Introduction
Glioma is the most common type of primary malignant
brain tumors in adults, accounting for 44.69% of intracranial
tumors. Despite intense therapy incorporating surgical resection,
radiation therapy and chemotherapy, the prognosis for glioblastoma
patients remains poor (1). Glioma
is a genetic disease that is caused by multiple oncogenic signaling
pathways and mutant genes (2). In
glioma cells, aberrant oncogenic factors cause activation of the
signaling pathways that are essential for cell proliferation and
survival. Therefore, selective degradation of such oncogenic
factors may be a promising approach to control glioma tumorigenesis
and progression.
One of the major protein degradation pathways is the
ubiquitin proteasome system (UPS) (3–7),
which is known as a cellular tool for selective degradation of
target proteins and has been shown to have an important role in
multiple physiological processes, such as signaling regulation,
transcriptional regulation, cell survival, migration, apoptosis and
DNA damage response (4,7). The UPS is composed of a 76-amino
acid protein ubiquitin (Ub), a multi-subunit protein organelle 26S
proteasome, and a three-step enzymatic cascade of Ub-activating
(E1), Ub-conjugating (E2) and Ub-ligase (E3) enzymes (6), which are essential for the
consistent recycling of a plethora of proteins with distinct
structural and functional roles within the cell, including cell
cycle regulation (3). A previous
study has confirmed the involvement of UPS in regulation of the
cell cycle in malignant gliomas (6).
The ubiquitin-associated protein 2-like (UBAP2L)
gene is located at human chromosome 1q21.3 and is involved in the
pathogenesis of several types of human cancer (8), including multiple myeloma (9), hepatocellular carcinoma (HCC)
(10) and malignant ovarian
tumors (11). Additionally,
UBAP2L contains a UBA domain near its N-terminus, which is a
recurring sequence of ~45 amino acids and is associated with the
UPS, DNA excision repair and cell signaling via protein kinases
(12). Thus, UBAP2L may have a
vital role in regulation of cell cycle and apoptosis by utilizing
the UPS. However, the function of UBAP2L in glioma has not been
investigated.
To study the potential role of UBAP2L in human
glioma, loss of function analysis using lentivirus-mediated short
hairpin RNA (shRNA) was performed in glioblastoma cell lines.
Subsequently, the effects of UBAP2L silencing were investigated on
cell proliferation, colony formation and cell cycle
progression.
Materials and methods
Cell culture
Human glioma U251, U87MG, U373, A172 and U-118MG
cell lines, as well as the human embryonic kidney (HEK) 293T cell
line, were purchased from the Cell Bank of Type Culture Collection
of Chinese Academy of Sciences (Shanghai, China). All the cell
lines were maintained in Dulbecco's modified Eagle's medium
(HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum
(HyClone) at 37°C in a humidified atmosphere of 5%
CO2.
Construction of UBAP2L shRNA lentivirus
and cell infection
The shRNA
(5′-GCCAATACTGATGATAACTATCTCGAGATAGTTATCATCAGTATTGGCTTTTT-3′)-targeting
human UBAP2L gene (NM_001127320.1) was synthesized, annealed and
ligated into the green fluorescent protein (GFP)-encoding pFH-L
vector (Shanghai Hollybio Co., Ltd., Shanghai, China). Scrambled
shRNA
(5′-CTAGCCCGGTTCTCCGAACGTGTCACGTATCTCGAGATACGTGACACGTTCGGAGAATTT
TTTTAAT-3′) was used as the control. The lentiviral-based
shRNA-expressing vector was verified by DNA sequencing. The
generated plasmid was termed pFH-L-shUBAP2L or -shCon. To avoid the
possible off-target effect, another shRNA
(5′-CGCAGCAGAATACCTTTCATCTCGAGATGAAAGGGTATTCTGCTGCGTTTTT-3′) was
used to obtain comparable results. Recombinant lentiviral vectors
and packaging vectors (pVSVG-I and pCMVΔR8.92) were subsequently
transfected into 293T cells using Lipofectamine 2000 (Invitrogen
Life Technologies, Carlsbad, CA, USA) according to the
manufacturer's instructions. Supernatants containing lentiviruses
expressing UBAP2L shRNA or control shRNA were harvested 3 days
after transfection. Subsequently, the lentiviruses were purified by
ultracentrifugation, and the titer of lentiviruses was determined
as described previously (13).
U251 and U373 cells were infected with the lentivirus constructs at
a multiplicity of infection of 10 and uninfected cells were used as
negative controls. The infection efficiency was determined by
counting GFP-expressing cells under an Olympus BX50
Brightfield/Fluorescence microscope (Olympus Corp., Tokyo, Japan)
96 h after infection.
