Introduction
Bladder cancer is the most frequent urological
malignancy in China and the second most common urological
malignancy in the USA (1).
Approximately 386,300 patients were diagnosed with bladder cancer
in 2008, and 150,200 deaths were caused by this disease worldwide
in the same year (2). The median
survival of patients with bladder cancer is 15 months, and the
5-year survival rate is only 15% (3). Therefore, it is critical to
understand the molecular mechanisms involved in the development and
progression of bladder cancer.
MicroRNAs (miRNAs or miRs) are a class of small
non-coding RNA molecules, approximately ~22 nucleotides in length.
They regulate target mRNAs at the post-transcriptional level and
play crucial roles in the regulation of diverse physiological and
pathological processes (4,5). A
number of miRNAs have been found to be linked with various types of
cancer (6,7). miR-485 is a functional miRNA which
has received much attention in recent years. miR-485 acts as an
important regulator of neural development and plasticity of the
hippocampus (8). Recently, it has
been demonstrated that miR-485 is associated with certain malignant
tumors, including hepatocellular carcinomas, breast cancer and
tumors of the central nervous system (9–12).
However, little is known about the expression of miR-485 or its
role in bladder cancer.
In the present study, we examined the expression of
miR-485 in human bladder cancer tissues and bladder cancer cell
lines, and examined the effects of miR-485-5p on cell metastasis
and epithelial-mesenchymal transition (EMT). In addition, we
explored the underlying mechanisms of the functions of miR-485 in
bladder cancer cells and identified a novel direct target of
miR-485-5p.
Materials and methods
Tumor samples
The bladder cancer tissue and the matched normal
tissue specimens were collected from 15 patients diagnosed with
bladder cancer who had undergone surgery, as the primary therapy,
in The First Affiliated Hospital of Bengbu Medical College (Bengbu,
China). None of the patients had received chemotherapy or
radiotherapy prior to surgery. Prior to their participation in our
study, all the patients provided written informed consent in
compliance with the code of ethics of the Declaration of Helsinki.
This study was approved by the Ethics Committee of The First
Affiliated Hospital of Bengbu Medical College. The specimens were
frozen in liquid nitrogen immediately after surgery.
Cell culture and transfection
Human bladder cancer cell lines (SW780, T24, HT1376
and HT5637) and human bladder epithelial cell lines HU609 and
HEK293 cells (all from the American Type Culture Collection,
Manassas, VA, USA) were cultured in Dulbecco's modified Eagle's
medium (DMEM; Gibco-BRL, Grand Island, NY, USA) containing 10%
newborn calf serum (Gibco-BRL) at 37°C with 5% CO2.
Prior to transfection, the cells were seeded in 6-well plates at
5×104 cells/well and allowed to grow for 24 h. The
miR-485-5p mimic (Bioneer, Shanghai, China), miR-485-5p inhibitor
(Bioneer), miR control, the pcDNA3.1 (Zhonghong Boyuan Biological
Technology Co., Ltd., Shenzhen, China) and high mobility group
AT-hook 2 (HMGA2)-pcDNA3.1 (Zhonghong Boyuan Biological Technology
Co., Ltd.) plasmids were transfected into the cells using a
Lipofectamine™ 2000 transfection kit (Invitrogen Life Technologies,
Carlsbad, CA, USA) according to the manufacturer's instructions.
Briefly, 2 µM miR-485-5p mimic or inhibitor or plasmids were
mixed with Lipofectamine 2000 to form lipid-DNA complexes. The
cells were cultured in serum-free medium with the lipid-DNA
complexes for 6 h, and the medium was then replaced with fresh DMEM
and the cells were maintained in the cultures for 24 h.
Luciferase reporter assay
We used microRNA.org (http://www.microrna.org/) and miRDB (http://mirdb.org/) to find the predicted target of
miR-485-5p. Then, the HMGA2 wild-type and mutant 3′ untranslated
region (UTR) were amplified and cloned into a psiCHECK-2 vector
(Promega Corp., Madison, WI, USA). The cells were transfected with
100 ng of the psiCHECK-2 vector and 100 nM of the miR-485-5p mimic
or miR control. After 24 h, the cells were lysed and assayed using
the Dual-Luciferase reporter assay system (Promega Corp.) according
to the manufacturer's instructions.
Cell invasion assay
Cell invasion was examined using Matrigel (BD
Biosciences, San Jose, CA, USA)-coated Transwell chambers (Corning
Inc., Corning, NY, USA). The cells seeded at a density of
5×104 cells/ml in serum-free medium were placed in the
1:8 diluted Matrigel-coated upper chamber. The lower chamber was
filled with 1 ml complete medium. Following incubation at 37°C for
24 h, the cells that had invaded the bottom of the membrane were
fixed in 95% ethanol for 20 min and then stained with hematoxylin.
The cells on the surface of the membrane were removed using a
cotton swab. The cells which had invaded were counted in 5 randomly
selected microscopic fields.
