Introduction
Hepatocellular carcinoma remains the most common
type of liver cancer, and usually develops secondary to cirrhosis
or viral hepatitis infection. The incidence of hepatocellular
carcinoma has gradually increased in certain developed and
developing countries. Despite advances in multidisciplinary
therapies, the median survival time of patients with hepatocellular
carcinoma is ~9 months (1).
Therefore, the characterization of molecular mechanisms underlying
the initiation and progress of hepatocellular carcinoma is
necessary. Certain factors, including inflammation, oxidative
stress and hypoxia, have been shown to facilitate the initiation,
progression and metastasis of hepatocellular carcinoma (2). The androgen receptor has been
reported to regulate cell adhesion and migration of hepatocellular
carcinoma via regulating the β1-integrin-AKT signaling pathway
(3). Overexpression of actopaxin
may lead to the increase of invasion and migration ability, and an
epithelial-mesenchymal transition process in hepatocellular
carcinoma (4). mTOR signaling was
also shown to have a critical role in the pathogenesis of
hepatocellular carcinoma (5).
Oxysterol-binding protein (OSBP)-related protein 8 (ORP8) markedly
inhibited tumor growth and induced cell apoptosis of hepatocellular
carcinoma cells through the Fas/FasL pathway in vivo and
in vitro (6).
MicroRNAs (miRNA) have been recognized as potential
therapeutic targets for hepatocellular carcinoma. miRNAs are a
class of small non-coding RNA molecules and post-transcriptionally
regulate protein expression by targeting the 3′ untranslated region
(3′UTR) of target mRNA, causing degradation or repression of
translation (7). Previous studies
have reported that miRNAs have important roles in the development
and maintenance of various diseases, such as cancer, heart
hypertrophy and fibrosis (8–11).
For example, in hearts, the upregulation of miR-23a induced
by isoproterenol or aldosterone caused hypertrophic growth of
cardiomyocytes, and the knockdown of miR-23a could attenuate
cardiomyocyte hypertrophy by regulating NFATc3 (12). Particularly in cancer, miRNAs have
emerged as new potential therapeutic molecules and targets
(13,14). The miR-543 level was
significantly reduced in hepatocellular carcinoma, and
overexpression of miR-543 enhanced the proliferation and
invasion of HepG2 via targeting PAQR3 in hepatocellular carcinoma
(13). Additionally,
miR-520e can inhibit the growth of hepatocellular carcinoma
cells, and the knockdown of miR-520e lead to increased cell
proliferation (14).
Overexpression of miR-375 reduced cell proliferation and
migration, and also induced G1 arrest and apoptosis in
hepatocellular carcinoma cells via targeting at AEG-1 (15). miR-1 has been shown to
promote apoptosis of HepG2 by targeting apoptosis inhibitor-5
(16). It was previously reported
that miR-1297 exerted antitumor effects on human lung
adenocarcinoma cell and colorectal cancer (17,18), and acted as a tumor suppressor.
However, whether miR-1297 has an inhibitory effect on
hepatocellular carcinoma has not been determined.
To the best of our knowledge, the present study
established for the first time, a potential association between
miR-1297 and the proliferation, apoptosis and migration of
hepatocellular carcinoma cells. The antitumor effects of
miR-1297 on HepG2 and SMMC7721 cells were investigated, and
the precise mechanism of miR-1297 was explored in inhibiting
liver cancer.
Materials and methods
Reagents
Dulbecco's modified Eagle's medium (DMEM), fetal
bovine serum (FBS) and phosphate-buffered saline (PBS) were
purchased from HyClone Laboratories. Lipofectamine™ 2000 was
purchased from Invitrogen (Carlsbad, CA, USA). Hsa-miR-1297
mimics (miR-1297), hsa-miR-1297 inhibitor (AMO-1297)
and its scramble negative control (control) were synthesized by
Sangon (Shanghai, China). Unless otherwise specified, all the other
reagents were purchased from Sigma Chemical Co. (St. Louis, MO,
USA).
