Introduction
Gilbert's syndrome, which was first reported by
Augustin Nicolas Gilbert in 1901, is a mild genetic liver disorder
characterized by unconjugated hyperbilirubinemia without overt
signs of hemolysis or structural liver disease (1). The clinical manifestation of
Gilbert's syndrome is an elevated level of serum bilirubin
(2). Moreover, some patients may
present with weakness, indigestion, abdominal pain in the liver
area and an intolerance to fat (1).
With the development of molecular biology, Gilbert's
syndrome has been investigated extensively. It has been found that
UDP-glucuronosyltransferase 1A1 (UGT1A1) plays a critical
role in the elimination pathway of bilirubin and defects in
UGT1A1 result in the development of Gilbert's syndrome
(3–5). Numerous mutations of the
UGT1A1 gene, in the regulatory region and the coding region
among others, have been detected to confirm the diagnosis of
Gilbert's syndrome (6–10,12,21). According to previous studies,
there are currently three common genotypes found in patients with
Gilbert's syndrome: the T-3279G mutation in the phenobarbital
responsive enhancer module (PBREM), the TA-insertion in the TATA
box, creating the A(TA)7TAA motif instead of A(TA)6TAA and the
G211A mutation in coding exon 1 of the UGT1A1 gene (11–13).
At present, the direct sequencing method is the
principal approach used to detect Gilbert's syndrome in the
clinical laboratory; however, the price of sequencing is expensive.
In order to diagnose Gilbert's syndrome, it is important to
establish a simple, effective and low cost method to detect
mutations of the UGT1A1 gene. In this study, to the best of
our knowledge, we applied the three-dimensional polyacrylamide
gel-based DNA microarray method for the first time, in order to
detect the T-3279G, A(TA)6/7TAA and G211A mutations of the
UGT1A1 gene to confirm the diagnosis of Gilbert's
syndrome.
Three-dimensional polyacrylamide gel-based DNA
microarray hybridized with dual-color fluorescent probes is a
rapid, simple and low coast approach used for gene mutation
analysis. This method relies on the co-polymerization of
acrylamide-modified PCR products with acrylamide monomers and
acryl-modified slides to prepare the gel-based microarray.
Acrylamide-modified PCR products from genomic DNA specimens are
spotted and immobilized onto acrylamide-modified glass slides to
fabricate a microarray. The slide is then transferred to a vacuum
chamber with N,N,N',N'-tetramethyl-ethylenediamine (TEMED), so that
TEMED is vaporized and diffuses into the spots to induce
polymerization. Following hybridization with the specific probes
labeled with Cy3 or Cy5, electrophoresis is performed to remove the
non-specifically bound targets and mismatches. Through two-color
fluorescent (green and red) scanning, images are captured to
determine the genotype of each sample (14).
In order to correctly diagnose Gilbert's syndrome
and to avoid side-effects from the adminstration of unecessary
therapeutic agents, in this study, we established a novel technique
(three-dimensional polyacrylamide gel-based DNA microarray) for the
first time, to the best of our knowledge, in order to identify
UGT1A1 gene mutations in 20 patients with hyperbilirubinemia
from the Chinese population.
Patients and methods
Study participants and DNA isolation
Twenty Chinese patients with hyperbilirubinemia were
recruited at the Second Hospital of Nanjing, Affiliated to the
Medical School of Southeast University, (Nanjing, China).
Peripheral blood samples were collected from all participants in
the morning following an overnight fast. Total DNA was extracted
using the QIAamp DNA Blood Midi kit (Qiagen, Hilden, Germany)
according to the standard protocol. All the participants provided
written informed consent prior to enrollment and all research
procedures were approved by the Ethics Committee of the Second
Hospital of Nanjing, Affiliated to the Medical School of Southeast
University.
