Introduction
Bone is a dynamic tissue that can be continuously
degraded and renewed. The processes of bone remodeling are
accomplished by the coordinated regulation of bone-resorbing
osteoclasts and bone-forming osteoblasts (1). It has been well documented that the
skeleton is a highly mechano-adaptive system (1), which can remodel its own structure
in response to external mechanical stimulation (2). By contrast, the lack of mechanical
stimuli to the weight-bearing regions of the skeleton leads to
lower bone formation and inferior bone quality (3). Studies have demonstrated the
capability of osteoblasts to respond to various forms of
physiological mechanical stimuli, such as fluid shear stress
(4,5), compressive force (6,7),
and cyclic stretch (8–12). It has been proven that cyclic
stretch is a potent mediator in promoting osteogenic mineralization
(10) and the expression of
osteoblastic differentiation markers, such as runt-related
transcription factor 2 (Runx2) (9,11)
and alkaline phosphatase (ALP) (8,9,11,12).
It has been demonstrated that the differentiation of
osteoblasts is also regulated by various growth factors, such as
transforming growth factor-β (TGF-β), insulin-like growth factors
and bone morphogenetic protein-2 (BMP-2) (13). BMP-2 is a bone-growth regulatory
factor that belongs to the TGF-β superfamily. In vivo
studies have demonstrated that BMP-2 plays a pivotal role in
stimulating bone regeneration and regulating bone remodeling
(14–17). Previous studies have also reported
that BMP-2 induces an increase in the expression of differentiation
markers (e.g., ALP and Runx2) and mineralized bone nodules in
osteoblasts in vitro (5,18).
Moreover, BMP-2-induced bone regeneration and ossification in
vivo can be enhanced by mechanical stimuli in distraction
osteogenesis or in models of bone segmental defects (19–21), revealing the therapeutic potential
of the combined application of BMP-2 and mechanical load in
clinical bone diseases. However, the underlying mechanisms through
which the combined application of mechanical load and BMP-2 promote
osteogenesis remain elusive. In addition, the mechanisms through
which mechanical load and BMP-2 regulate osteoblastic
differentiation remain poorly understood.
Hes-related family bHLH transcription factor with
YRPW motif 1 (Hey1), a member of the basic helix-loop-helix family
(22), is a downstream mediator
of Notch signaling (23) which
regulates bone remodeling and osteoblastic differentiation
(24,25). Previous studies have revealed that
Hey1 negatively regulates bone regeneration in vivo
(26) and osteoblastic
differentiation in vitro (18). Furthermore, BMP-2 induces an
increase in the expression of Hey1 in osteoblasts (18), suggesting that Hey1 serves as a
negative regulatory factor in BMP-2-induced osteoblastic
differentiation. In addition, substantial evidence has demonstrated
the regulatory role of cyclic stretch in the expression of Hey1 in
vascular smooth muscle cells and human umbilical vein endothelial
cells (27–30). However, the role of Hey1 in the
regulation of mechanically-induced osteoblastic differentiation
remains unclear. It also remains unknown whether Hey1 expression is
affected by cyclic stretch in the presence or absence of BMP-2 in
osteoblasts.
Therefore, in the present study, the effects and
potential mechanisms of cyclic stretch in the regulation of
BMP-2-induced osteoblastic differentiation were investigated in
osteoblast-like MC3T3-E1 cells. Firstly, we investigated the
effects of mechanical load or BMP-2 on osteoblastic differentiation
markers (ALP and Runx2). We then evaluated the effects of cyclic
stretch on the expression of osteoblastic differentiation markers
and Hey1 in the presence or absence of BMP-2 in MC3T3-E1 cells.
Finally, the expression levels of osteoblastic differentiation
markers under the combined stimulation of cyclic stretch and BMP-2
were measured following the overexpression of Hey1 by the transient
transfection of a Hey1 expression plasmid in MC3T3-E1 cells. Our
findings provide a novel molecular mechanism through which cyclic
stretch enhances BMP-2-induced osteoblastic differentiation through
the inhibition of Hey1.
Materials and methods
Reagents
Recombinant BMP-2 was purchased from Sigma-Aldrich
(St. Louis, MO, USA). Rabbit anti-GAPDH monoclonal antibody (#2118)
was obtained from Cell Signaling Technology (Danvers, MA, USA).
Rabbit anti-Runx2 (sc-10758) and anti-His-probe (sc-803) polyclonal
antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa
Cruz, CA, USA). Rabbit anti-Hey1 polyclonal antibody (ab22614) was
purchased from Abcam (Cambridge, MA, USA). HRP-conjugated goat
secondary antibody (AP307P) was obtained from Millipore (Billerica,
MA, USA). Alexa Fluor® 594, 488-conjugated secondary
antibodies (A11037 and A27034) and the pcDNA3.1 vector were
obtained from Invitrogen (Carlsbad, CA, USA).
Cell culture and cyclic stretch
stimulation
The MC3T3-E1 cells were obtained from the American
Type Culture Collection (ATCC; Manassas, VA, USA). The MC3T3-E1
cells were cultured in a humidified atmosphere of 5% CO2
at 37°C in alpha minimum essential medium (α-MEM) supplemented with
10% fetal bovine serum (FBS) (both from HyClone, Logan, UT, USA).