Cell culture
Human hepatocellular carcinoma HepG2 and SMMC7721
cell lines were purchased from the China Center for Type Culture
Collection (Wuhan, China). These two cell lines were cultured in
DMEM supplemented with 10% FBS and penicillin (100
U/ml)/streptomycin (100 µg/ml), and were subsequently
transferred into plastic culture flasks and cultured at 37°C in
humid air with 5% CO2.
miRNAs transfection
miRNAs were transected into HepG2 and SMMC7721 at
50–70% confluence using Lipofectamine 2000 reagent according to the
manufacturer's instructions. The final oligonucleotide
concentration of miR-1297, AMO-1297 or its scramble reached
50 nmol/l. Three hours after miRNAs transfection, the culture
medium was changed to fresh DMEM with 10% FBS. Subsequently, HepG2
and SMMC7721 were incubated for an additional 48 h and harvested
for further investigation. These transfected oligonucleotides were
divided into four groups: Control, hsa-miR-1297 mimics
(miR-1297), hsa-miR-1297 + hsa-miR-1297
inhibitor (miR-1297 + AMO-1297) and its scramble negative control
(scramble).
Cell viability assay
Cell viability of HepG2 and SMMC7721 was determined
by the MTT assay, according to the methods described in a previous
study (14). In brief, HepG2 and
SMMC7721 cells were seeded in 96-well plates, and were transfected
with miR-1297, miR-1297 + AMO-1297 or its scramble.
After transfection, the cells were exposed to 10 µl of MTT
solution (5 mg/ml) for 2–4 h at 37°C. After the supernatant was
removed from formazan crystals, and dimethyl sulfoxide (DMSO) was
added. The cell viability was calculated according to the
absorbance at 570 nm using an OPTImax microplate reader.
Terminal
deoxynucleotidyltransferase-mediated dUTP nick end labelling
(TUNEL) assay
Apoptosis of HepG2 and SMMC7721 cells was determined
using a TUNEL detection kit (Roche, Penzberg, Germany) according to
the manufacturer's instructions. After transfection with
miR-1297, miR-1297 + AMO-1297 or its scramble, HepG2
and SMMC7721 cells were fixed with 4% paraformaldehyde in PBS for 1
h at room temperature, and permeabilized with 0.1% Triton X-100 in
0.1% sodium citrate for 2 min on ice. Following washing in PBS,
sections were incubated with the TUNEL reaction mixture for 1 h at
37°C. After washing with PBS, the stained cells were visualized
using a fluorescence microscope (Eclipse TE300; Nikon, Tokyo,
Japan).
Wound healing assay
HepG2 and SMMC7721 cells were cultured in a 6-well
plate for 24 h. The wound healing assay was carried out by
introducing a small linear scratch with a pipette tip.
Subsequently, the cells in each well were incubated with serum-free
DMEM medium. Images of 48 h following the scratch in cultured cells
were captured under a phase-contrast microscope (magnification,
×200) to monitor the cell migration process.
Transwell assay
The Transwell assay was performed using a Transwell
chamber with pore size of 8.0 µm (Millipore, Billerica, MA,
USA). The cells were resuspended in serum-free medium and were
placed in the upper chamber in 5% CO2 at 37°C. Following
transfection, cultured HepG2 and SMMC7721 cells in the upper
chamber were removed, and the attached cells in the lower section
were stained with 0.1% crystal violet. The migration rate was
quantified by counting the migration cells in six random fields
under a light microscope.
Bioinformatics predication
The potential target gene of miR-1297 was
predicted by computer-aided algorithms using TargetScan (http://www.targetscan.org).
Luciferase reporter assay
After plating in a 24-well plate, HEK293 cells were
transfected with the constructed reporter plasmid containing 3′UTR
of high-mobility group AT-hook 2 (HMGA2) plus miR-1297,
miR-1297 + AMO-1297 or its scramble. Luciferase assays were
performed using the Dual-Luciferase Reporter Assay system (Promega,
Madison, WI, USA) 24 h after transfection.