Reverse transcription-polymerase chain
reaction (RT-PCR)
Total RNA was extracted from the cultured cells
using TRIzol reagent (Invitrogen Life Technologies). cDNA was
synthesized using M-MLV Reverse Transcriptase (Promega Corp.,
Madison, WI, USA) according to the manufacturer's instructions. In
brief, a mixture including 1.5 µg of total RNA, 0.75
µg oligo(dT) primer (Sangon Biotech Co., Ltd., Shanghai,
China) and nuclease-free water was heated at 70°C for 5 min and
subsequently cooled on ice for a further 5 min. In addition, 4
µl M-MLV buffer, 1.25 µl dNTP, 0.5 µl RNasin
and 0.75 µl M-MLV-RT were supplemented to the mixture up to
a final volume of 20 µl, followed by incubation at 42°C for
60 min.
RT-quantitative PCR (RT-qPCR)
RT-qPCR analysis was performed using SYBR-Green
Master mix kits (Takara Bio, Inc., Tokyo, Japan) according to the
manufacturer's instructions for the Bio-Rad Connect Real-Time PCR
platform (Bio-Rad, Hercules, CA, USA). Briefly, each PCR reaction
mixture, containing 10 µl of 2X SYBR-Green Master mix, 0.8
µl of sense and antisense primers (2.5 µM) and 5
µl of cDNA (10 ng), was run for 40 cycles with an initial
denaturation at 95°C for 60 sec, and subsequent denaturation at
95°C for 5 sec, and annealing and extension at 60°C for 30 sec. The
relative quantification of gene expression was calculated using the
2−ΔΔCT method (14).
The primer sequences for PCR amplification of the UBAP2L gene were
forward, 5′-ACACAATCCCCATCACTGGT-3′ and reverse,
5′-CAGAGGAGAAGACGGAGGTG-3′. β-actin was used as an internal
control. The primer sequences of β-actin were forward,
5′-GTGGACATCCGCAAAGAC-3′ and reverse,
5′-AAAGGGTGTAACGCAACTA-3′.
Western blot analysis
U251 and U373 cells were respectively collected 5
days after infection and total proteins were isolated from the
cells and quantitated by the bicinchoninic acid method. Proteins
were separated by SDS-PAGE and transferred onto polyvinylidene
difluoride membrane (Millipore, Billerica, MA, USA). Proteins were
probed overnight at 4°C with the rabbit anti-UBAP2L (1:2,000
dilution; cat. no. ab70319; Abcam, Cambridge, UK) or rabbit
anti-GAPDH (1:50,000 dilution; cat. no. 10494-1-AP; Proteintech
Group, Inc., Chicago, IL, USA) primary antibodies, followed by
incubation with horseradish peroxidase-conjugated goat anti-rabbit
immunoglobulin G (1:5,000 dilution; cat. no. sc-2054; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) at room temperature for 2 h.
GAPDH was used as the internal standard. Subsequently, proteins
were detected by the respective antibodies according to the
manufacturer's instructions for the ECL kit (Pierce Biotechnology,
Inc., Rockford, IL, USA) and exposed to X-ray films that were
quantified using the Amersham Image Scanner with LabScan ImageQuant
TL Software (Amersham Pharmacia Biotech, Piscataway, NJ, USA).
MTT assay
After 4 days of lentivirus infection, U251 and U373
cells were trypsinized, resuspended and seeded in 96-well plates at
a concentration of 2,000 and 2,500 cells/well, respectively. On the
following day, the cultured cells were incubated with 1, 2, 3, 4, 5
and 10 µl of 5 mg/ml MTT (Sigma-Aldrich, St. Louis, MO,
USA), and incubation was continued for 3 h. Following the careful
removal of the remaining medium, 100 µl of acidified
isopropanol was added to each well at the end of the incubation.
The absorbance was measured at 595 nm on the spectrophotometer.
Colony formation assay
After 4 days of lentivirus infection, U251 and U373
cells were incubated in 6-well plates at a concentration of 200 and
400 cells/well, respectively. The medium was changed at regular
time intervals. After 11 days of cultivation at 37°C, the natural
colonies were washed by phosphate-buffered saline (PBS) and fixed
with 4% paraformaldehyde (Sigma-Aldrich) for 30 min at room
temperature. The colonies were stained using crystal violet
(Sigma-Aldrich) for 10 min, were subsequently washed with water and
were air-dried. The total number of colonies including >50 cells
was counted under fluorescence microscopy.
Fluorescence-activated cell sorting
analysis
The cell cycle distribution was analyzed using flow
cytometry with propidium iodide (PI) staining. Briefly,
lentivirus-transduced cells (3×105 cells/dish) were
seeded in 6-cm dishes and incubated at 37°C for 40 h. Cells were
harvested following trypsinization, washed with PBS and fixed in
70% cold ethanol overnight at 4°C. Cells were subsequently
collected by centrifugation and resuspended in PBS containing 100
µg/ml of DNase-free RNase and 50 µg/ml PI
(Sigma-Aldrich), and incubated in the dark at room temperature for
1 h. Finally, the suspension was subjected to FACSCalibur flow
cytometer analysis (BD Biosciences, San Jose, CA, USA). The
fractions of the cells in the G0/G1, S and
G2/M phases were analyzed with the ModFit LT DNA
analysis program (Verity Software House, Topsham, ME, USA).