Cell adhesion assay
The 96-well plates were coated with fibronectin
(Sigma-Aldrich, St. Louis, MO, USA) and blocked with 1% bovine
serum albumin (BSA; Sigma-Aldrich) in phosphate-buffered saline
(PBS) at 37°C for 2 h. The cells were suspended in serum-free
medium and then seeded in the 96-well plates. Following incubation
at 37°C for 2 h, the non-adherent cells were removed by washing
with PBS. The adherent cells were fixed with 4% paraformaldehyde
and stained with 0.5% crystal violet.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using an RNeasy/miRNeasy
Mini kit (Qiagen, Limburg, The Netherlands) according to the
manufacturer's instructions. A reverse transcriptase kit
(Fermentas, Vilnius, Lithuania) was used to synthesize the cDNA.
Quantitative PCR (qPCR) was carried out using SYBR-Green PCR Master
Mix (Applied Biosystems, Foster City, CA, USA) on an ABI 7500
System (Applied Biosystems). The expression of miR-485-3p and
miR-485-5p was examined and normalized to the internal control
(GAPDH), and relative induction was calculated using the
2−ΔΔCt method.
Western blot analysis
Whole cell extracts were isolated using a Total
Protein Extraction kit (Applygen Technologies, Inc., Beijing,
China) according to the manufacturer's instructions. Proteins (40
µg) were resolved by sodium dodecyl sulfate polyacrylamide
gel electrophoresis (SDS-PAGE) and transferred onto a
polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA).
The membrane was blocked with 5% BSA and probed with the primary
antibodies [rabbit polyclonal to HMGA2 (ab97276), mouse monoclonal
to E-cadherin (ab45990), rabbit polyclonal to vimentin (ab45939),
mouse monoclonal to N-cadherin (ab98952) all from Abcam, Cambridge,
MA, USA; mouse monoclonal to GAPDH (A01020), Amyjet Scientific,
Inc., Wuhan, China]. The GAPDH antibody was used as an internal
control. Antibody binding was detected with horseradish peroxidase
(HRP)-linked secondary antibody [goat anti-mouse IgG (sc-2005),
goat anti-rabbit IgG (sc-2004), Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA] and the chemiluminescence (ECL western blotting
kit; Pierce Biotechnology, Inc., Rockford, IL, USA).
Statistical analysis
SPSS 19.0 software (IBM, Armonk, NY, USA) was used
to perform the statistical analysis. Data are expressed as the
means ± SD. The student's t-test was used to evaluate the
differences between 2 groups. A P-value <0.05 was considered to
indicate a statistically significant difference.
Results
Expression of miR-485 and HMGA2 in human
bladder cancer cell lines
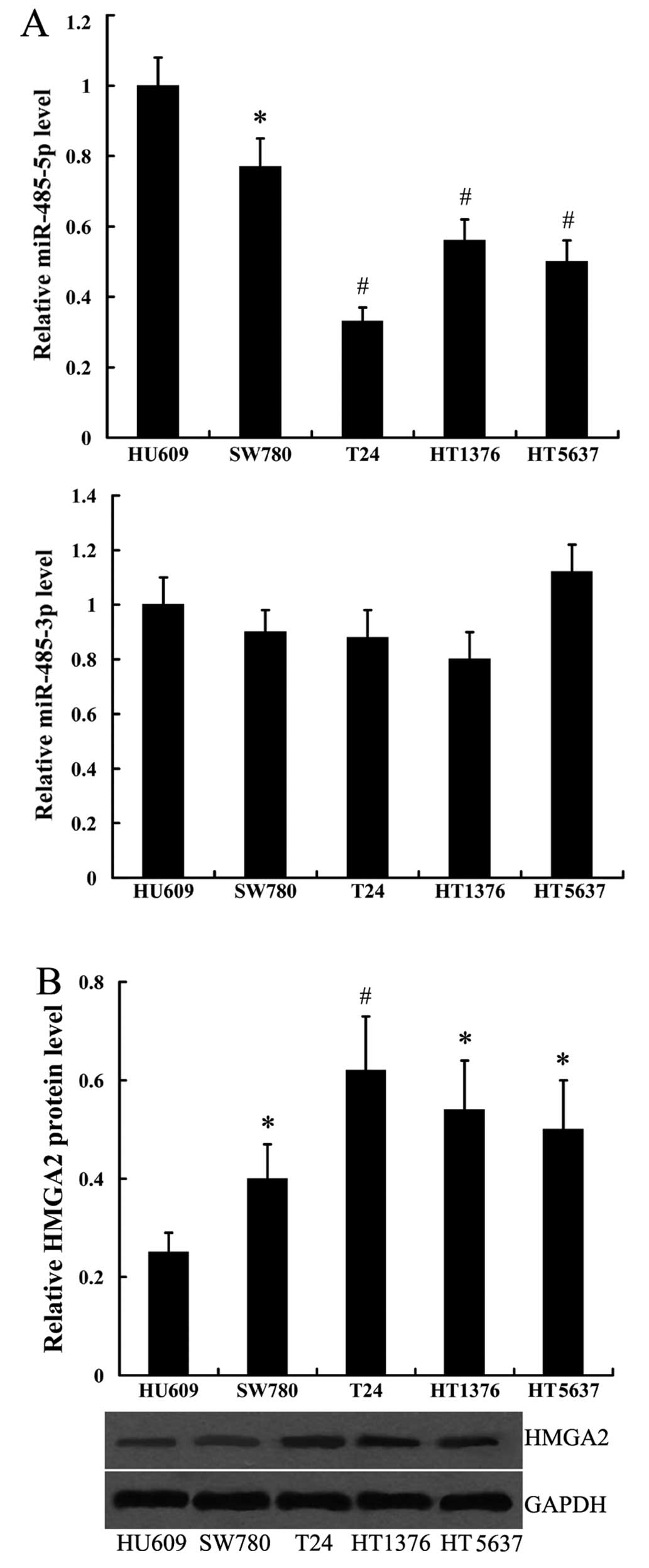
To detect the expression of miR-485-3p, miR-485-5p
and HMGA2 in human bladder cancer cell lines, we used 4 cell lines,
SW780, T24, HT1376 and HT5637 from different grades of bladder
cancer, and the human bladder epithelial cell line, HU609, was used
as the normal control. The expression of miR-485-5p was highest in
the HU609 cells, relatively lower in the SW780, HT1376 and HT5637
cells, and it was lowest in the poorly differentiated cell line,
T24 (Fig. 1A). On the contrary,
HMGA2 protein expression was lowest in the HU609 cells, a moderate
expression was noted in the SW780, HT1376 and HT5637 cells, and a
high expression was noted in the T24 cells (Fig. 1B). However, the expression level
of miR-485-3p in the 4 bladder cancer cell lines did not differ
significantly from that in the HU609 cells (Fig. 1A).