RNA extraction and reverse
transcripion-quantitative polymerase chain reaction (RT-qPCR)
analysis
Total RNA was extracted from the cells using TRIzol
according to the manufacturer's instructions. RT-qPCR was conducted
using an RT-PCR kit (Takara, Dalian, China). The primers used were
all synthesized by Sangon. Semi-quantitative RT-PCR was performed
using a thermal cycler (Bio-Rad, Hercules, CA, USA). The primer
sequences for human HMGA2 gene expression were as follows:
HMGA2 forward, 5′-TGGGAGGA GCGAAATCTAAA-3′ and reverse,
5′-AAGCACCTTG GTCAACCATC-3′; and β-actin forward, 5′-TGAAGATCAA
GATCATTGCTCC-3′ and reverse, 5′-GCCATGCCAAT CTCATCTTG-3′. The
relative HMGA2 mRNA levels were normalized to those of
β-actin mRNA levels using Quality One analysis software
(Bio-Rad).
Western blot analysis
The protein samples were extracted from the cultured
HepG2 and SMMC7721 cells. After 12,000 × g centrifugation at 4°C
for 10 min, the concentration of extracted protein was determined
by the bicinchoninic acid method (Beyotime, Jiangsu, China). The
extracted proteins were separated using 10–12% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis and subsequently
electrophoretically transferred to the PVDF membrane (Millipore).
The membranes were blocked in blocking buffer (5% non-fat milk
dissolved in PBS-Tween-20) overnight at 4°C. Subsequently, the
membranes were incubated with goat polyclonal antibodies against
HMGA2 (sc-23684) and mouse monoclonal anti-glyceraldehyde
3-phosphate dehydrogenase (GAPDH) antibody (sc-32233; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA). The bands were visualized by
ECL (Santa Cruz Biotechnology, Inc.) and detected by ECL Detection
Systems (Thermo Scientific, Waltham, MA, USA). The GAPDH protein
was used as an internal control.
Statistical analysis
Statistical data are shown as mean ± standard error
of the mean. All the statistical analysis was performed by SPSS
13.0 software (SPSS, Inc., Chicago, IL, USA). Differences between
two groups were defined with a t-test, and the significant
differences among three groups were determined using analysis of
variance. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-1297 reduces the cellular viability
of HepG2 and SMMC7721 cells
To evaluate the effects of miR-1297 on the
proliferation of HepG2 and SMMC7721 cells, the MTT assay was used
to measure the cellular viability of HepG2 and SMMC7721. The
overexpression of miR-1297 resulted in a significant
reduction of cellular viability in HepG2 cells compared with the
control (P<0.05) (Fig. 1A).
Co-transfection with AMO-1297 may abolish the inhibitory effects of
miR-1297 on the viability of HepG2 cells (P<0.05)
(Fig. 1A). The scrambled negative
control did not affect the viability of HepG2 cells. A similar
anti-proliferation action of miR-1297 was also observed in
SMMC7721 cells (Fig. 1B). These
results suggest that miR-1297 has an inhibitory role in the
proliferation of HepG2 and SMMC7721 cells.
miR-1297 induces the apoptosis of HepG2
and SMMC7721 cells
Whether miR-1297 overexpression was able to
induce apoptosis of HepG2 and SMMC7721 cells was further studied.
The TUNEL assay was carried out to investigate the influence of
miR-1297 on the apoptosis of HepG2 and SMMC7721 cells.
Fig. 2A demonstrates that
miR-1297 over-expression induced a significant increase of
HepG2 cells that were positive for TUNEL staining compared with the
control (P<0.05). Similarly, the number of HepG2 cells positive
for TUNEL staining was also increased by miR-1297
overexpression in SMMC7721 cells (P<0.05) (Fig. 2C). Co-transfection with AMO-1297
may suppress the increase of apoptosis by miR-1297 in HepG2
and SMMC7721 cells (Fig. 2). The
scrambled negative control did not significantly induce the
apoptosis of HepG2 and SMMC7721 cells.
miR-1297 inhibits the migration of HepG2
and SMMC7721 cells
In addition to cellular proliferation, the effect of
miR-1297 on the migration activity of HepG2 and SMMC7721
cells was examined. The wound healing assay in vitro was
carried out to determine cell migration of HepG2 and SMMC7721.