PCR amplification
A pair of primers F1, 5′-CACCTCCTCCTTATTCTCTT-3′ and
R1, 5′-acrylamide-CTCATTCCTCCTCTCTAGCC-3′, whose design was based
on published DNA sequences (GenBank no. AF297093.1), was used for
PCR to obtain the PBREM region of the UGT1A1 gene. The
cycling conditions were as follows: 94°C for 3 min, 32 cycles of
94°C for 30 sec, 54.2°C for 45 sec, and 72°C for 45 sec, and then
72°C for 10 min. The region containing the TATA-box and the 211
site of the UGT1A1 gene was generated by PCR using Ex Taq
(Takara, Otsu, Japan) with two primers F2,
5′-CCCTGCTACCTTTGTGGACT-3′ and R2,
5′-acrylamide-CATTATGCCCGAGACTAACAAA-3′. The reaction conditions
were as follows: 94°C for 3 min followed by 32 cycles of 94°C for
30 sec, 57°C for 45 sec, and 72°C for 45 sec, and then a final
elongation step at 72°C for 10 min. Following agarose gel
electrophoresis, the acrylamide-modified PCR products were
processed by ethanol precipitation overnight at −20°C. The
acrylamide-modified PCR products were subsquently harvested by
centrifugation at 14,000 × g for 20 min and diluted in water.
Immobilization of acrylamide-modified PCR
products
Preparation of the acrylamide-modified slides is the
first step of PCR product immobilization. The protocol of
acryl-modified slides fabrication was performed as previously
described (15). Solutions
containing acrylamide-modified PCR products, acrylamide monomer
(29:1, acrylamide:bis-acrylamide), glycerol and ammonium persulfate
(APS) were then prepared at the desired concentrations and spotted
on the modified glass slide. After spotting, the slide was placed
into a humid sealed chamber in which a well containing TEMED had
been deposited in advance. The pressure in the sealed chamber was
reduced to approximately 1,000 Pascal (Pa), and this pressure was
maintained for 30 min at room temperature. Under this pressure,
TEMED was vaporized and diffused into the spots and onto the slide
surfaces to induce the co-polymerization of the acrylamide groups
and the acryl groups.
Hybridization with the corresponding
probes
Following the immobilization of the
acrylamide-modified PCR products, double-stranded DNA (dsDNA) on
the slide was denatured in 0.1 M sodium hydroxide solution for 10
min to obtain single-stranded DNA (ssDNA), and then subjected to
electrophoresis in 1X TBE buffer for 10 min to remove sodium
hydroxide. Finally, hybridization was performed in a humid glass
chamber with the corresponding probes (Table I) at 37°C for 2 h. A schematic
outline of the gel immobilization micro-array approach for
high-throughput genotyping is illustrated in Fig. 1.
 | Table IProbe sequences used in this
study. |
Table I
Probe sequences used in this
study.
| Probe | Probe sequences |
|---|
| −3279T |
5′-Cy3-TTCAGTTTGAACA-3′ |
| −3279G |
5′-Cy5-TTCAGTGTGAACA-3′ |
| A(TA)6TAA |
5′-Cy3-GCCATATATATATATATAAG-3′ |
| A(TA)7TAA |
5′-Cy5-GCCATATATATATATATATAAG-3′ |
| 211G |
5′-Cy3-AGAGACGGAGCAT-3′ |
| 211A |
5′-Cy5-AGAGACAGAGCAT-3′ |
Image scanning
Following hybridization, in order to remove the
non-specifically bound targets and mismatches, the slide was
subjected to electrophoresis under 38 V/cm for 25 min in 1X TBE
buffer at room temperature. The slide was then rinsed in water and
dried under a stream of nitrogen. Images of the hybridization slide
were scanned using a confocal scanner (LuxScan-10K/A; CapitalBio
Corp., Beijing, China) and analyzed with LuxScan 3.0 software.
Sequencing the PBREM region and the
region containing the TATA-box and the 211 site of the UGT1A1
gene
Two pairs of primers F1 and R3,
5′-CTCATTCCTCCTCTCTAGCC-3′, and F2 and R4,
5′-CATTATGCCCGAGACTAACAAA-3′, were used for PCR to obtain the PBREM
region and the region containing the TATA-box and the 211 site of
the UGT1A1 gene, respectively. The PCR products were
sequenced directly with the use of an BigDye Terminator v1.1 Cycle
Sequencing kit (Applied Biosystems, Foster City, CA, USA) with the
appropriate primers.