Statistical analysis
All the data are expressed as the mean ± standard
deviation of at least triplicate determination. The Student's
t-test was used to evaluate the differences between groups using
the SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
Lentivirus-mediated shRNA inhibits UBAP2L
mRNA and protein expression in U251 and U373 cells
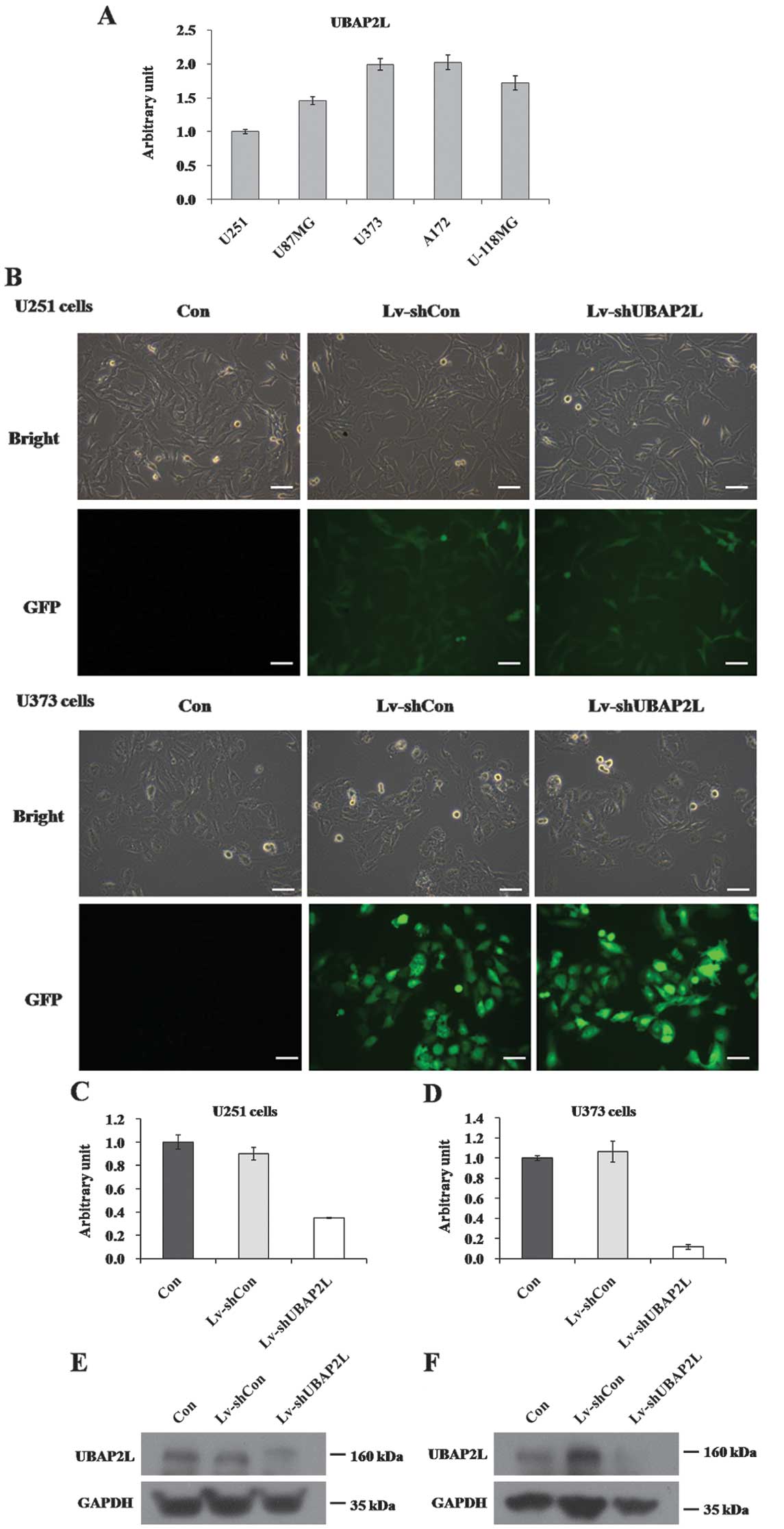
The expression of UBAP2L mRNA was first detected in
five glioma cell lines, U251, U87MG, A172, U373 and U-118MG, using
RT-qPCR. As shown in Fig. 1A,
UBAP2L mRNA was widely expressed in all five cell lines.
Subsequently, U251, U373 and A172 cell lines were applied for the
loss-of-function investigation in the following study. A lentiviral
vector system was constructed to express shRNA targeting of UBAP2L
with a GFP reporter gene. To determine whether the recombinant
lentiviruses could infect glioma cell lines, cells infected with
Lv-shUBAP2L and Lv-shCon were observed under light and fluorescence
microscopes. Four days post-infection, >90% of U251 and U373
cells expressed GFP (Fig. 1B),
suggesting high-efficiency infection by the lentivirus. There was
no significant difference concerning cell morphology and cell
growth between the Lv-shCon and Con groups. To verify the knockdown
efficiency, the transcription and translation levels of UBAP2L in
U251 and U373 cells were assessed by RT-qPCR and western blot
analysis following 7 days of lentivirus infection. Compared with
the Lv-shCon groups, the mRNA levels of UBAP2L in the Lv-shUBAP2L
groups were markedly reduced by 61.0% in U251 cells and 88.9% in
U373 cells (Fig. 1C and D). The
protein levels of UBAP2L were concomitantly decreased in U251 and
U373 cells (Fig. 1E and F). These
results indicated that the constructed lentiviruses could
successfully knockdown UBAP2L expression in glioma cells.