Expression of miR-485 and HMGA2 in human
bladder cancer tissues
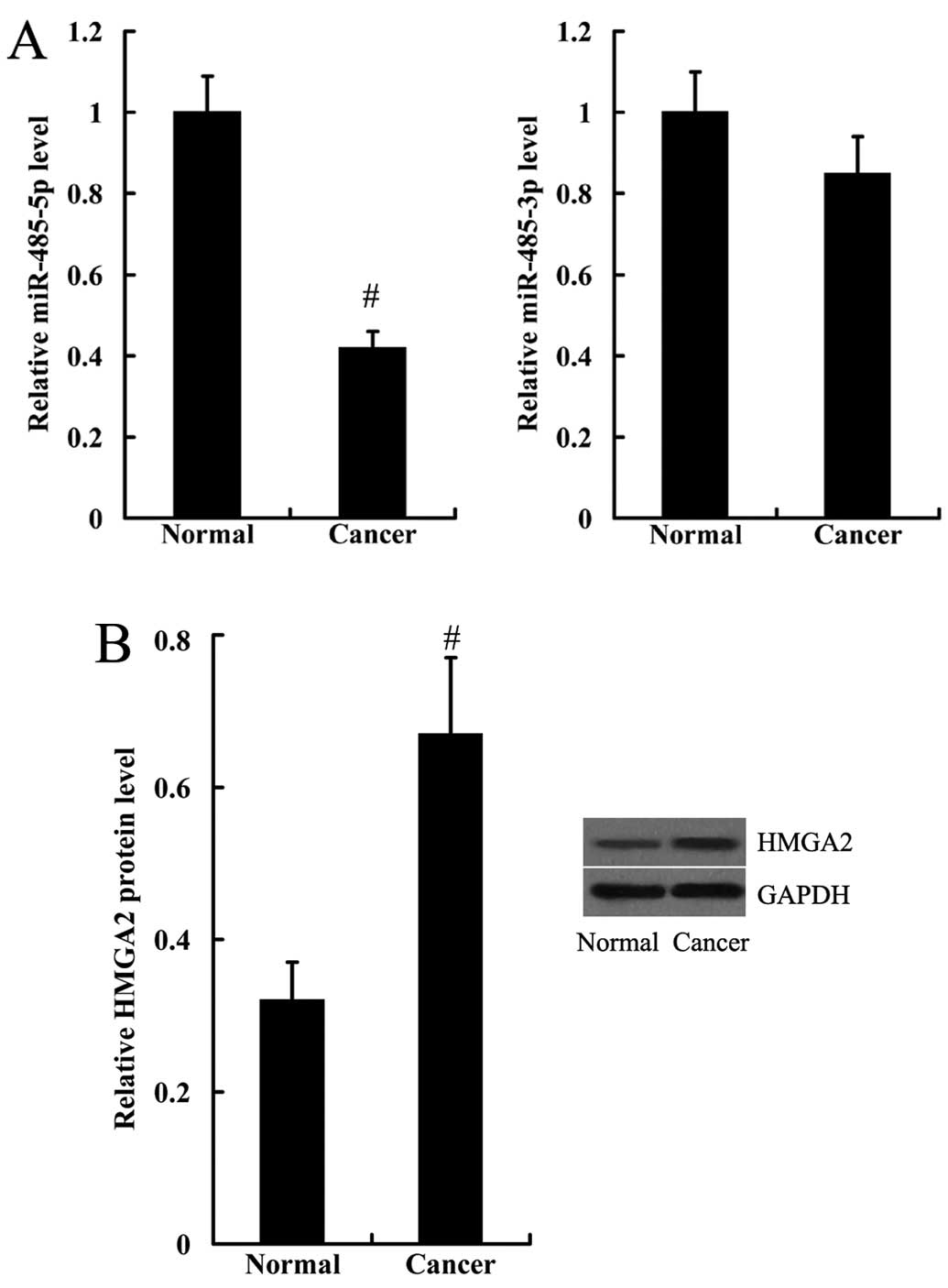
The expression levels of miR-485-3p, miR-485-5p and
HMGA2 were detected in the human bladder cancer tissues. Consistent
with the results obtained from the human bladder cancer cell lines,
the expression level of miR-485-5p was significantly decreased,
whereas HMGA2 protein expression was significantly increased in the
bladder cancer tissues, compared with the normal tissues (Fig. 2). The expression levels of
miR-485-3p in the human bladder cancer tissues and normal tissues
did not differ significantly (Fig.