Fig. 3A shows the quantified
wound closure in cultured HepG2 cells following miR-1297,
co-transfection with AMO-1297 and its scramble transfection. Wound
closure was significantly slowed in miR-1297-treated HepG2
and SMMC7721 cells at 48 h after the scratch, compared with the
control group (Fig. 3B and C).
Consistently, the Transwell migration assay was also performed in
HepG2 and SMMC7721 cells with miR-1297, co-transfection with
AMO-1297 and its scramble transfection. In agreement with the wound
healing assay, transfection with miR-1297 overexpression
produced an inhibition of migration of HepG2 (Fig. 4A and B) and SMMC7721 cells
(Fig. 4C and D). This data
demonstrated that the suppression of migration of hepatocellular
carcinoma cells was induced by miR-1297.
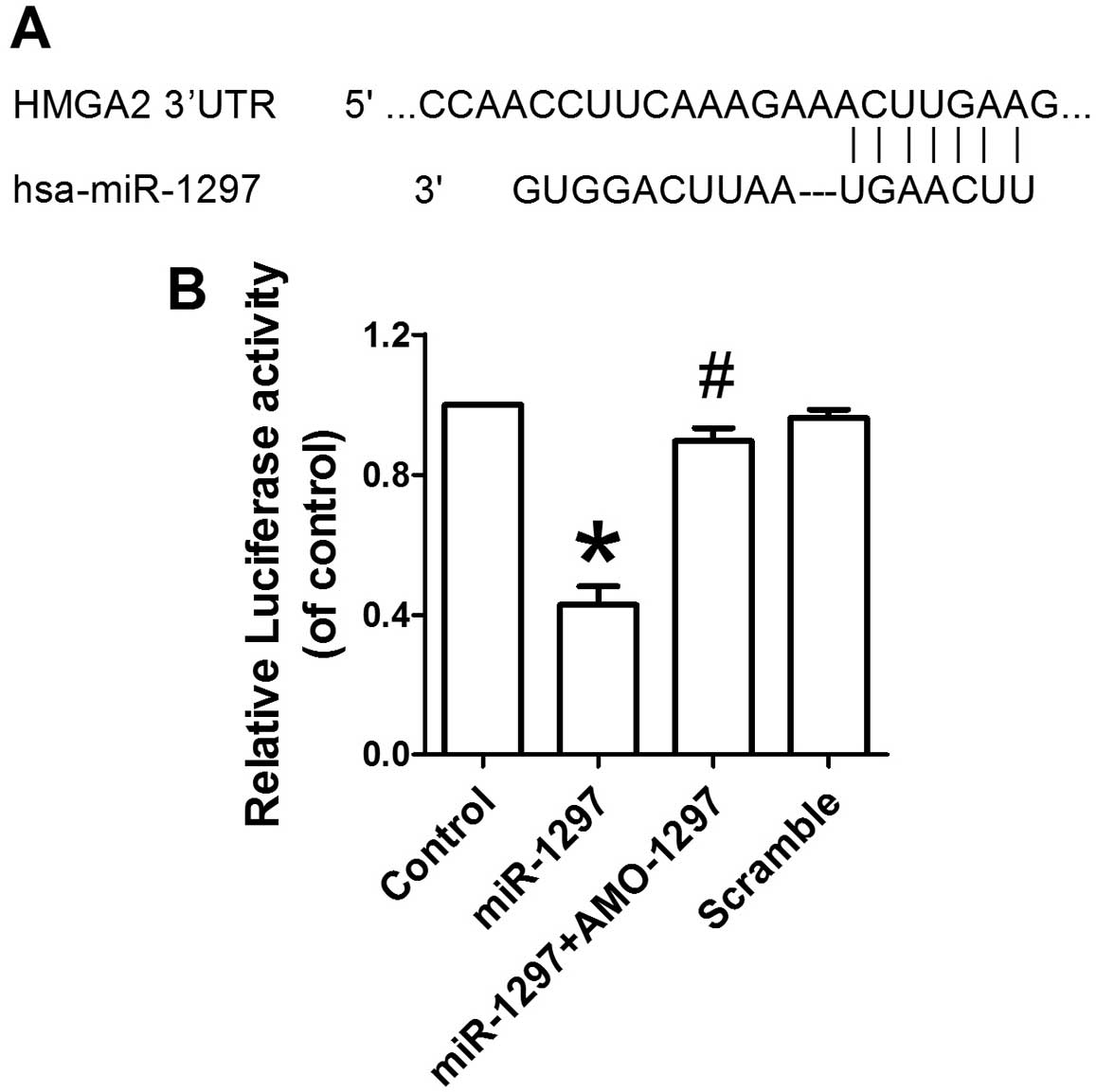
miR-1297 targets the 3′UTR of HMGA2
A bioinformatics database of miRNAs was used
(http://www.targetscan.org/) to predict
the potential target gene of miR-1297. miR-1297 was
shown to have a putative binding site to the 3′UTR region of
HMGA2 (Fig. 5A). A highly
conservative miR-1297 binding site at the HMGA2 3′UTR
1867–1873 base position was predicted in humans. Subsequently, the
Dual-Luciferase reporter assay was performed to confirm that
miR-1297 directly targeted the 3′UTR of HMGA2.
Overexpression of miR-1297 decreased the luciferase activity
of the reporter plasmid harboring HMGA2 3′UTR, suggesting
that miR-1297 inhibits the translation of HMGA2 by
binding to its 3′UTR (Fig. 5B).
However, co-transfection of AMO-1297 alleviated the reduction in
luciferase activity caused by miR-1297 in HEK293. These
results indicate that HMGA2 is a direct target of
miR-1297.
miR-1297 regulates HMGA2 expression in
hepatocellular carcinoma
Regulatory effects of miR-1297 on
HMGA2 expression were further confirmed. miR-1297,
miR-1297 + AMO-1297 or its scramble were transfected into
HepG2 and SMMC7721 cells, and western blotting was subsequently
used to determine the level of HMGA2 protein. Fig. 6 demonstrates that the
overexpression of miR-1297 caused a significant reduction of