Effects of UBAP2L knockdown on
proliferation of U251 and U373 cells
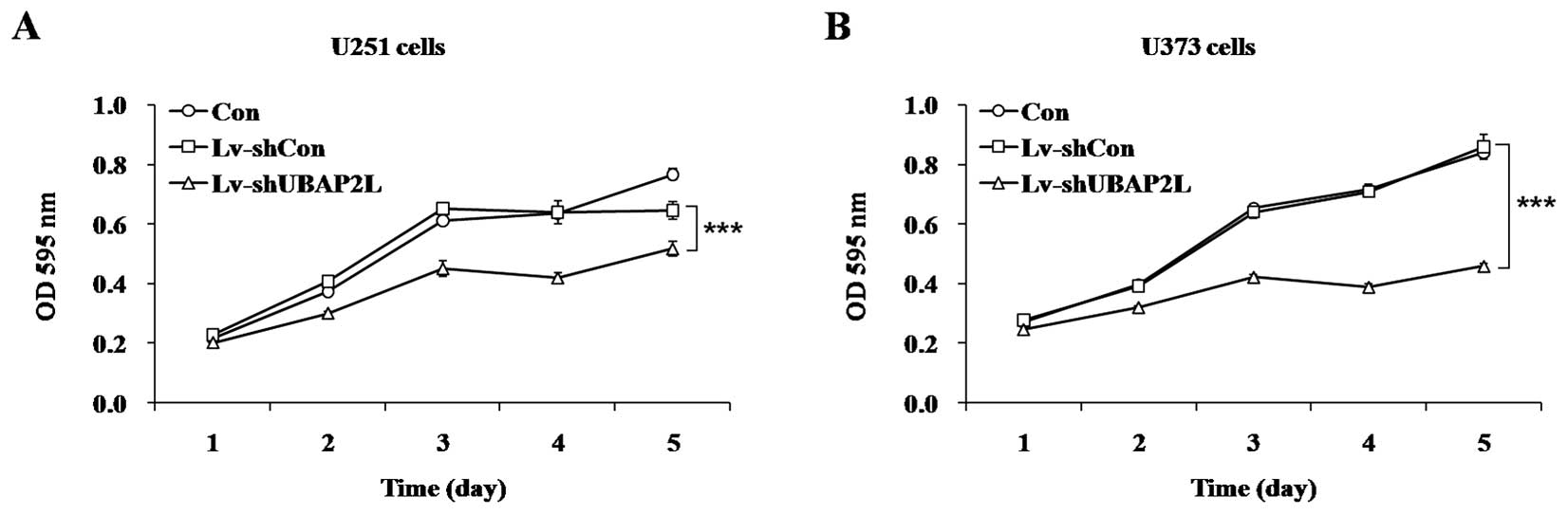
To investigate the effect of UBAP2L knockdown on
cell proliferation, the MTT assay was performed in U251 and U373
cells following 7 days of lentivirus infection. In comparison with
Lv-shCon groups, depletion of UBAP2L significantly inhibited the
proliferation of glioma cells (P<0.001) by a 19.8% reduction in
U251 cells (Fig. 2A) and 46.5%
reduction in U373 cells (Fig.
2B). These results suggested that UBAP2L accelerated glioma
cell proliferation.
Effects of UBAP2L knockdown on colony
formation of U251 and U373 cells
To detect whether UBAP2L has an influence on the
colony-forming capacity of glioma cells, the colony formation assay
was performed in U251 and U373 cells. The size of the single colony
and the number of colonies formed in the Lv-shUBAP2L groups were
significantly decreased when compared with the Lv-shCon and Con
groups [(PU251<0.01; Fig. 3A and C);
(PU373<0.001; Fig. 3B
and D)], suggesting that the reduced expression of UBAP2L could
significantly inhibit colony formation in glioma cells.