2).
Effect of miR-485-5p on cell invasion and
adhesion
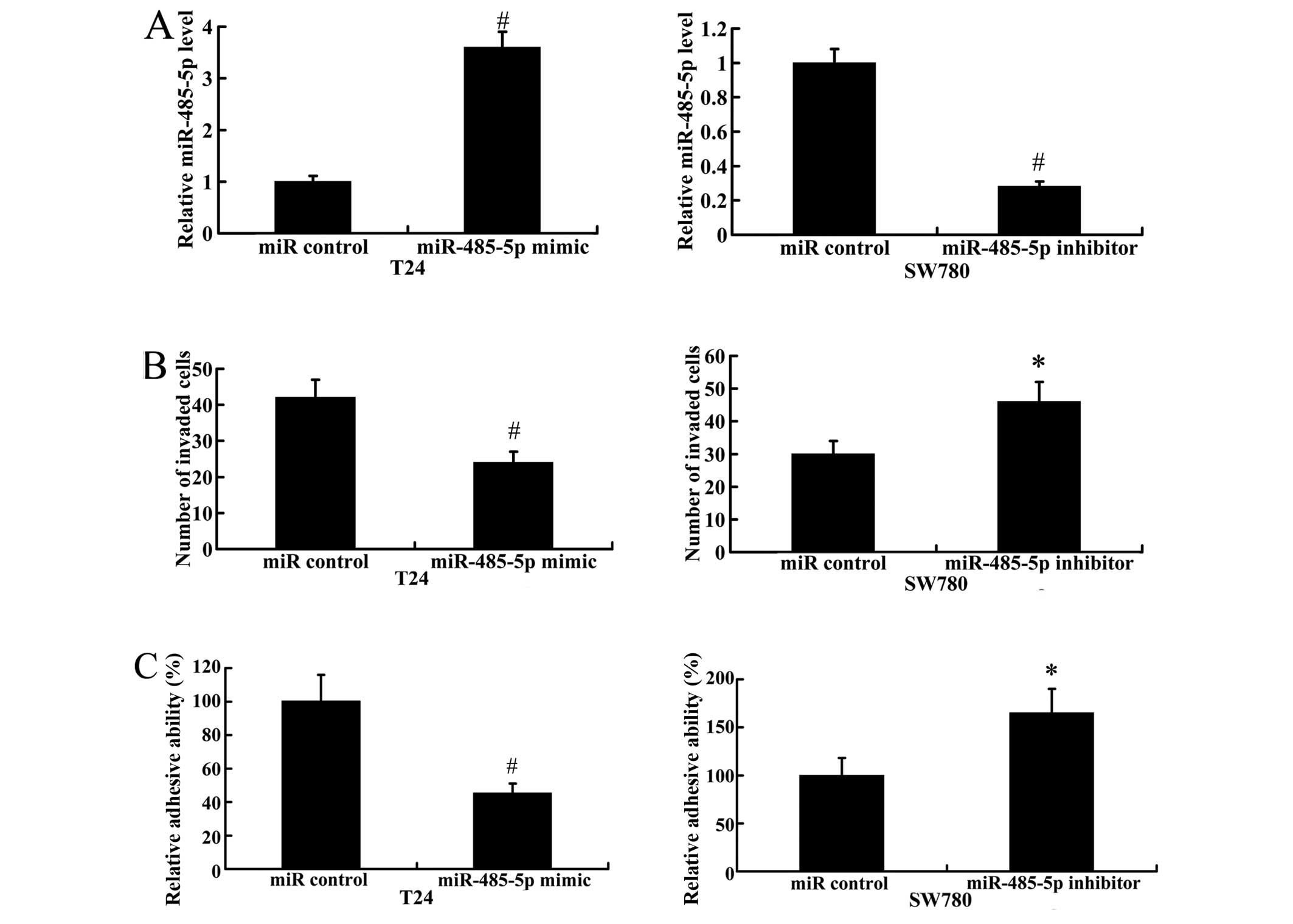
To explore the effects of miR-485-5p on cell
invasion and adhesion, T24 cells (which exhibited the lowest levels
of miR-485-5p expression of the 4 bladder cancer cell lines
examined) were transfected with the miR-485-5p mimic to induce the
over-expression of miR-485-5p. We suppressed the expression of
miR-485-5p in the SW780 cells, which exhibited a high miR-485-5p
expression, by transfection with the miR-485-5p inhibitor. As
demonstrated in Fig. 3A,
transfection with the miR-485-5p mimic significantly increased the
expression level of miR-485-5p, while transfection with the
miR-485-5p inhibitor led to a significantly decrease in the
expression of miR-485-5p. The results of Transwell-Matrigel assay
revealed that the number of invaded cells was significantly
decreased significantly when miR-485-5p was overexpressed.
Moreover, the inhibition of miR-485-5p (by transfection with
miR-485-5p inhibitor) promoted cell invasion compared with the
cells transfected with the miR control (Fig. 3B). A cell adhesion assay revealed
that adhesive ability decreased in the T24 cells transfected with
the miR-485-5p mimic, but increased in the SW780 cells transfected
with the miR-485-5p-inhibitor, compared with the cells transfected
with the miR control (Fig.
