrhosis and HCC by
expressing HCV core protein in transgenic zebrafish Danio rerio.
Toxicology. 243:11–22. 2008. View Article : Google Scholar
|
|
9
|
Wallace KN and Pack M: Unique and
conserved aspects of gut development in zebrafish. Dev Biol.
255:12–29. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Amatruda JF, Shepard JL, Stern HM and Zon
LI: Zebrafish as a cancer model system. Cancer Cell. 1:229–231.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lam SH, Wu YL, Vega VB, et al:
Conservation of gene expression signatures between zebrafish and
human liver tumors and tumor progression. Nat Biotechnol. 24:73–75.
2006. View
Article : Google Scholar
|
|
12
|
Ding CB, Zhang JP, Zhao Y, Peng ZG, Song
DQ and Jiang JD: Zebrafish as a potential model organism for drug
test against hepatitis C virus. PLoS One. 6:e229212011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Thisse C and Thisse B: High-resolution in
situ hybridization to whole-mount zebrafish embryos. Nat Protoc.
3:59–69. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Quasdorff M, Hösel M, Odenthal M, et al: A
concerted action of HNF4alpha and HNF1alpha links hepatitis B virus
replication to hepatocyte differentiation. Cell Microbiol.
10:1478–1490. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qadri I, Iwahashi M, Kullak-Ublick GA and
Simon FR: Hepatocyte nuclear factor (HNF) 1 and HNF4 mediate
hepatic multidrug resistance protein 2 up-regulation during
hepatitis C virus gene expression. Mol Pharmacol. 70:627–636. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bailly A, Elena M, Torres-Padilla, Tinel
AP and Weiss MC: An enhancer element 6 kb upstream of the mouse
HNF4α1 promoter is activated by glucocorticoids and liver-enriched
transcription factors. Nucleic Acids Res. 29:3495–3505. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nishimura-Sakurai Y, Sakamoto N, Mogushi
K, et al: Comparison of HCV-associated gene expression and cell
signaling pathways in cells with or without HCV replicon and in
replicon-cured cells. J Gastroenterol. 45:523–536. 2010. View Article : Google Scholar
|
|
18
|
Yilmaz Y, Yonal O, Kurt R, et al: Serum
fetuin A/α2HS-glycoprotein levels in patients with non-alcoholic
fatty liver disease: relation with liver fibrosis. Ann Clin
Biochem. 47:549–553. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Parent R, Qu X, Petit MA and Beretta L:
The heat shock cognate protein 70 is associated with hepatitis C
virus particles and modulates virus infectivity. Hepatology.
49:1798–1809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Helbig KJ, Ruszkiewicz A, Semendric L,
Harley HA, McColl SR and Beard MR: Expression of the CXCR3 ligand
I-TAC by hepatocytes in chronic hepatitis C and its correlation
with hepatic inflammation. Hepatology. 39:1220–1229. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chang ML, Yeh CT, Chen JC, et al: Altered
expression patterns of lipid metabolism genes in an animal model of
HCV core-related, nonobese, modest hepatic steatosis. BMC Genomics.
9:1092008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Blais DR, Brûlotte M, Qian Y, Bélanger S,
Yao SQ and Pezacki JP: Activity-based proteome profiling of
hepatoma cells during hepatitis C virus replication using protease
substrate probes. J Proteome Res. 9:912–923. 2010. View Article : Google Scholar
|
|
23
|
Shieh YS, Chang YS, Hong JR, et al:
Increase of hepatic fat accumulation by liver specific expression
of hepatitis B virus X protein in zebrafish. Biochim Biophys Acta.
1801:721–730. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheng SP, Chi CW, Tzen CY, et al:
Clinicopathologic significance of leptin and leptin receptor
expressions in papillary thyroid carcinoma. Surgery. 147:847–853.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tsiperson V, Goldshmidt O, Ilan N, et al:
Heparanase enhances early hepatocyte inclusion in the recipient
liver after transplantation in partially hepatectomized rats.
Tissue Eng Part A. 14:449–458. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma AC, Chung MI, Liang R and Leung AY: The
role of survivin2 in primitive hematopoiesis during zebrafish
development. Leukemia. 23:712–720. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ploss A and Rice CM: Towards a small
animal model for hepatitis C. EMBO Rep. 10:1220–1227. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Park IW, Ndjomou J, Fan Y, Henao-Mejia J
and He JJ: Hepatitis C virus is restricted at both entry and
replication in mouse hepatocytes. Biochem Biophys Res Commun.
387:489–493. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lerat H, Kammoun HL, Hainault I, et al:
Hepatitis C virus proteins induce lipogenesis and defective
triglyceride secretion in transgenic mice. J Biol Chem.
284:33466–33474. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guévin C, Lamarre A and Labonté P: Novel
HCV replication mouse model using human hepatocellular carcinoma
xenografts. Antiviral Res. 84:14–22. 2009. View Article : Google Scholar : PubMed/NCBI
|