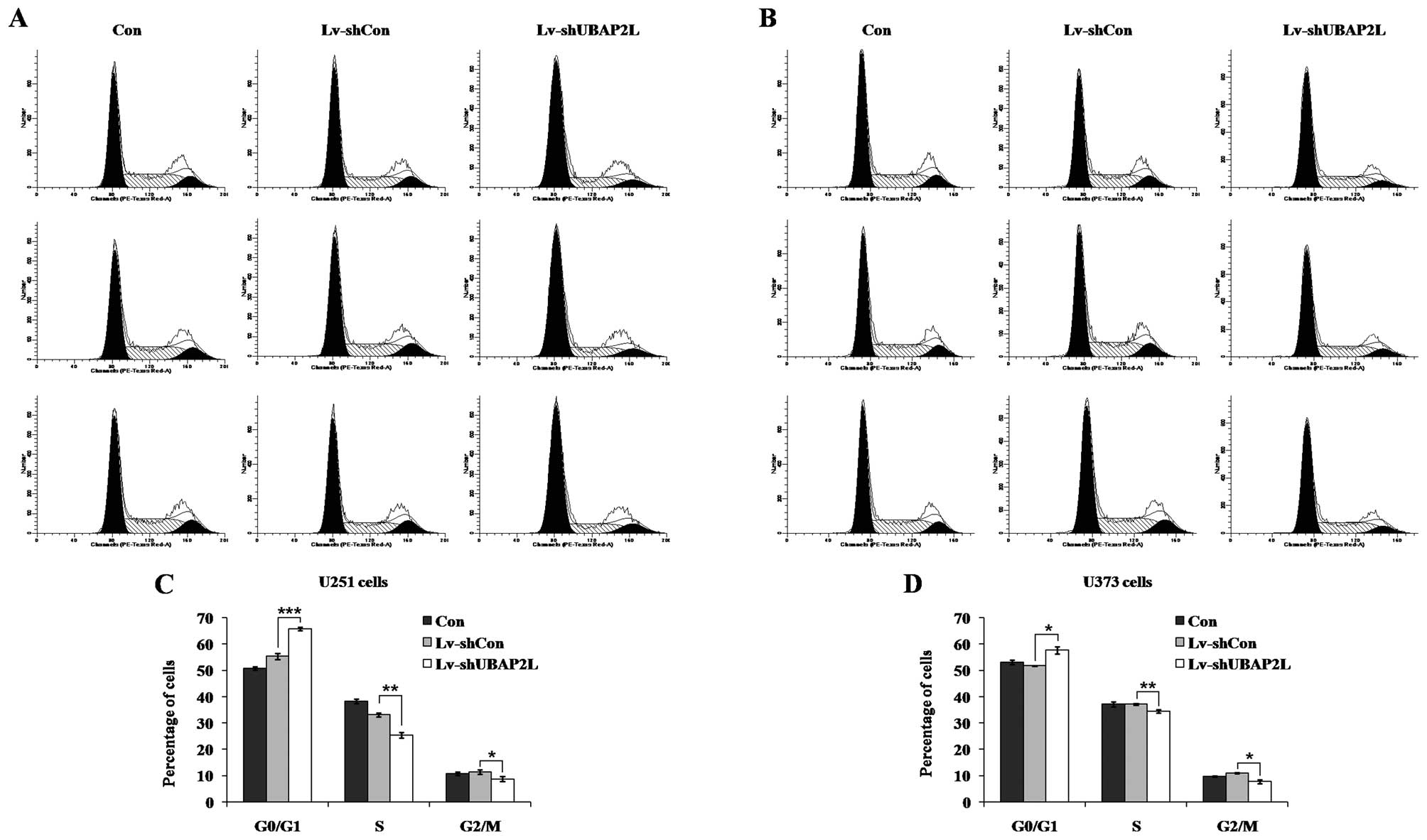
Effects of UBAP2L knockdown on cell cycle
distribution of U251 and U373 cells
To elucidate the mechanism underlying cell
proliferation inhibition, the cell cycle distribution of all three
groups was analyzed by flow cytometry in U251 and U373 cells
following lentivirus infection (Fig.
4A and B). Compared to the Lv-shCon groups (U251, 55.34±1.13%;
U373, 51.81±0.11%), the cell percentages of
G0/G1 phase in the Lv-shUBAP2L groups (U251,
65.80±0.72%; U373, 57.66±1.36%) were significantly increased in the
U251 and U373 cells (Fig. 4C and
D). There was no evident difference between the Lv-shCon and
Con groups in all the cell lines. These results indicated that
knockdown of UBAP2L could inhibit glioma cell growth via blockade
of cell cycle progression.