HMGA2 protein in HepG2 and SMMC7721 cells compared with control
(P<0.05). By contrast, co-transfection with AMO-1297
significantly reversed the reduction of the HMGA2 protein following
miR-1297 transfection (P<0.05). Transfection of scramble
sequence did not affect the expression of HMGA2 protein in HepG2
and SMMC7721 cells. This finding indicates that miR-1297
inhibits HMGA2 protein expression in hepatocellular carcinoma.
Discussion
In the present study, miR-1297 inhibited the
proliferation and migration, and promoted the apoptosis of
hepatocellular carcinoma, and the direct target of miR-1297
is the HMGA2 gene. This finding provides a new therapeutic
approach for liver cancer.
Hepatocellular carcinoma is the most common type of
liver cancer, accounting for ~90% liver cancers (19). Currently, hepatocellular carcinoma
has become the third leading cause of cancer fatalities worldwide
(19). Numerous studies have
revealed that liver cirrhosis and viral hepatitis infection are two
important risk factors of hepatocellular carcinoma (20). In addition, inflammation,
necrosis, fibrosis and ongoing regeneration also contribute to
cirrhotic liver and lead to the development of hepatocellular
carcinoma (2). Recent studies
have characterized the altered molecular pathways, such as p53,
PIKCA and β-catenin genes during carcinogenesis, and the mutations
on these genes was most frequently reported in patients with
hepatocellular carcinoma (21–24). The abnormal activation of
Wnt/β-catenin appears to be frequently altered in hepatocellular
carcinoma (23,24). mTOR signaling was also shown to
have a critical role in the pathogenesis of hepatocellular
carcinoma (5). Androgen receptor
has been reported to regulate cell adhesion and migration of
hepatocel-lular carcinoma via regulating the β1-integrin-AKT
signaling pathway (3). Recently,
miRNAs have been recognized as a potential therapeutic target for
hepatocellular carcinoma. In the present study, the molecular
mechanism of antitumor activity of miR-1297 on
hepatocellular carcinoma was explored.