3C).
Effect of miR-485-5p on EMT
To examine the effect of miR-485-5p on EMT, the
levels of the epithelial cell marker, E-cadherin, as well as the
mesenchymal cell markers, vimentin and N-cadherin, were detected in
T24 and SW780 cells following transfection with the miR-485-5p
mimic or miR-485-5p inhibitor. As shown in Fig. 4, transfection with the miR-485-5p
mimic suppressed the protein expression of vimentin and N-cadherin,
whereas it promoted the expression of E-cadherin protein in the T24
cells. However, the inhibition of miR-485-5p with the miR-485-5p
inhibitor in the SW780 cells induced EMT, evidenced by the
increased protein expression of vimentin and N-cadherin, and the
decreased protein expression of E-cadherin.
miR-485-5p directly targets the 3′UTR of
HMGA2
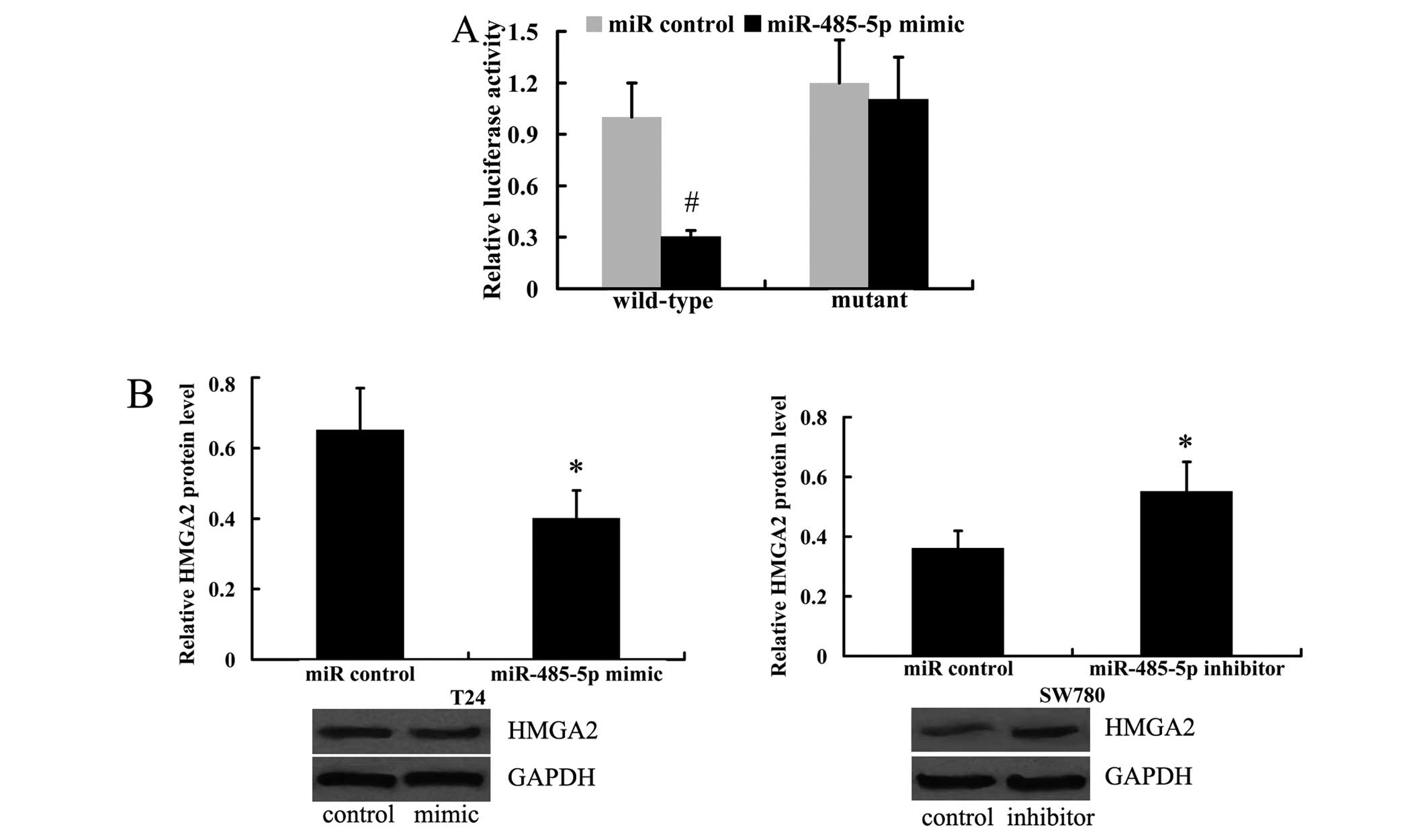
Using the prediction algorithm, microRNA.org
(http://www.microrna.org/) and miRDB (http://mirdb.org/), HMGA2 was predicted to be a target
of miR-485-5p. To determine the association between miR-485-5p and
HMGA2, we performed a luciferase reporter assay to determine
whether miR-485-5p directly targets the 3′UTR of HMGA2. The HEK293
cells were transfected with the HMGA2 wild-type 3′UTR or mutant
3′UTR vector and then with the miR-485-5p mimic or miR control. The
results revealed that the luciferase activity of wild-type 3′UTR
was significantly decreased by transfection with the miR-485-5p
mimic. However, the luciferase activity of mutant 3′UTR was not
altered substantially (Fig.