Effects of UBAP2L knockdown on
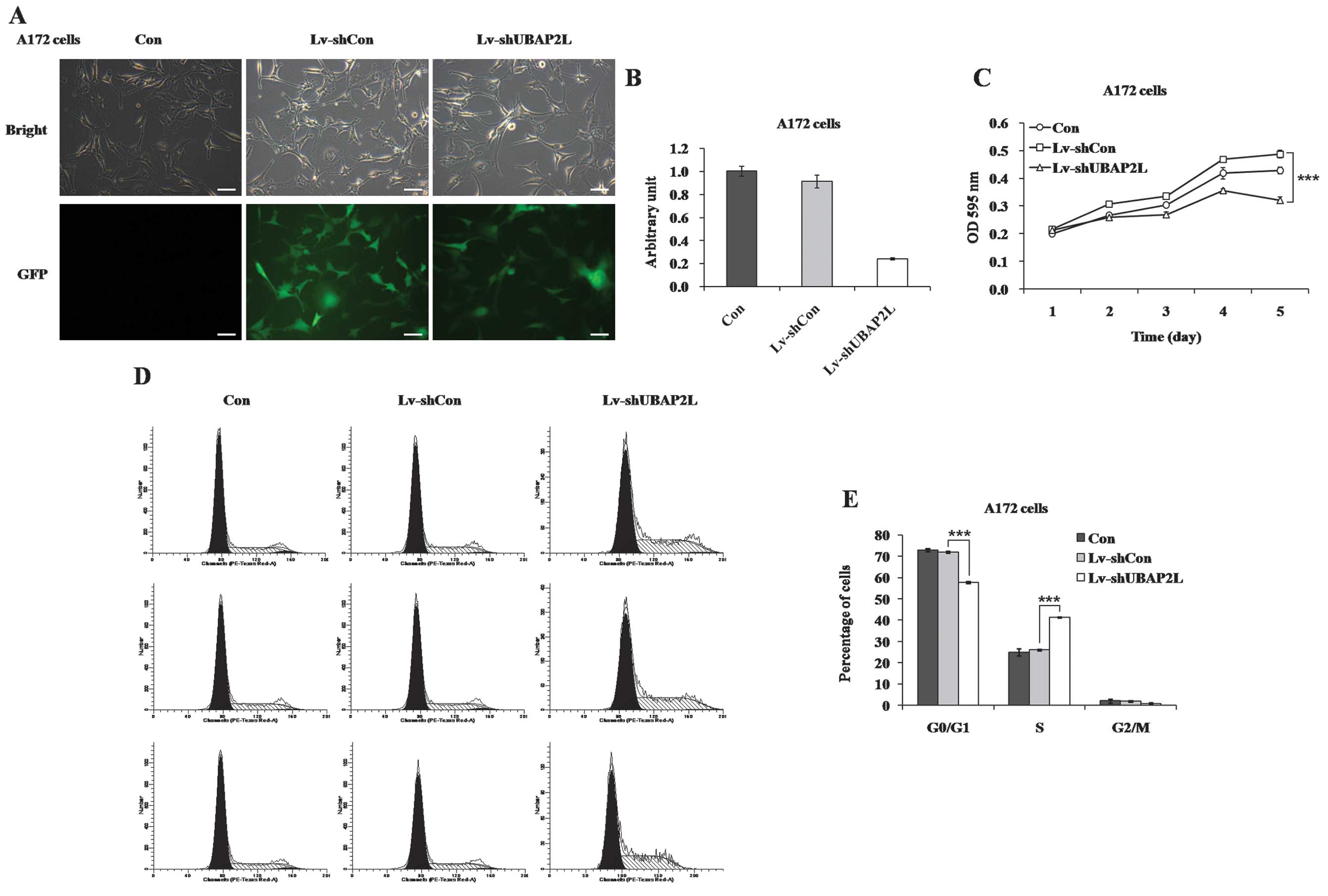
proliferation and cell cycle distribution of A172 cells
Furthermore, the effects of UBAP2L knockdown on cell
growth were confirmed in A172 cells. More than 80% of A172 cells
expressed GFP following infection for 4 days (Fig. 5A). RT-qPCR showed that UBAP2L
expression was reduced by 73.8% in A172 cells (Fig. 5B). The proliferation rate of A172
cells was also markedly decreased in the Lv-shUBAP2L group
(Fig. 5C). In addition, the cell
cycle distribution of A172 cells was examined following lentivirus
infection (Fig. 5D). A clear
increase of cell percentage in the S phase was observed in A172
cells (Lv-shUBAP2L, 41.41±0.24% vs. Lv-shCon, 26.04±0.41%; Fig. 5E), which could be due to the
specific cell type.
Evaluation of lentivirus-mediated
shRNA-targeting UBAP2L specificity
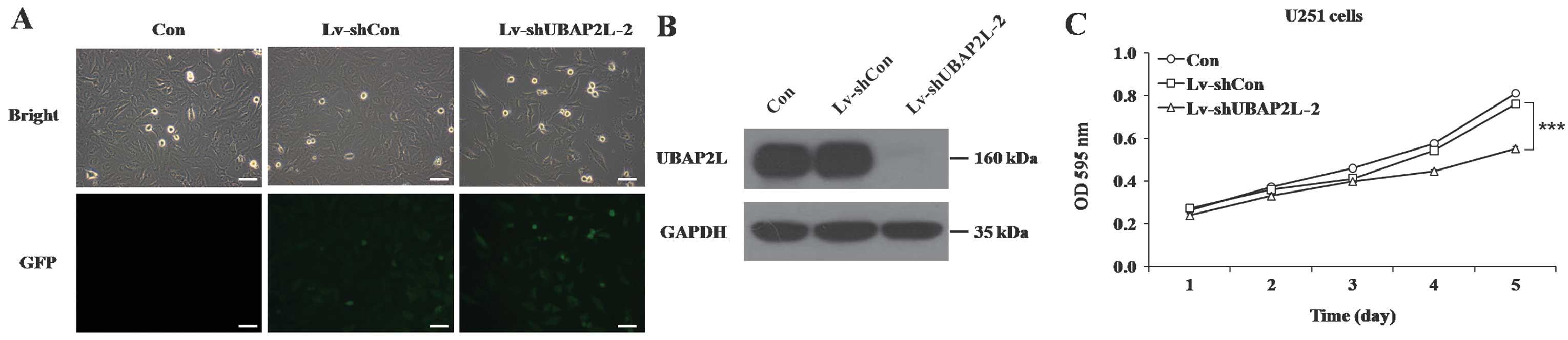
To exclude the off-target effect of shRNA, another
shRNA against UBAP2L was constructed and transduced into U251
cells. Similarly, >90% of U251 cells expressed GFP following
infection with another Lv-shUBAP2L (Lv-shUBAP2L-2) (Fig. 6A), suggesting high-efficiency and
high-specificity infection by the recombinant lentivirus. In
addition, the level of endogenous UBAP2L expression was evidently
downregulated in U251 cells infected with Lv-shUBAP2L-2 (Fig. 6B). Cell proliferation was also
significantly impeded in response to Lv-shUBAP2L-2 infection
(P<0.001; Fig. 6C).