To investigate whether miR-1297 has an
antitumor role in HepG2 and SMMC7721, the MTT assay was employed to
observe the effects of miR-1297 on cellular viability of
HepG2 and SMMC7721. The results showed that miR-1297 can
significantly inhibit the proliferation of HepG2 and SMMC7721
cells, and co-transfection with AMO-1297 can abolish the inhibition
of cell viability in HepG2 and SMMC7721 cells. Additionally, the
effects of miR-1297 overexpression were assessed on the
apoptosis of HepG2 and SMMC7721, and the results showed that forced
expression of miR-1297 may lead to the apoptosis of HepG2
and SMMC7721. In agreement with the present findings, previous
studies also reported that miR-1297 inhibited the growth of
human lung adenocarcinoma cell and colorectal cancer (17,18). COX2 and TRIB2 were identified as
the target genes of miR-1297 in two types of cells, and have been
shown to be associated with the inhibition of proliferation.
However, whether COX2 and TRIB2 are involved in the miR-1297
inhibition of liver cancer cell proliferation remains unknown. It
has been reported that TRIB2 contributes to the inhibition of
proliferation and the induction of apoptosis of HepG2 cells
(25). It is thus proposed that
TRIB2 is a possible target of miR-1297 in hepatocellualr carcinoma
cells.
Migration of cancer cells is an important factor for
cancer metastasis. Inhibition of migration contributed to the
treatment of hepatocellular carcinoma (3). The impact of miR-1297
overexpression on the migration of hepatocellular carcinoma was
also observed in the present study. Overexpression of
miR-1297 significantly inhibited the migration of HepG2 and
SMMC7721 cells, and co-transfection with AMO-1297 may attenuate the
slowed migration of hepatocellular carcinoma by miR-1297.
Consistently, it has been reported that miR-1297 inhibited
the migration and invasion of human colorectal cancer in
vivo and in vitro (17).
Bioinformatics predication on a microRNA database
(TargetScan) was used to screen the possible candidate gene of
miR-1297. A binding site of miR-1297 was identified
in the 3′UTR of HMGA2, suggesting HMGA2 as a
potential target of miR-1297. Activation of the HMGA2
gene was observed in the cells of numerous human malignancies, such
as breast cancer, lung cancer and pancreatic carcinoma (26–30). The inhibition or silencing of
HMGA2 contributed to the inhibition of tumor growth, the
induction of apoptosis and the suppression of tumor metastasis
(31). In the present study, the
luciferase assay showed that miR-1297 overexpression
markedly inhibited the luciferase activity of the reporter plasmid
harboring the 3′UTR of HMGA2 gene. To the best of our
knowledge, this is the first study to uncover the inhibition
effects of miR-1297 on HMGA2 in hepatocellular
carcinoma cells.
To further confirm that miR-1297
overexpression inhibits the translation of HMGA2 in
hepatocellular carcinoma, the effects of miR-1297 were
studied on the expression of the HMGA2 protein in HepG2 and
SMMC7721. Overexpression of miR-1297 also reduced the
expression of HMGA2 protein in the HepG2 and SMMC7721 cells.
Whereas, the mRNA level of HMGA2 was not affected by
overexpression of miR-1297. These results suggest that
miR-1297 directly acts on the 3′UTR of HMGA2
gene.
In conclusion, this is the first study to show that
miR-1297 inhibited the proliferation and migration, and
promoted the apoptosis of hepatocellular carcinoma cells via
directly targeting the HMGA2 gene. miR-1297 is a
potential therapeutic target of hepatocellular carcinoma.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (grant no. 81100483), and the
Foundation of Building Key Department of Heilongjiang Province
Universities.