For the application of cyclic stretch, the MC3T3-E1 cells were
seeded at 2×105 cells/well (1×105 cells/ml)
on 6-well BioFlex culture plates coated with type I collagen
(Flexcell International Corp., Hillsborough, NC, USA) and incubated
until they reached 70% confluence. The cells were then cultured in
serum-free α-MEM for 24 h to be synchronized prior to mechanical
stimulation. The medium was then replaced with fresh α-MEM
containing 10% FBS with or without various concentrations of BMP-2
(0, 50, 100, 150, 200 or 250 ng/ml). The cells were then subjected
to sine-wave stretch with different peak magnitudes of elongation
(0, 5, 10 or 15%) at 0.1 Hz (5-sec stretch/5-sec relaxation) for 24
h using an FX-4000 Tension System (Flexcell International Corp.).
The control cells were maintained under the same experimental
conditions, but were not exposed to mechanical stretch.
Plasmid construction and transient
transfection
The ORF of the mouse Hey1 cDNA was amplified by
RT-PCR using specific primers (sense, 5′-CGG AAT TCA TGG AGA GAG
CTC ACC C-3′ and antisense, 5′-TTG CGG CCG CTT AGA AAG CTC CGA
TC-3′) that were designed based on the Hey1 gene (GenBank ID:
NM_010423.2) by Takara (Shiga, Japan). The gel-purified PCR
products were digested with the restriction enzymes, EcoRI
and NotI (Takara), and cloned into the eukaryotic expression
vector, pcDNA3.1 (Invitrogen), which contained a 6xHis-tag (~5.5
kDa), to yield pcDNA3.1-Hey1. The inserted sequence was confirmed
by DNA sequencing. Transient transfection was carrried out using
Lipofectamine 2000 reagent (Invitrogen) following the
manufacturer's instructions. Following 24 h of transfection, the
cells were harvested and analyzed for the expression of Hey1 and
His-tag. Untransfected cells were used as controls and cells
transfected with the empty pcDNA3.1 vector served as the
mock-transfected cells.
Determination of ALP activity
ALP activity in the medium of MC3T3-E1 cells were
determined by colorimetric assay using an Alkaline Phosphatase
assay kit (Jiancheng Bioengineering Institute, Nanjing, China)
according to the manufacturer's instructions. In brief, 20
µl of cell culture medium mixed with 1 ml of reaction
solution containing 4-nitrophenyl phosphate (18 mM) and
2-amino-2-methyl-1-propanol (0.5 M) were incubated in the dark for
15 min at 37°C in microcentrifuge tubes. The absorbance values were
recorded at 405 nm using the Synergy 2 Multi-Mode Microplate Reader
(BioTek, Winooski, VT, USA). Every sample was measured in
triplicate and the assays were performed 4 times.
Immunofluorescence staining
Cells in the BioFlex culture plate were fixed with
4% paraformaldehyde for 30 min and subsequently permeabilized with
0.1% Triton X-100 for 5 min. After being blocked with 2% goat serum
in phosphate-buffered saline (PBS) at 37°C for 1 h, the cells were
incubated overnight at 4°C with primary antibodies to Hey1 (1:200)
and Runx2 (1:50). The cells were then incubated with Alexa
Fluor® 594, 488-conjugated goat anti-rabbit antibodies
(1:400) at 37°C for 1 h and counterstained with
4′,6-diamidino-2-phenylindole (DAPI) for 5 min at room temperature.
Images were obtained using a confocal laser scanning microscope
(FV1000; Olympus, Tokyo, Japan). Fluorescence intensity was
determined by Image-Pro Plus software (Media Cybernetics, Inc.,
Rockville, MD, USA).
Total RNA isolation and RT-qPCR
Total RNA was isolated from the MC3T3-E1 cells using
TRizol reagent (Invitrogen) according to the instructions provided
by the manufacturer and quantified by spectrophotometry (NanoDrop
2000c spectrophotometer; Thermo Fisher Scientific, Rockford, IL,
USA). RNA (1 µg) was reverse transcribed into cDNA in a 20
µl reaction with oligo(dT)18 as a primer using a
First Strand cDNA Synthesis kit (Thermo Fisher Scientific,
Pittsburgh, PA, USA) according to the manufacturer's instructions.
qPCR was performed on 1 µl of cDNA in a 20 µl
reaction with SYBR Premix Ex Taq II (Takara) using the Bio-Rad
CFX96 real-time PCR detection system (Bio-Rad, Philadelphia, PA,
USA). The sense and antisense primers were: 5′-CGA CGA GAC CGA ATC
AAT AAC-3′ and 5′-CAA ACT CCG ATA GTC CAT AGC C-3′ for Hey1
(GenBank ID: NM_010423.2); 5′-GGG CAT TGT GAC TAC CAC TCG-3′ and
5′-CCT CTG GTG GCA TCT CGT TAT-3′ for ALP (GenBank ID:
NM_007431.2); 5′-GAC ACT GCC ACC TCT GAC TTC T-3′ and 5′-ATG AAA
TGC TTG GGA ACT GC-3′ for Runx2 (GenBank ID: NM_001145920.2); and
5′-GGT GAA GGT CGG TGT GAA CG-3′ and 5′-CTC GCT CCT GGA AGA TGG
TG-3′ for GAPDH (GenBank ID: NM_008084.2). The protocol for the
RT-qPCR reactions was as follows: an initial denaturation at 95°C
for 30 sec followed by 45-cycle denaturation at 95°C for 15 sec,
annealing at 60°C for 15 sec, and extension at 72°C for 15 sec.