Results
Firstly, the acrylamide-modified PCR products of
different sizes were obtained and analyzed by electrophoresis on a
1% agarose gel (Fig. 2). In
principle, the homozygous wild-type yielded strongly fluorescent
Cy3 spots (green fluorescence), while the homozygous mutant yielded
strongly fluorescent Cy5 spots (red fluorescence). Moreover,
fluorescent Cy3 and Cy5 spots were shown for the heterozygote, and
after overlap-ping, a strong 'yellow' fluorescence was shown.
For the T-3279T homozygote, the probe-3279T labeled
with Cy3 was perfectly matched with the immobilized ssDNA, while
the probe-3279G labeled with Cy5 had a mismatched base in the
middle of the sequence to the ssDNA. Thus, only a Cy3 fluorescent
signal (green fluorescence) was obtained in the dual-color
fluorescence hybridization (Fig.
3A, line 2). The fluorescence scores of Cy3 and Cy5 in the
homozygote T-3279T were 11,328 and 682, respectively, and Cy3/Cy5
was 17 (second row in Fig. 3B).
In the same way, for the G-3279G homozygote, only the Cy5
fluorescent signal (red fluorescence) was shown (Fig. 3A, line 3). The fluorescence scores
of Cy5 and Cy3 in the homozygote G-3279G were 10,027 and 515,
respectively, and Cy5/Cy3 was 20 (Fig. 3B, third row). Moreover, for the
T-3279G heterozygote, both the Cy3 and Cy5 fluorescent signals
(green fluorescence and red fluorescence) were detected, and after
overlapping, a strong 'yellow' fluorescence was shown (Fig. 3A, line 4). The fluorescence scores
of Cy3 and Cy5 in the heterozygote T-3279G, were 9,106 and 8,544,
respectively, and Cy3/Cy5 was 1.07 (Fig. 3B, fourth row). In order to
evaluate the reliability of this technique, we compared the results
obtained by sequencing (Fig.
3C).
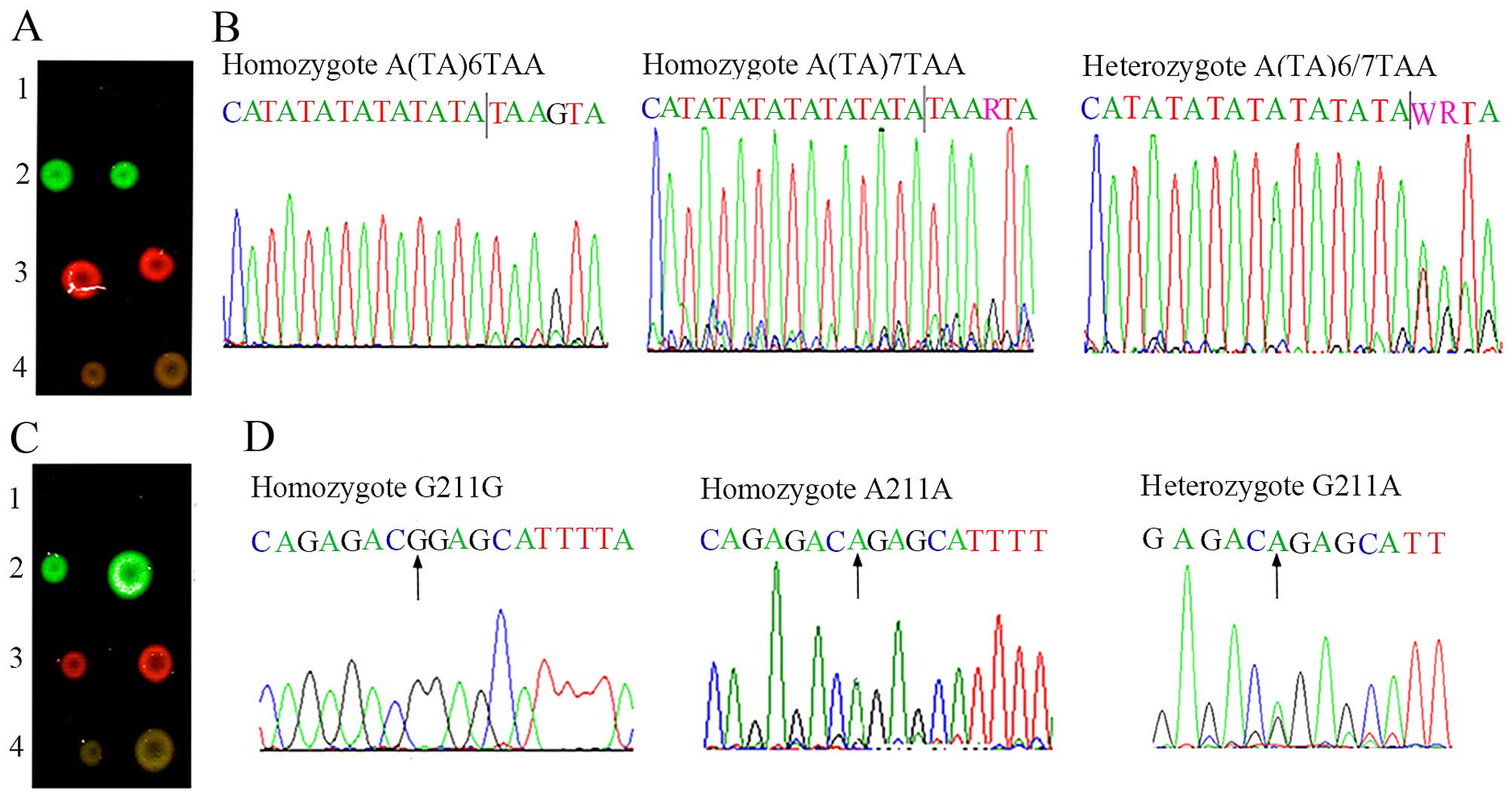
In the same way, for the A(TA)6TAA homozygote and
G211G homozygote, only the Cy3 fluorescent signal (green
fluorescence) was obtained in the dual-color fluorescence
hybridization (Fig. 4A and C,
line 2). For the A(TA)7TAA homozygote and A211A homozygote, only
the Cy5 fluorescent signal (red fluorescence) was shown (Fig. 4A and C, line 3). Moreover, for the
A(TA)6/7TAA heterozygote and G211A heterozygote, both the Cy3 and
Cy5 fluorescent signals (green fluorescence and red fluorescence)
were detected, and after overlapping, a strong 'yellow'
fluorescence was shown (Fig. 4A and
C, line 4). The above-mentioned results were further validated
by sequencing (Fig. 4B and