References
|
1
|
Zhu AX, Blaszkowsky LS, Ryan DP, Clark JW,
Muzikansky A, Horgan K, Sheehan S, Hale KE, Enzinger PC, Bhargava
P, et al: Phase II study of gemcitabine and oxaliplatin in
combination with bevacizumab in patients with advanced
hepatocellular carcinoma. J Clin Oncol. 24:1898–1903. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Aravalli RN, Cressman EN and Steer CJ:
Cellular and molecular mechanisms of hepatocellular carcinoma: An
update. Arch Toxicol. 87:227–247. 2013. View Article : Google Scholar
|
|
3
|
Ma WL, Jeng LB, Lai HC, Liao PY and Chang
C: Androgen receptor enhances cell adhesion and decreases cell
migration via modulating β1-integrin-AKT signaling in
hepatocellular carcinoma cells. Cancer Lett. 351:64–71. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ng L, Tung-Ping Poon R, Yau S, Chow A, Lam
C, Li HS, Chung-Cheung Yau T, Law WL and Pang R: Suppression of
actopaxin impairs hepatocellular carcinoma metastasis through
modulation of cell migration and invasion. Hepatology. 58:667–679.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Villanueva A, Chiang DY, Newell P, et al:
Pivotal role of mTOR signaling in hepatocellular carcinoma.
Gastroenterology. 135:1972–1983. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhong W, Qin S, Zhu B, Pu M, Liu F, Wang
L, Ye G, Yi Q and Yan D: Oxysterol-binding protein (OSBP)-related
protein 8 (ORP8) increases sensitivity of hepatocellular carcinoma
cells to Fas-mediated apoptosis. J Biol Chem. Jan 16–2015.Epub
ahead of print. View Article : Google Scholar
|
|
7
|
Williams AH, Liu N, van Rooij E and Olson
EN: MicroRNA control of muscle development and disease. Curr Opin
Cell Biol. 21:461–469. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Divakaran V and Mann DL: The emerging role
of microRNAs in cardiac remodeling and heart failure. Circ Res.
103:1072–1083. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van Rooij E, Sutherland LB, Thatcher JE,
DiMaio JM, Naseem RH, Marshall WS, Hill JA and Olson EN:
Dysregulation of microRNAs after myocardial infarction reveals a
role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA.
105:13027–13032. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van Rooij E, Liu N and Olson EN: MicroRNAs
flex their muscles. Trends Genet. 24:159–166. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Frost RJ and van Rooij E: miRNAs as
therapeutic targets in ischemic heart disease. J Cardiovasc Transl
Res. 3:280–289. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lin Z, Murtaza I, Wang K, Jiao J, Gao J
and Li PF: miR-23a functions downstream of NFATc3 to regulate
cardiac hypertrophy. Proc Natl Acad Sci USA. 106:12103–12108. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu L, Zhou L, Cheng Y, Sun L, Fan J, Liang
J, Guo M, Liu N and Zhu L: MicroRNA-543 acts as an oncogene by
targeting PAQR3 in hepatocellular carcinoma. Am J Cancer Res.
4:897–906. 2014.PubMed/NCBI
|
|
14
|
Zhang S, Shan C, Kong G, Du Y, Ye L and
Zhang X: MicroRNA-520e suppresses growth of hepatoma cells by
targeting the NF-κB-inducing kinase (NIK). Oncogene. 31:3607–3620.