GAPDH was used as an internal control for normalization. The
relative quantity of mRNA was calculated (2−ΔΔCt
analysis). All RT-qPCR reactions were performed in triplicate.
Western blot analysis
The cells were washed with ice-cold PBS and lysed to
release the whole proteins using RIPA buffer with 1 mM PMSF. The
cell lysates were transferred into a pre-cooled microcentrifuge
tube and constant agitation was maintained for 30 min at 4°C. The
protein extracts were then centrifuged at 4°C for 20 min at 12,000
rpm. The protein content of the supernatant was collected and the
protein concentration was determined by BCA assay (Pierce Chemical
Co., Rockford, IL, USA). The protein extracts (30 µg/sample)
were subjected to electrophoretic separation by 10% Tris-glycine
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE), and transferred onto PVDF membranes (Millipore), after
being mixed with 5X loading buffer and boiled for 8 min. The PVDF
membranes were blocked in Tris-buffered saline with 0.5% Tween-20
(TBST) containing 5% BSA for 2 h, and incubated overnight at 4°C
with primary antibodies to His-probe (1:500), GAPDH (1:1,000) and
Runx2 (1:400) in TBST containing 5% BSA. The membranes were then
incubated with a 1:5,000 dilution of HRP-conjugated goat
anti-rabbit secondary antibody for 1 h at room temperature, and
then visualized using an ECL system (GE ImageQuant 350; GE
Healthcare, Piscataway, NJ, USA). GAPDH was used as an internal
control for normalization. Semi-quantitative analyses of the bands
were performed by using the Quantity One software (Bio-Rad).
Statistical analysis
All data presented in this study are expressed as
the means ± standard deviation (SD). Statistical analyses were
performed using Microsoft SPSS version 13.0 software (SPSS, Inc.,
Chicago, IL, USA). One-way analysis of variance (ANOVA) with Tukey
post hoc analysis was used to determine the differences between 2
groups. A value of P<0.05 was considered to indicate a
statistically significant difference.
Results
Cyclic stretch stimulation or BMP-2
induces an increase in the expression of differentiation markers in
osteoblasts
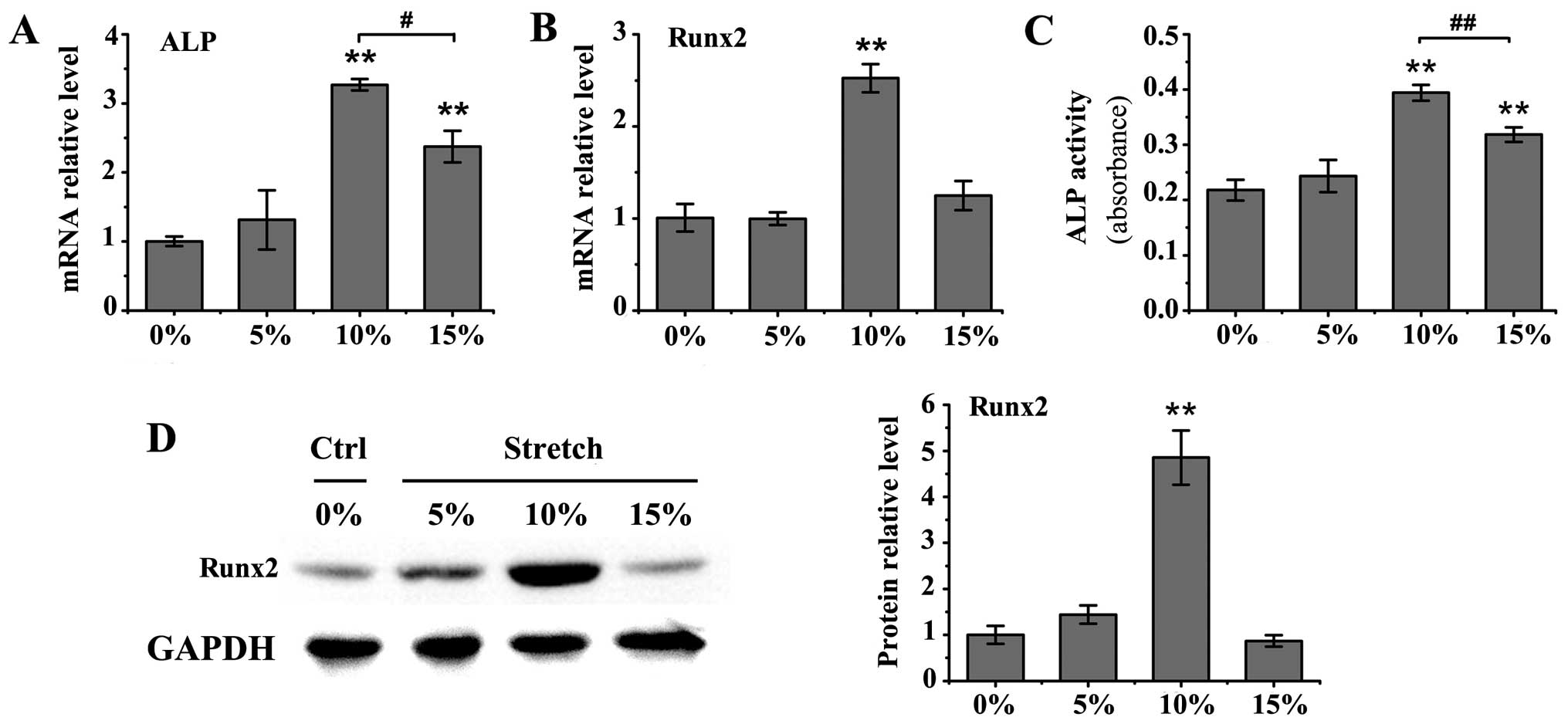
Cyclic stretch with 10 or 15% elongation induced a
significant increase in ALP mRNA levels in the MC3T3-E1 cells
(P<0.01), as well as an increase in ALP activity in the medium
(P<0.01; Fig. 1A and C)
compared to the untreated controls. The upregulation in ALP mRNA
expression and activity was more prominent under mechanical stretch