D).
Samples from 20 patients with hyperbilirubinemia
were analyzed for the presence of the T-3279G locus, the
TA-insertion locus (A(TA)6/7TAA) and the G211A locus. All possible
genotypes of the 20 samples from the patients enlisted were
successfully identified and are shown in Fig. 5. In addition, all results obtained
by three-dimensional polyacrylamide gel-based DNA microarray method
were further validated by sequencing.
Discussion
Gilbert's syndrome is a mild genetic liver disorder
characterized by unconjugated hyperbilirubinemia without overt
signs of hemolysis or structural liver disease (1). Its estimated prevalence is
approximately 3–7% in the general population (16). In general, Gilbert's syndrome is
considered a benign condition and does not require therapy since it
does not cause chronic liver dysfunction or fibrosis (17,18). However, this mild
hyperbilirubinemia may be mistaken for hepatic jaundice, hemolytic
jaundice or obstructive jaundice. Thus, patients may suffer from
unwarranted anxiety and unexpected toxicity from therapeutic
agents. For these reasons, it is important to make the correct
diagnosis in time.
Currently, the direct sequencing method, the TaqMan
MGB SNP genotyping assay, DNA melting curve analysis and the
restriction fragment length polymorphism (RFLP) method have been
used to detect mutations of the UGT1A1 gene and thereby
diagnose Gilbert's syndrome (19–21). In this study, we established a
novel method (three-dimensional polyacrylamide gel-based DNA
microarray) for the first time, to the best of our knowledge, in
order to detect UGT1A1 gene mutations in 20 patients with
hyperbilirubinemia from the Chinese population.
The three-dimensional polyacrylamide gel-based DNA
microarray method is a rapid, simple and low cost approach with
which to carry out gene mutation analysis. It has been widely used
in the genotyping of a number of genes, such as the oxidized
low-density lipoprotein receptor 1 (OLR-1) gene, the brain-derived
neurotrophic factor (BDNF) gene, and the gamma-aminobutyric acid
receptor beta 3 subunit (GABRB3) gene (14,22,23). Three-dimensional polyacrylamide
gel-based DNA microarray only requires a small quantity of
expensive fluorescent-labeled probes which can be used for
genotyping an unlimited number of samples. Furthermore, this method
is time-saving and increases efficiency by assaying thousands of
samples in one experiment.