Discussion
Previous studies have demonstrated that UBAP2L is
involved in the pathogenesis of several human malignant tumors. The
study by Sawyer (9) identified
UBAP2L as a candidate gene that showed amplification expression in
myeloma. A recent study (10)
based on correlating expression arrays and array comparative
genomic hybridization (CGH) data also found that UBAP2L showed
amplification expression in HCC, which is one of the early genomic
events associated with HCC development. Naz and Dhandapani
(11) reported that UBAP2L showed
97% homology at the nucleotide and amino acid sequences with
ZPC-interacting protein involved in malignant ovarian tumors
(15). Furthermore, Sudhir et
al (16) identified UBAP2L as
a novel target of mitogen-activated protein kinase (MAPK) family
kinases that acts as a downstream component of Ras-mediated
signaling and has an important role in the pathogenesis of certain
types of human cancer, such as lung cancer and glioma (17). However, whether UBAP2L has a role
in the tumorigenesis and progression of glioma via regulation of
the MAPK pathway, has not yet been investigated.
In the present study, UBAP2L was ubiquitously
expressed in five human glioma cell lines. To explore the function
of UBAP2L in glioma, a loss-of-function analysis was performed via
an shRNA-expressing lentivirus system, which is a safe non-toxic
shRNA delivery method that ensures a long-lasting stable silencing
effect (18,19). U251 and U373 cells infected with
Lv-shUBAP2L exhibited significant reductions in cell proliferation
and colony formation, and an increase of cell population in the
G0/G1 phase. Knockdown of UBAP2L in A172
cells also inhibited cell proliferation along with S phase arrest,
which showed a different regulatory mechanism of cell growth
inhibition in specific glioma cell type.
In cancer cells, cell cycle is a critical mechanism
of development, progression and resistance to treatment (2). Aberrant function of cell cycle
regulators generally alters the properties of growth,
differentiation and apoptosis in cancer cells (20). Previous studies have shown that
the involvement of UPS in cell cycle regulation of glioma cells is
critical. Piva et al (21)
found that the cyclin-dependent kinase inhibitor (p27) was degraded
in a proteasome-dependent manner, which provides evidence
indicative of an association between the stability of cell cycle
proteins and UPS in gliomas. Pamarthy et al (22) further showed that S-phase
kinase-associated protein 2 (Skp2), which belongs to the Ub ligase
F-box family, could promote G1-S transition through
targeting of p27 for degradation. Amador et al (23) demonstrated that the Ub ligase
APC/C (Cdc20) contributed to activation of CDK1 in early M phase in
gliomas by controlling the UPS-dependent degradation of cell
cycle-related protein (p21). An et al (24) suggested that UPS exerted an
indirect role in the cell cycle of glioma cells by regulation of
the oncoprotein c-Myc stability, which is known as an activator of
cell cycle acceleration and involved in the G1 phase. In
addition, Rb and p53, which have crucial roles in cell cycle
regulation, could be regulated by UPS (25,26). In the present study, UBAP2L, which
contains a UBA involved in UPS, was found to facilitate cell growth
by regulating G0/G1 to S phase progression in
glioma. Therefore, previous studies that focus on the function and
mechanism of UPS can provide a foundation for further studies
regarding UBAP2L in malignant glioma.
In conclusion, the present study indicates that
UBAP2L has a key role in glioma cell growth, suggesting that UBAP2L
may act as an oncogene to promote the development of glioma via
cell proliferation and cell cycle regulation. Further investigation
is required to elucidate the precise molecular mechanisms by which
UBAP2L affects human glioma progression.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (grant no. 81072066).