with 10% elongation than 15% elongation (mRNA expression,
P<0.05; activity, P<0.01). Furthermore, the mRNA and protein
levels of Runx2 were significantly increased only by cyclic stretch
with 10% elongation (P<0.01 vs. control group; Fig. 1B and D). Treatment with BMP-2 at
various concentrations (50, 100, 150, 200 or 250 ng/ml) induced an
increase in the mRNA levels of ALP in the MC3T3-E1 cells compared
to the untreated controls (P<0.01; Fig. 2A), and all concentrations of BMP-2
significantly enhanced ALP activity (P<0.01 vs. control group)
apart from the concentration of 50 ng/ml (Fig. 2C). Moreover, western blot analysis
revealed a significant increase in Runx2 protein expression
following treatment with 200 ng/ml or 250 ng/ml BMP-2 (P<0.01
vs. control group; Fig. 2D).
BMP-2 at the concentrations of 150, 200 and 250 ng/ml also
significantly upregulated the mRNA levels of Runx2 in the MC3T3-E1
cells (150 ng/ml, P<0.05; 200 and 250 ng/ml, P<0.01 vs.
control group; Fig. 2B).
Treatment with BMP-2 at the concentration of 200 ng/ml exhibited
the most prominent effects on the expression of osteoblastic
differentiation markers.
Cyclic stretch enhances the BMP-2-induced
upregulation in the expression of differentiation markers in
osteoblasts
Cyclic stretch stimulation (10%, 0.1 Hz) and
treatment with BMP-2 (200 ng/ml) separately induced a significant
increase in the mRNA levels of ALP and Runx2 compared to the
untreated controls (ALP: stretch, P<0.05; BMP-2, P<0.01;
Runx2: stretch, P<0.01; BMP-2, P<0.01; Fig. 3A and B). Moreover, the combined
application of cyclic stretch and BMP-2 significantly enhanced the
upregulation of the mRNA levels of ALP and Runx2 in the MC3T3-E1
cells as compared with the stretch group or BMP-2 group (P<0.01;
Fig. 3A and B). Cyclic stretch
also enhanced ALP activity in the medium in the BMP-2-stimulated
MC3T3-E1 cells (P<0.01; Fig.
3C). Furthermore, western blot analysis revealed that the
combined application of cyclic stretch and BMP-2 further promoted
the upregulation of Runx2 protein expression in the MC3T3-E1 cells
compared with either cyclic stretch or BMP-2 stimulation alone
(P<0.01; Fig. 3D). The
promotional role of cyclic stretch in the BMP-2-induced increase in
Runx2 protein expression in the MC3T3-E1 cells was further
confirmed by immunofluorescence staining, which revealed a higher
Runx2 protein expression under the combined application of cyclic
stretch and BMP-2 compared with either cyclic stretch or BMP-2
stimulation alone (P<0.01; Fig.
3E).