Immobilization and electrophoresis are two critical
steps in the three-dimensional polyacrylamide gel-based DNA
microarray method. Firstly, immobilization relies on the
co-polymerization of acrylamide-modified PCR products with
acrylamide monomers and acryl-modified slides to prepare the
gel-based microarray. Thus, in the present study, reverse primers
(R1 and R2) were modified with an acrylamide group at the
5′-terminal in order to covalently attach to the polyacryl-amide
gel. TEMED is a volatile alkali and is easily vaporized at room
temperature. When the pressure in the sealed chamber was reduced to
approximately 1,000 Pa, TEMED was vaporized and diffused into the
spots and onto the slide surfaces to induce co-polymerization of
the acrylamide groups and the acryl groups. Subsequently, the array
was hybridized with specific fluorescent-labeled probes. The
removal of the non-specifically bound targets and mismatches is the
most important procedure. However, polyacrylamide gel has a porous
structure which intensively adsorbs the non-specifically labeled
probes during hybridization. Thus, the conventional washing steps
fail to remove the non-specifically adsorbed probes, resulting in
high background signals. As nucleic acids in PCR products carry the
negative charges, electrophoresis is an effective method with which
to effectively remove the non-specifically adsorbed probes. If the
voltage is too high or the duration of electrophoresis is too long,
specifical probes will be removed. Through repeated tests, the
slide was subjected to electrophoresis under 38 V/cm for 25 min in
1X TBE buffer at room temperature to remove the non-specifically
bound targets and mismatches. Finally, genotyping was based on the
images captured through two-color fluorescent scanning. The T-3279T
homozygote, the A(TA)6TAA homozygote and the G211G homozygote all
yielded strong fluorescent Cy3 spots (green fluorescence), while
the G-3279G homozygote, the A(TA)7TAA homozygote and the A211A
homozygote all yielded strong fluorescent Cy5 spots (red
fluorescence) (Figs. 3A and
4A and C). Moreover, both
fluorescing the Cy3 and Cy5 spots were shown for the T-3279G
heterozygote, the A(TA)6/7TAA heterozygote and the G211A
heterozygote, and after overlapping, a strong 'yellow' fluorescence
was shown.
In conclusion, in the present study, we successfully
detected the UGT1A1 gene mutations in 20 Chinese patients
with hyperbilirubinemia with the use of the three-dimensional
polyacrylamide gel-based DNA microarray method. This method holds
significant promise for future applications in the diagnosis of
Gilbert's syndrome.
Acknowledgments
The present study was supported by the Medical
Science and Technology Development Foundation, Nanjing Department
of Health (no. JQX14007) and by the National Natural Science
Foundation of China (no. 81301938).
References
|
1
|
Fretzayas A, Moustaki M, Liapi O and
Karpathios T: Gilbert syndrome. Eur J Pediatr. 171:11–15. 2012.
View Article : Google Scholar
|
|
2
|
Teich N, Lehmann I, Rosendahl J, Tröltzsch
M, Mössner J and Schiefke I: The inverse starving test is not a
suitable provocation test for Gilbert's syndrome. BMC Res Notes.
1:352008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bosma PJ, Seppen J, Goldhoorn B, Bakker C,
Oude Elferink RP, Chowdhury JR, Chowdhury NR and Jansen PL:
Bilirubin UDP-glucuronosyltransferase 1 is the only relevant
bilirubin glucuronidating isoform in man. J Biol Chem.