5A).
Subsequently, we investigated whether miR-485-5p
regulates the expression of HsxMGA2 in the T24 and SW780 cells. As
shown in Fig. 5B, miR-485-5p
overexpression induced by transfection with miR-485-5p mimic
resulted in a decrease in the HMGA2 protein level in the T24 cells.
Moreover, the inhibition of miR-485-5p led to the upregulated
protein expression of HMGA2 in the SW780 cells (Fig. 5B).
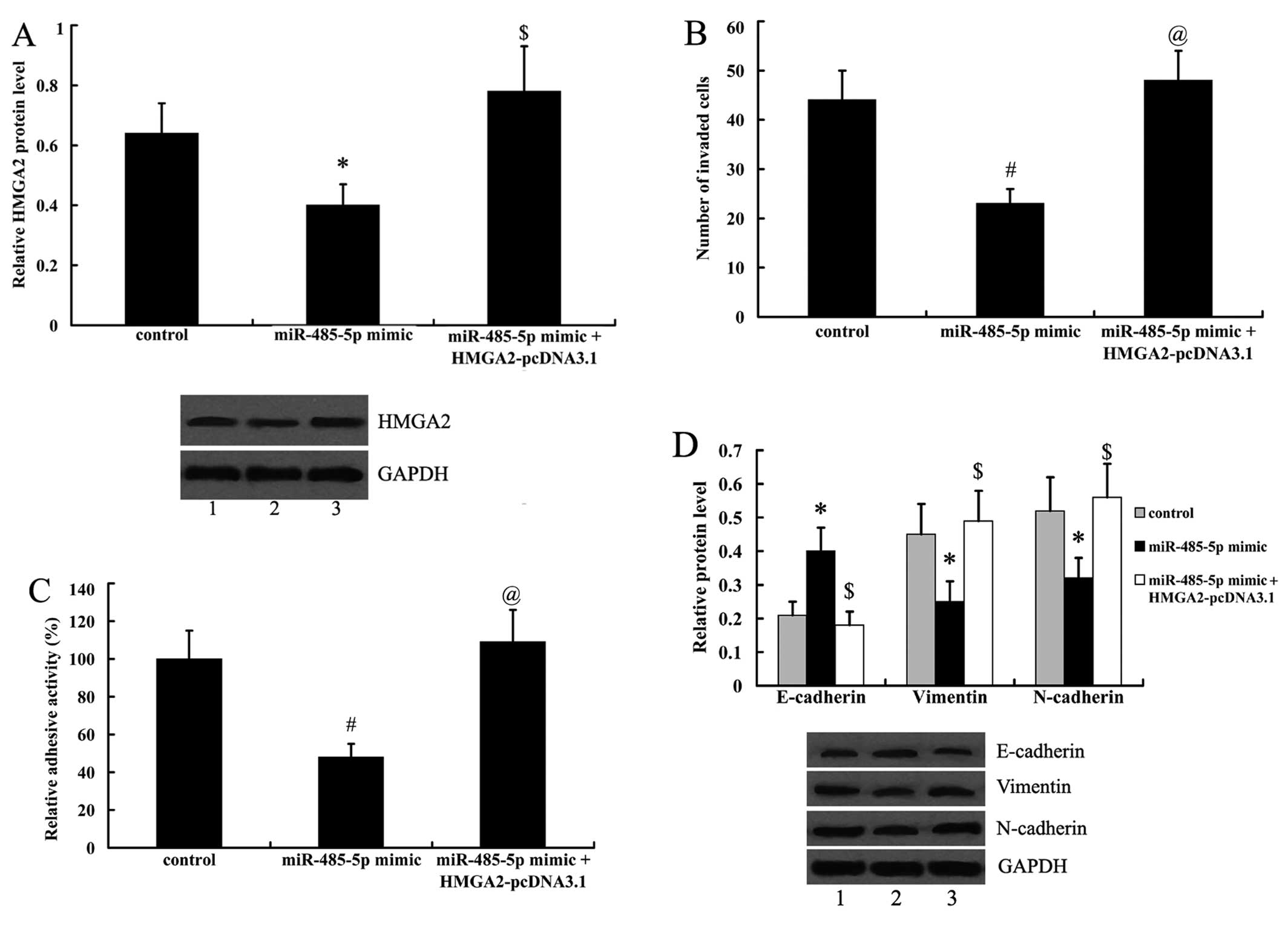
HMGA2 overexpression reverses the effects
of miR-485-5p on cell metastasis and EMT
To investigate whether the effects of miR-485-5p on
cell metastasis and ETM are related to HMGA2, the T24 cells
overexpressing miR-485-5p were transfected with HMGA2-pcDNA3.1. As
shown in Fig. 6A, the expression
of HMGA2 was suppressed by the miR-485-5p mimic in T24 cells, and
was significantly upregulated by transfection with HMGA2-pcDNA3.1.
Moreover, we found that the inhibitory effects of miR-485-5p on
cell invasion, adhesion and EMT were reversed by transfection with
HMGA2-pcDNA3.1 (Fig. 6B–D).