Cyclic stretch inhibits the BMP-2-induced
upregulation of Hey1 in osteoblasts
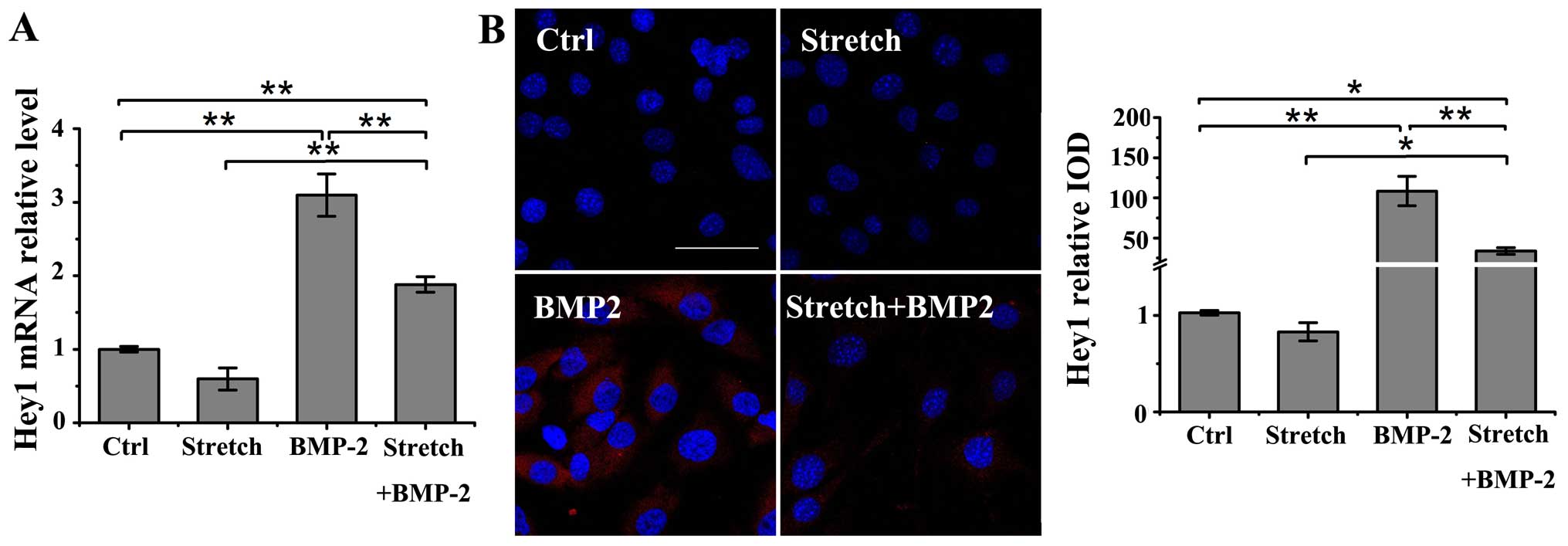
The expression of Hey1, a potent regulator of
osteogenesis (18,26), was evaluated under the following
conditions: stimulation with either cyclic stretch (10%, 0.1 Hz) or
BMP-2 (200 ng/ml) alone, or with cyclic stretch plus BMP-2 in the
MC3T3-E1 cells. Treatment with BMP-2 alone induced a significant
increase in the mRNA and protein levels of Hey1 compared to the
untreated controls as shown by RT-qPCR and immunofluorescence
staining (P<0.01); however, mechanical loading alone did not
affect the Hey1 mRNA and protein expression (Fig. 4A and B). However, cyclic stretch
stimulation significantly suppressed the BMP-2-induced upregulation
in the mRNA and protein levels of Hey1 in the MC3T3-E1 cells
(P<0.01). Nonetheless, cyclic stretch did not completely
neutralize the BMP-2-induced increase in Hey1 mRNA or protein
expression compared to the control group (mRNA, P<0.01; protein,
P<0.05).
Transient Hey1 overexpression reverses
the effects of cyclic stretch on the BMP-2-induced upregulation of
differentiation markers in osteoblasts
To establish the role of Hey1 in the stretch-induced
upregulation of differentiation markers in BMP-2-stimulated
MC3T3-E1 cells, we first constructed a Hey1 expression plasmid
using the eukaryotic expression vector, pcDNA3.1, and following
transient transfection into the MC3T3-E1 cells for 24 h, the mRNA
and protein expression levels of Hey1 were measured by RT-qPCR and
western blot analysis. The results of RT-qPCR revealed that the
MC3T3-E1 cells transfected with pcDNA3.1-Hey1 expressed
significantly higher mRNA levels of Hey1 than the controls
(untransfected cells) and mock-transfected cells (transfected with
the empty control vector, pcDNA3.1) (P<0.01; Fig. 5A). Furthermore, western blot
analysis with an anti-His-probe antibody was performed to verify
the effectiveness of the plasmid to induce Hey1 overexpression
(Fig. 5B).
The cells in medium containing BMP-2 (200 ng/ml)
were subjected to cyclic stretch (10%, 0.1 Hz) for 24 h following
24 h of transfection. The results of RT-qPCR revealed that the
overexpression of Hey1 inhibited the stretch-induced increase in
the mRNA levels of ALP and Runx2 in the BMP-2-stimulated MC3T3-E1
cells (P<0.01; Fig. 5C and D).
Moreover, the stretch-induced upregulation of ALP activity in the
medium was also suppressed by the overexpression of Hey1 in the
BMP-2-stimulated MC3T3-E1 cells (P<0.01; Fig. 5E). Furthermore, the results from
western blot anlaysis and immunofluorescence staining revealed that
Hey1 overexpression inhibited the stretch-induced increase in Runx2
protein expression in the MC3T3-E1 cells treated with BMP-2
(P<0.01; Fig. 5F and G).