2012. View Article : Google Scholar
|
|
15
|
He XX, Chang Y, Meng FY, Wang MY, Xie QH,
Tang F, Li PY, Song YH and Lin JS: MicroRNA-375 targets AEG-1 in
hepatocellular carcinoma and suppresses liver cancer cell growth in
vitro and in vivo. Oncogene. 31:3357–3369. 2012. View Article : Google Scholar
|
|
16
|
Li D, Liu Y, Li H, Peng JJ, Tan Y, Zou Q,
Song XF, Du M, Yang ZH, Tan Y, et al: MicroRNA-1 promotes apoptosis
of hepatocarcinoma cells by targeting apoptosis inhibitor-5
(API-5). FEBS Lett. 589:68–76. 2015. View Article : Google Scholar
|
|
17
|
Chen P, Wang BL, Pan BS and Guo W:
MiR-1297 regulates the growth, migration and invasion of colorectal
cancer cells by targeting cyclo-oxygenase-2. Asian Pac J Cancer
Prev. 15:9185–9190. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang C, Chi YL, Wang PY, Wang YQ, Zhang
YX, Deng J, Lv CJ and Xie SY: miR-511 and miR-1297 inhibit human
lung adenocarcinoma cell proliferation by targeting oncogene TRIB2.
PLoS One. 7:e460902012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Perz JF, Armstrong GL, Farrington LA,
Hutin YJ and Bell BP: The contributions of hepatitis B virus and
hepatitis C virus infections to cirrhosis and primary liver cancer
worldwide. J Hepatol. 45:529–538. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Meng X, Franklin DA, Dong J and Zhang Y:
MDM2-p53 pathway in hepatocellular carcinoma. Cancer Res.
74:7161–7167. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ying TH, Tsai JH, Wu TT, Tsai MT, Su WW,
Hsieh YS and Liu JY: Immunochemical localization of protein kinase
Calpha in the biopsies of human hepatocellular carcinoma. Chin J
Physiol. 51:269–274. 2008.
|
|
23
|
Muche S, Kirschnick M, Schwarz M and
Braeuning A: Synergistic effects of β-catenin inhibitors and
sorafenib in hepatoma cells. Anticancer Res. 34:4677–4683.
2014.PubMed/NCBI
|
|
24
|
Mokkapati S, Niopek K, Huang L, Cunniff
KJ, Ruteshouser EC, deCaestecker M, Finegold MJ and Huff V:
β-catenin activation in a novel liver progenitor cell type is
sufficient to cause hepatocellular carcinoma and hepatoblastoma.
Cancer Res. 74:4515–4525. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang J, Park JS, Wei Y, Rajurkar M, Cotton
JL, Fan Q, Lewis BC, Ji H and Mao J: TRIB2 acts downstream of
Wnt/TCF in liver cancer cells to regulate YAP and C/EBPα function.
Mol Cell. 51:211–225. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Morishita A, Zaidi MR, Mitoro A,
Sankarasharma D, Szabolcs M, Okada Y, D'Armiento J and Chada K:
HMGA2 is a driver of tumor metastasis. Cancer Res. 73:4289–4299.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun M, Song CX, Huang H, Frankenberger CA,
Sankarasharma D, Gomes S, Chen P, Chen J, Chada KK, He C, et al:
HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth
and metastasis. Proc Natl Acad Sci USA. 110:9920–9925. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zaidi MR, Okada Y and Chada KK:
Misexpression of full-length HMGA2 induces benign mesenchymal
tumors in mice. Cancer Res. 66:7453–7459. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li Y, Zhao Z, Xu C, Zhou Z, Zhu Z and You
T: HMGA2 induces transcription factor Slug expression to promote
epithelial-to-mesenchymal transition and contributes to colon
cancer progression. Cancer Lett. 355:130–140. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Luo Y, Li W and Liao H: HMGA2 induces
epithelial-to-mesenchymal transition in human hepatocellular
carcinoma cells. Oncol Lett. 5:1353–1356. 2013.PubMed/NCBI
|
|
31
|
Pentimalli F, Dentice M, Fedele M,
Pierantoni GM, Cito L, Pallante P, Santoro M, Viglietto G, Dal Cin
P and Fusco A: Suppression of HMGA2 protein synthesis could be a
tool for the therapy of well differentiated liposarcomas
overexpressing HMGA2. Cancer Res. 63:7423–7427. 2003.PubMed/NCBI
|