References
|
1
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sakamoto KM, Kim KB, Verma R, Ransick A,
Stein B, Crews CM and Deshaies RJ: Development of Protacs to target
cancer-promoting proteins for ubiquitination and degradation. Mol
Cell Proteomics. 2:1350–1358. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wei W and Lin H-K: The key role of
ubiquitination and sumoylation in signaling and cancer: A research
topic. Front Oncol. 2:1872012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yamaguchi H, Hsu JL and Hung MC:
Regulation of ubiqui-tination-mediated protein degradation by
survival kinases in cancer. Front Oncol. 2:152012. View Article : Google Scholar
|
|
6
|
Vlachostergios PJ, Voutsadakis IA and
Papandreou CN: The ubiquitin-proteasome system in glioma cell cycle
control. Cell Div. 7:182012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ciechanover A: Intracellular protein
degradation: From a vague idea through the lysosome and the
ubiquitin-proteasome system and onto human diseases and drug
targeting. Bioorg Med Chem. 21:3400–3410. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wilde IB, Brack M, Winget JM and Mayor T:
Proteomic char-acterization of aggregating proteins after the
inhibition of the ubiquitin proteasome system. J Proteome Res.
10:1062–1072. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sawyer JR: The prognostic significance of
cytogenetics and molecular profiling in multiple myeloma. Cancer
Genet. 204:3–12. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhu Z-Z, Wang D, Cong W-M, Jiang H, Yu Y,
Wen BJ, Dong H, Zhang X, Liu SF, Wang AZ, et al: Sex-related
differences in DNA copy number alterations in hepatitis B
virus-associated hepatocellular carcinoma. Asian Pac J Cancer Prev.
13:225–229. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Naz RK and Dhandapani L: Identification of
human sperm proteins that interact with human zona pellucida3 (ZP3)
using yeast two-hybrid system. J Reprod Immunol. 84:24–31. 2010.
View Article : Google Scholar :
|
|
12
|
Madura K: The ubiquitin-associated (UBA)
domain: On the path from prudence to prurience. Cell Cycle.
1:235–244. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sakoda T, Kasahara N, Hamamori Y and Kedes
L: A high-titer lentiviral production system mediates efficient
transduction of differentiated cells including beating cardiac
myocytes. J Mol Cell Cardiol. 31:2037–2047. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
15
|
Rahman NA, Bennink HJTC, Chrusciel M,
Sharp V, Zimmerman Y, Dina R, Li X, Ellonen A, Rivero-Müller A,
Dilworth S, et al: A novel treatment strategy for ovarian cancer
based on immunization against zona pellucida protein (ZP) 3. FASEB
J. 26:324–333. 2012. View Article : Google Scholar
|
|
16
|
Sudhir PR, Hsu CL, Wang MJ, Wang YT, Chen
YJ, Sung TY, Hsu WL, Yang UC and Chen JY: Phosphoproteomics
identifies oncogenic Ras signaling targets and their involvement in
lung adenocarcinomas. PLoS One. 6:e201992011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Olsen BB, Svenstrup TH and Guerra B:
Downregulation of protein kinase CK2 induces autophagic cell death
through modulation of the mTOR and MAPK signaling pathways in human
glioblastoma cells. Int J Oncol. 41:1967–1976. 2012.PubMed/NCBI
|
|
18
|
Bank A, Dorazio R and Leboulch P: A phase
I/II clinical trial of beta-globin gene therapy for
beta-thalassemia. Ann N Y Acad Sci. 1054:308–316. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fang J, Yu Z, Lian M, Ma H, Tai J, Zhang L
and Han D: Knockdown of zinc finger protein, X-linked (ZFX)
inhibits cell proliferation and induces apoptosis in human
laryngeal squamous cell carcinoma. Mol Cell Biochem. 360:301–307.
2012. View Article : Google Scholar
|
|
20
|
Sherr CJ: The Pezcoller lecture: Cancer
cell cycles revisited. Cancer Res. 60:3689–3695. 2000.PubMed/NCBI
|
|
21
|
Piva R, Cancelli I, Cavalla P, Bortolotto
S, Dominguez J, Draetta GF and Schiffer D: Proteasome-dependent
degradation of p27/kip1 in gliomas. J Neuropathol Exp Neurol.
58:691–696. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pamarthy D, Tan M, Wu M, Chen J, Yang D,
Wang S, Zhang H and Sun Y: p27 degradation by an ellipticinium
series of compound via ubiquitin-proteasome pathway. Cancer Biol
Ther. 6:360–366. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Amador V, Ge S, Santamaría PG,
Guardavaccaro D and Pagano M: APC/C(Cdc20) controls the
ubiquitin-mediated degradation of p21 in prometaphase. Mol Cell.
27:462–473. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
An J, Yang DY, Xu QZ, Zhang SM, Huo YY,
Shang ZF, Wang Y, Wu DC and Zhou PK: DNA-dependent protein kinase
catalytic subunit modulates the stability of c-Myc oncoprotein. Mol
Cancer. 7:322008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ying H and Xiao ZX: Targeting
retinoblastoma protein for degradation by proteasomes. Cell Cycle.
5:506–508. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ohgaki H and Kleihues P: Genetic pathways
to primary and secondary glioblastoma. Am J Pathol. 170:1445–1453.
2007. View Article : Google Scholar : PubMed/NCBI
|