Discussion
Numerous studies have demonstrated the involvement
of cyclic stretch or BMP-2 in the regulation of osteoblastic
differentiation (8–12,14–18). However, the relevant mechanisms
remain elusive. Previous in vivo studies have demonstrated
that BMP-2-induced bone regeneration and ossification can be
enhanced by mechanical loading (19,21). The present in vitro study
revealed that cyclic stretch enhanced the BMP-2-induced
upregulation of osteoblastic differentiation markers (ALP and
Runx2) through the inhibition of Hey1, and highlighted that Hey1
serves as a potent negative regulator of osteoblastic
differentiation.
During distraction osteogenesis, cancellous and
cortical bones undergo mechanical stimulation and osteoblasts
located on the surface of unmineralized matrix of cancellous and
cortical bones are subjected to mechanical loads. In this study, to
imitate the synergistic action of cyclic stretch and BMP-2 during
distraction osteogenesis for an in vitro investigation,
MC3T3-E1 cells were cultured as a monolayer on flexible substrate
surfaces and subjected to mechanical stretch in the presence of
BMP-2. Our results revealed that cyclic stretch of 10% elongation
or treatment with BMP-2 at a concentration of 200 ng/ml exhibited
the most obvious effects on osteoblastic differentiation markers
(ALP and Runx2). Moreover, previous in vitro studies have
also used similar magnitudes of mechanical stretch to investigate
the mechanotransduction of bone cells (31,32) and the same concentrations of BMP-2
have been applied to research osteoblastic differentiation
(5,33). Furthermore, our findings also
demonstrated that treatment with BMP-2 at a lower concentration (50
ng/ml) induced an increase in ALP mRNA levels, but not in ALP
activity in the medium. Treatment with BMP-2 at 150 ng/ml induced
an increase in Runx2 mRNA levels, but not in Runx2 protein
expression, suggesting that osteoblastic gene transcription may be
more sensitive to BMP-2 concentration than translation.
A hierarchy of transcription factors and osteogenic
markers are expressed during osteoblastic differentiation (34). Runx2, a bone-specific
transcription factor, plays an essential role in bone formation
in vivo and osteoblastic differentiation in vitro
(34,35). Runx2 forms a heterodimeric complex
with the transcriptional co-activator core binding factor β, and
binds to osteoblast-specific cis-element 2 sites to modulate
the transcription of osteoblast-related genes (34,36), such as ALP, an early-stage marker
of osteoblastic differentiation (33). Therefore, in this study, we
investigated the synergistic effects of cyclic stretch and BMP-2 on
ALP and Runx2 expression to demonstrate osteogenic differentiation.
Our results revealed that cyclic stretch enhanced the BMP-2-induced
increase in the mRNA levels of ALP and Runx2, and ALP activity in
the medium and Runx2 protein expression. These data suggest that
cyclic stretch improves osteoblastic differentiation in response to
BMP-2 in vitro, and they reveal the significant promotional
effects of the combined application of BMP-2 and mechanical stretch
on osteoblastic differentiation, which is consistent with previous
in vivo studies (19,21).
Notch signaling has been proven to play a crucial
role in bone remodeling and osteoblast function (24,25). Hey1, as a downstream target gene
of Notch signaling, can be stimulated by BMP-2 and negatively
regulates bone remodeling and osteoblastic differentiation by
suppressing the transcriptional activation of Runx2 (18,26). Moreover, previous studies have
shown that cyclic strain inhibits the mRNA and protein levels of
Hey1 in vascular smooth muscle cells (28,30). The above-mentioned evidence
supports the important role of cyclic stretch in the regulation of
Hey1 expression. Therefore, in the present study, we investigated
the effects of cyclic stretch on Hey1 expression in
BMP-2-stimulated MC3T3-E1 cells. We suggest that mechanical stretch
enhances BMP-2-induced osteoblastic differentiation by suppressing
Hey1. Our results revealed that BMP-2 induced a significant
increase in Hey1 expression, and that cyclic stretch alone did not
affect Hey1 expression. Nevertheless, this study also revealed that
cyclic stretch inhibited the BMP-2-induced upregulation of Hey1 in
osteoblasts. These results indicate that the inhibitory effect of
mechanical stretch on the upregulation of Hey1 under BMP-2
stimulation may be an underlying mechanism through which cyclic
stretch promotes BMP-2-induced osteoblastic differentiation.
However, mechanical stretch was not able to completely neutralize
the BMP-2-induced increase in Hey1 expression in the present study.
A possible reason for this is that mechanical load increases
osteoblastic BMP-2 expression (5,9)
and Hey1 may be upregulated by BMP-2, which eliminate the
inhibitory effect of cyclic stretch on Hey1 to a certain
extent.
To further confirm the role of Hey1 in the
regulation of osteoblastic differentiation under the combined
application of BMP-2 and stretch stimulation, we constructed a Hey1
expression plasmid using the vector pcDNA3.1 and transiently
transfected the plasmid into MC3T3-E1 cells. The cells in the
presence of BMP-2 were subjected to cyclic stretch for 24 h. As a
result, our findings revealed that Hey1 overexpression reversed the
role of cyclic stretch in the BMP-2-induced upregulation of ALP
mRNA levels and activity, and Runx2 expression. Thus, our findings
reveal the importance of Hey1 in the regulation of BMP-2-induced
osteoblastic differentiation in response to cyclic stretch. The
present study also highlighted that Hey1 functions as a key
negative regulator of osteoblastic differentiation induced by the
combined application of BMP-2 and mechanical stretch.