Discussion
The differential expression profile of miRNAs has
been widely reported between bladder cancer and normal cells
(13). In the present study, we
firstly demonstrated that the expression of miR-485-5p was
downregulated in human bladder cancer tissues and various bladder
cancer cell lines. These results suggest that miR-485-5p plays an
important role in the development and progression of bladder
cancer.
It has been demonstrated that miR-485-5p responds to
hypoxic stress in soft tissue sarcoma cells (14). The expression of miR-485-5p was
downregulated in Alzheimer's disease and malignant serous ovarian
tumors, and correlated with FIGO grade in the latter (14), suggesting that miR-485-5p may be
of potential importance as a diagnostic biomarker. Consistently,
miR-485-3p is downregulated in metastatic cell lines as compared
with normal prostatic epithelial cells (11). In addition, it has been found that
miR-485 suppresses the proliferation and migration of breast
carcinoma T47D cells (12). In
the present study, we performed cell invasion and adhesion assays
to investigate the effects of miR-485-5p on bladder cancer cell
metastasis. It is well known that EMT is a well-coordinated process
for metastasis (15,16); therefore, the effects of
miR-485-5p on EMT were also examined. The results demonstrated that
miR-485-5p inhibited bladder cancer cell metastasis and EMT.
In the present study, we examined HMGA2 as a novel
direct target of miR-485-5p. It has been demonstrated elsewhere
that HMGA2, an architectural transcription factor,plays a role in
certain cellular biological processes, such as cell proliferation,
differentiation, cell transformation and cell cycle control
(17–19). HMGA2 is expressed predominantly in
various undifferentiated tissues during embryonic development,
whereas it is usually not detectable in normal adult tissues
(20–23). The heightened expression of HMGA2
in adult tissues is commonly associated with various human cancers
(24–28), such as lung, breast and ovarian
cancer. It has been reported that HMGA2 is upregulated in bladder
cancer at both the transcriptional and translational level, and it
has been noted that HMGA2 expression is a potential prognostic
marker for both tumor recurrence and tumor progression (29). Moreover, a it has been previously
demonstrated that the expression of HMGA2 is closely associated
with EMT in bladder cancer (30).
Consistent with previous reports, in the present study, we found
that the protein expression of HMGA2 was upregulated in human
bladder cancer tissues and bladder cancer cell lines. To examine
the association between miR-485-5p and HMGA2, we searched for the
potential target genes of miR-485-5p using the prediction
algorithm, microRNA.org (http://www.microrna.org/) and miRDB (http://mirdb.org/), and HMGA2 was predicted to be a
target gene. Using a luciferase reporter assay and western blot
analysis, we demonstrated that miR-485-5p suppressed HMGA2
expression by directly targeting the 3′UTR of HMGA2. In addition,
using the HMGA2 overexpression plasmid, HMGA2-pcDNA3.1, we found
that HMGA2 reversed the effects of miR-485-5p on cell metastasis
and EMT. These results suggest that HMGA2 plays an important role
in mediating the effects of miR-485-5p.
Taken together, to the best of our knowledge, the
present study is the fist to identify miR-485-5p as a suppressive
miRNA in human bladder cancer. In addition, we demonstrated that
miR-485-5p exerts its effect at least partly through the
suppression of HMGA2. Our study further expanded on the function of
miR-485 and provided a novel potential target for the treatment of
bladder cancer.
References
|
1
|
Zhu Z, Shen Z and Xu C: Inflammatory
pathways as promising targets to increase chemotherapy response in
bladder cancer. Mediators Inflamm. 2012:5286902012.PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
von der Maase H, Sengelov L, Roberts JT,
Ricci S, Dogliotti L, Oliver T, Moore MJ, Zimmermann A and Arning
M: Long-term survival results of a randomized trial comparing
gemcitabine plus cisplatin, with methotrexate, vinblastine,
doxorubicin, plus cisplatin in patients with bladder cancer. J Clin
Oncol. 23:4602–4608. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kusenda B, Mraz M, Mayer J and Pospisilova
S: MicroRNA biogenesis, functionality and cancer relevance. Biomed
Pap Med Fac Univ Palacky Olomouc Czech Repub. 150:205–215. 2006.
View Article : Google Scholar
|
|
7
|
Mraz M, Pospisilova S, Malinova K, Slapak
I and Mayer J: MicroRNAs in chronic lymphocytic leukemia
pathogenesis and disease subtypes. Leuk Lymphoma. 50:506–509. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cohen JE, Lee PR and Fields RD: Systematic
identification of 3′-UTR regulatory elements in activity-dependent
mRNA stability in hippocampal neurons. Philos Trans R Soc Lond B
Biol Sci. 369:201305092014. View Article : Google Scholar
|
|
9
|
Yang H, Cho ME, Li TW, Peng H, Ko KS, Mato
JM and Lu SC: MicroRNAs regulate methionine adenosyltransferase 1A
expression in hepatocellular carcinoma. J Clin Invest. 123:285–298.
2013. View
Article : Google Scholar :
|
|
10
|
Costa FF, Bischof JM, Vanin EF, Lulla RR,
Wang M, Sredni ST, Rajaram V, Bonaldo MF, Wang D, Goldman S, et al:
Identification of microRNAs as potential prognostic markers in
ependymoma. PLoS One. 6:e251142011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Formosa A, Markert EK, Lena AM, Italiano
D, Finazzi-Agro' E, Levine AJ, Bernardini S, Garabadgiu AV, Melino
G and Candi E: MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c,
miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p,
mapped to the 14q32.31 locus, regulate proliferation, apoptosis,
migration and invasion in metastatic prostate cancer cells.
Oncogene. 33:5173–5182. 2014. View Article : Google Scholar
|
|
12
|
Anaya-Ruiz M, Bandala C and Perez-Santos
JL: miR-485 acts as a tumor suppressor by inhibiting cell growth
and migration in breast carcinoma T47D cells. Asian Pac J Cancer
Prev. 14:3757–3760. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Catto JW, Alcaraz A, Bjartell AS, De Vere
White R, Evans CP, Fussel S, Hamdy FC, Kallioniemi O, Mengual L,
Schlomm T and Visakorpi T: MicroRNA in prostate, bladder, and
kidney cancer: a systematic review. Eur Urol. 59:671–681. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gits CM, van Kuijk PF, de Rijck JC,
Muskens N, Jonkers MB, van IJcken WF, Mathijssen RH, Verweij J,
Sleijfer S and Wiemer EA: MicroRNA response to hypoxic stress in
soft tissue sarcoma cells: microRNA mediated regulation of HIF3α.
BMC Cancer. 14:4292014. View Article : Google Scholar
|
|
15
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brabletz T, Hlubek F, Spaderna S,
Schmalhofer O, Hiendlmeyer E, Jung A and Kirchner T: Invasion and
metastasis in colorectal cancer: epithelial-mesenchymal transition,
mesenchymal-epithelial transition, stem cells and beta-catenin.
Cells Tissues Organs. 179:56–65. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Di Cello F, Hillion J, Hristov A, Wood LJ,
Mukherjee M, Schuldenfrei A, Kowalski J, Bhattacharya R, Ashfaq R
and Resar LM: HMGA2 participates in transformation in human lung
cancer. Mol Cancer Res. 6:743–750. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lanahan A, Williams JB, Sanders LK and
Nathans D: Growth factor-induced delayed early response genes. Mol
Cell Biol. 12:3919–3929. 1992.PubMed/NCBI
|
|
19
|
Narita M, Narita M, Krizhanovsky V, Nuñez
S, Chicas A, Hearn SA, Myers MP and Lowe SW: A novel role for
high-mobility group a proteins in cellular senescence and
heterochromatin formation. Cell. 126:503–514. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gattas GJ, Quade BJ, Nowak RA and Morton
CC: HMGIC expression in human adult and fetal tissues and in
uterine leiomyomata. Genes Chromosomes Cancer. 25:316–322. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou X, Benson KF, Ashar HR and Chada K:
Mutation responsible for the mouse pygmy phenotype in the
developmentally regulated factor HMGI-C. Nature. 376:771–774. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ashar HR, Chouinard RA Jr, Dokur M and
Chada K: In vivo modulation of HMGA2 expression. Biochim Biophys
Acta. 1799:55–61. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pfannkuche K, Summer H, Li O, Hescheler J
and Dröge P: The high mobility group protein HMGA2: A co-regulator
of chromatin structure and pluripotency in stem cells? Stem Cell
Rev. 5:224–230. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Piscuoglio S, Zlobec I, Pallante P, Sepe
R, Esposito F, Zimmermann A, Diamantis I, Terracciano L, Fusco A
and Karamitopoulou E: HMGA1 and HMGA2 protein expression correlates
with advanced tumour grade and lymph node metastasis in pancreatic
adenocarcinoma. Histopathology. 60:397–404. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu Y, Song Y and Liu H: Expression and its
clinical significance of HMGA2 in the patients with non-small cell
lung cancer. Zhongguo Fei Ai Za Zhi. 11:377–381. 2008.In Chinese.
PubMed/NCBI
|
|
26
|
Langelotz C, Schmid P, Jakob C, Heider U,
Wernecke KD, Possinger K and Sezer O: Expression of
high-mobility-group-protein HMGI-C mRNA in the peripheral blood is
an independent poor prognostic indicator for survival in metastatic
breast cancer. Br J Cancer. 88:1406–1410. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Malek A, Bakhidze E, Noske A, Sers C,
Aigner A, Schäfer R and Tchernitsa O: HMGA2 gene is a promising
target for ovarian cancer silencing therapy. Int J Cancer.
123:348–356. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang X, Liu X, Li AY, Chen L, Lai L, Lin
HH, Hu S, Yao L, Peng J, Loera S, et al: Overexpression of HMGA2
promotes metastasis and impacts survival of colorectal cancers.
Clin Cancer Res. 17:2570–2580. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang GL, Zhang LH, Bo JJ, Hou KL, Cai X,
Chen YY, Li H, Liu DM and Huang YR: Overexpression of HMGA2 in
bladder cancer and its association with clinicopathologic features
and prognosis HMGA2 as a prognostic marker of bladder cancer. Eur J
Surg Oncol. 37:265–271. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ding X, Wang Y, Ma X, Guo H, Yan X, Chi Q,
Li J, Hou Y and Wang C: Expression of HMGA2 in bladder cancer and
its association with epithelial-to-mesenchymal transition. Cell
Prolif. 47:146–151. 2014. View Article : Google Scholar : PubMed/NCBI
|