In conclusion, the present study demonstrated that
cyclic stretch enhanced the BMP-2-induced upregulation of
osteoblastic differentiation markers in MC3T3-E1 cells through the
inhibition of Hey1. Our findings indicate that Hey1 is an essential
regulator of osteoblastic differentiation under stimulation with
mechanical stretch and BMP-2. The present study broadens our
fundamental knowledge of osteoblastic mechanotransduction and also
sheds new insight into the mechanisms through which the combined
application of BMP-2 and mechanical load promotes osteogenesis.
Acknowledgments
The present study was supported by grants from the
National Natural Science Foundation of China (nos. 31070836 and
81100750).
References
|
1
|
Thompson WR, Rubin CT and Rubin J:
Mechanical regulation of signaling pathways in bone. Gene.
503:179–193. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bolam KA, van Uffelen JG and Taaffe DR:
The effect of physical exercise on bone density in middle-aged and
older men: a systematic review. Osteoporos Int. 24:2749–2762. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jing D, Cai J, Wu Y, Shen G, Li F, Xu Q,
Xie K, Tang C, Liu J, Guo W, et al: Pulsed electromagnetic fields
partially preserve bone mass, microarchitecture, and strength by
promoting bone formation in hindlimb-suspended rats. J Bone Miner
Res. 29:2250–2261. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jing D, Lu XL, Luo E, Sajda P, Leong PL
and Guo XE: Spatiotemporal properties of intracellular calcium
signaling in osteocytic and osteoblastic cell networks under fluid
flow. Bone. 53:531–540. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mai Z, Peng Z, Wu S, Zhang J, Chen L,
Liang H, Bai D, Yan G and Ai H: Single bout short duration fluid
shear stress induces osteogenic differentiation of MC3T3-E1 cells
via integrin β1 and BMP2 signaling cross-talk. PLoS One.
8:e616002013. View Article : Google Scholar
|
|
6
|
Rath B, Nam J, Deschner J, Schaumburger J,
Tingart M, Grässel S, Grifka J and Agarwal S: Biomechanical forces
exert anabolic effects on osteoblasts by activation of SMAD 1/5/8
through type 1 BMP receptor. Biorheology. 48:37–48. 2011.PubMed/NCBI
|
|
7
|
Rath B, Nam J, Knobloch TJ, Lannutti JJ
and Agarwal S: Compressive forces induce osteogenic gene expression
in calvarial osteoblasts. J Biomech. 41:1095–1103. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang L, Li JY, Zhang XZ, Liu L, Wan ZM, Li
RX and Guo Y: Involvement of p38MAPK/NF-κB signaling pathways in
osteoblasts differentiation in response to mechanical stretch. Ann
Biomed Eng. 40:1884–1894. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guo Y, Zhang CQ, Zeng QC, Li RX, Liu L,
Hao QX, Shi CH, Zhang XZ and Yan YX: Mechanical strain promotes
osteoblast ECM formation and improves its osteoinductive potential.
Biomed Eng Online. 11(80)2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jansen JH, Eijken M, Jahr H, Chiba H,
Verhaar JA, van Leeuwen JP and Weinans H: Stretch-induced
inhibition of Wnt/beta-catenin signaling in mineralizing
osteoblasts. J Orthop Res. 28:390–396. 2010.
|
|
11
|
Kanno T, Takahashi T, Tsujisawa T,
Ariyoshi W and Nishihara T: Mechanical stress-mediated Runx2
activation is dependent on Ras/ERK1/2 MAPK signaling in
osteoblasts. J Cell Biochem. 101:1266–1277. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nishioka S, Fukuda K and Tanaka S: Cyclic
stretch increases alkaline phosphatase activity of osteoblast-like
cells: a role for prostaglandin E2. Bone Miner. 21:141–150. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Papachroni KK, Karatzas DN, Papavassiliou
KA, Basdra EK and Papavassiliou AG: Mechanotransduction in
osteoblast regulation and bone disease. Trends Mol Med. 15:208–216.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kirker-Head C, Karageorgiou V, Hofmann S,
Fajardo R, Betz O, Merkle HP, Hilbe M, von Rechenberg B, McCool J,
Abrahamsen L, et al: BMP-silk composite matrices heal critically
sized femoral defects. Bone. 41:247–255. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chu TM, Warden SJ, Turner CH and Stewart
RL: Segmental bone regeneration using a load-bearing biodegradable
carrier of bone morphogenetic protein-2. Biomaterials. 28:459–467.
2007. View Article : Google Scholar :
|
|
16
|
Chatakun P, Núñez-Toldrà R, Díaz López EJ,
Gil-Recio C, Martínez-Sarrà E, Hernández-Alfaro F, Ferrés-Padró E,
Giner-Tarrida L and Atari M: The effect of five proteins on stem
cells used for osteoblast differentiation and proliferation: a
current review of the literature. Cell Mol Life Sci. 71:113–142.
2014. View Article : Google Scholar
|
|
17
|
Kamiya N and Mishina Y: New insights on
the roles of BMP signaling in bone-A review of recent mouse genetic
studies. Biofactors. 37:75–82. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zamurovic N, Cappellen D, Rohner D and
Susa M: Coordinated activation of notch, Wnt, and transforming
growth factor-beta signaling pathways in bone morphogenic protein
2-induced osteogenesis. Notch target gene Hey1 inhibits
mineralization and Runx2 transcriptional activity. J Biol Chem.
279:37704–37715. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schwarz C, Wulsten D, Ellinghaus A, Lienau
J, Willie BM and Duda GN: Mechanical load modulates the stimulatory
effect of BMP2 in a rat nonunion model. Tissue Eng Part A.
19:247–254. 2013. View Article : Google Scholar :
|
|
20
|
Yonezawa H, Harada K, Ikebe T, Shinohara M
and Enomoto S: Effect of recombinant human bone morphogenetic
protein-2 (rhBMP-2) on bone consolidation on distraction
osteogenesis: a preliminary study in rabbit mandibles. J
Craniomaxillofac Surg. 34:270–276. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cheung LK and Zheng LW: Effect of
recombinant human bone morphogenetic protein-2 on mandibular
distraction at different rates in an experimental model. J
Craniofac Surg. 17:100–108; discussion 109–110. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Maier MM and Gessler M: Comparative
analysis of the human and mouse Hey1 promoter: Hey genes are new
Notch target genes. Biochem Biophys Res Commun. 275:652–660. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Belandia B, Powell SM, García-Pedrero JM,
Walker MM, Bevan CL and Parker MG: Hey1, a mediator of notch
signaling, is an androgen receptor corepressor. Mol Cell Biol.
25:1425–1436. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zanotti S and Canalis E: Notch signaling
in skeletal health and disease. Eur J Endocrinol. 168:R95–R103.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Regan J and Long F: Notch signaling and
bone remodeling. Curr Osteoporos Rep. 11:126–129. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Salie R, Kneissel M, Vukevic M, Zamurovic
N, Kramer I, Evans G, Gerwin N, Mueller M, Kinzel B and Susa M:
Ubiquitous overexpression of Hey1 transcription factor leads to
osteopenia and chondrocyte hypertrophy in bone. Bone. 46:680–694.
2010. View Article : Google Scholar
|
|
27
|
Zhu JH, Chen CL, Flavahan S, Harr J, Su B
and Flavahan NA: Cyclic stretch stimulates vascular smooth muscle
cell alignment by redox-dependent activation of Notch3. Am J
Physiol Heart Circ Physiol. 300:H1770–H1780. 2011. View Article : Google Scholar :
|
|
28
|
Guha S, Cullen JP, Morrow D, Colombo A,
Lally C, Walls D, Redmond EM and Cahill PA: Glycogen synthase
kinase 3 beta positively regulates Notch signaling in vascular
smooth muscle cells: role in cell proliferation and survival. Basic
Res Cardiol. 106:773–785. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Morrow D, Cullen JP, Cahill PA and Redmond
EM: Cyclic strain regulates the Notch/CBF-1 signaling pathway in
endothelial cells: role in angiogenic activity. Arterioscler Thromb
Vasc Biol. 27:1289–1296. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Morrow D, Sweeney C, Birney YA, Cummins
PM, Walls D, Redmond EM and Cahill PA: Cyclic strain inhibits Notch
receptor signaling in vascular smooth muscle cells in vitro. Circ
Res. 96:567–575. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tang L, Lin Z and Li YM: Effects of
different magnitudes of mechanical strain on osteoblasts in vitro.
Biochem Biophys Res Commun. 344:122–128. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li FF, Chen FL, Wang H, Yu SB, Cui JH,
Ding Y and Feng X: Proteomics based detection of differentially
expressed proteins in human osteoblasts subjected to mechanical
stress. Biochem Cell Biol. 91:109–115. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Park JK, Jang H, Hwang S, Kim EJ, Kim DE,
Oh KB, Kwon DJ, Koh JT, Kimura K, Inoue H, et al: ER
stress-inducible ATF3 suppresses BMP2-induced ALP expression and
activation in MC3T3-E1 cells. Biochem Biophys Res Commun.
443:333–338. 2014. View Article : Google Scholar
|
|
34
|
Komori T: Regulation of osteoblast
differentiation by transcription factors. J Cell Biochem.
99:1233–1239. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ziros PG, Basdra EK and Papavassiliou AG:
Runx2: of bone and stretch. Int J Biochem Cell Biol. 40:1659–1663.
2008. View Article : Google Scholar
|
|
36
|
Ducy P, Zhang R, Geoffroy V, Ridall AL and
Karsenty G: Osf2/Cbfa1: a transcriptional activator of osteoblast
differentiation. Cell. 89:747–754. 1997. View Article : Google Scholar : PubMed/NCBI
|