269:17960–17964. 1994.PubMed/NCBI
|
|
4
|
Bosma PJ, Chowdhury JR, Bakker C, Gantla
S, de Boer A, Oostra BA, Lindhout D, Tytgat GN, Jansen PL, Oude
Elferink RP, et al: The genetic basis of the reduced expression of
bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. N
Engl J Med. 333:1171–1175. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Raijmakers MT, Jansen PL, Steegers EA and
Peters WH: Association of human liver bilirubin
UDP-glucuronyltransferase activity with a polymorphism in the
promoter region of the UGT1A1 gene. J Hepatol. 33:348–351. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sato H, Adachi Y and Koiwai O: The genetic
basis of Gilbert's syndrome. Lancet. 34:557–558. 1996. View Article : Google Scholar
|
|
7
|
Sugatani J, Yamakawa K, Yoshinari K,
Machida T, Takagi H, Mori M, Kakizaki S, Sueyoshi T, Negishi M and
Miwa M: Identification of a defect in the UGT1A1 gene promoter and
its association with hyperbilirubinemia. Biochem Biophys Res
Commun. 292:492–497. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Maruo Y, Sato H, Yamano T, Doida Y and
Shimada M: Gilbert syndrome caused by a homozygous missense
mutation (Tyr486Asp) of bilirubin UDP-glucuronosyltransferase gene.
J Pediatr. 132:1045–1047. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Maruo Y, Nishizawa K, Sato H, Doida Y and
Shimada M: Association of neonatal hyperbilirubinemia with
bilirubin UDP-glucuronosyltransferase polymorphism. Pediatrics.
103:1224–1227. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Canu G, Minucci A, Zuppi C and Capoluongo
E: Gilbert and Crigler Najjar syndromes: An update of the
UDP-glucuronosyltransferase1A1 (UGT1A1) gene mutation database.
Blood Cells Mol Dis. 50:273–280. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Maruo Y, D'Addario C, Mori A, Iwai M,
Takahashi H, Sato H and Takeuchi Y: Two linked polymorphic
mutations (A(TA)7TAA and T-3279G) of UGT1A1 as the principal cause
of Gilbert syndrome. Hum Genet. 115:525–526. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Matsui K, Maruo Y, Sato H and Takeuchi Y:
Combined effect of regulatory polymorphisms on transcription of
UGT1A1 as a cause of Gilbert syndrome. BMC Gastroenterol.
10:572010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kalotychou V, Karakosta M, Tzanetea R,
Stamoulakatou A, Konstantopoulos K and Rombos Y: Contribution of
G71R mutation to Gilbert's syndrome phenotype in a Greek patient: a
case report. World J Gastrointest Pharmacol Ther. 2:42–45. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xiao PF, Cheng L, Wan Y, Sun BL, Chen ZZ,
Zhang SY, Zhang CZ, Zhou GH and Lu ZH: An improved gel-based DNA
microarray method for detecting single nucleotide mismatch.
Electrophoresis. 27:3904–3915. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rehman FN, Audeh M, Abrams ES, Hammond PW,
Kenney M and Boles TC: Immobilization of acrylamide-modified
oligonu-cleotides by co-polymerization. Nucleic Acids Res.
27:649–655. 1999. View Article : Google Scholar
|
|
16
|
Kim YH, Yeon JE, Jung GM, Kim HJ, Kim JS,
Byun KS, Bak YT and Lee CH: A study of polymorphism in
UDP-glucuronosyltransferase 1 (UGT-1A1) promoter gene in Korean
patients with Gilbert's syndrome. Taehan Kan Hakhoe Chi. 8:132–138.
2002.In Korean. PubMed/NCBI
|
|
17
|
Tukey RH and Strassburg CP: Human
UDP-glucurono-syltransferases: Metabolism, expression, and disease.
Annu Rev Pharmacol Toxicol. 40:581–616. 2000. View Article : Google Scholar
|
|
18
|
Minucci A, Concolino P, Giardina B, Zuppi
C and Capoluongo E: Rapid UGT1A1 (TA)(n) genotyping by high
resolution melting curve analysis for Gilbert's syndrome diagnosis.
Clin Chim Acta. 411:246–249. 2010. View Article : Google Scholar
|
|
19
|
Wong FL, Wang MK, Boo NY, Hamidah NH and
Ainoon BO: Rapid detection of the UGT1A1 single nucleotide
polymorphism G211A using real-time PCR with Taqman minor groove
binder probes. J Clin Lab Anal. 21:167–172. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hsieh TY, Shiu TY, Chu NF, Chao TY, Chu
HC, Chang WK, Chao YC and Huang HH: Rapid molecular diagnosis of
the Gilbert's syndrome-associated exon 1 mutation within the UGT1A1
gene. Genet Mol Res. 13:670–679. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shiu TY, Huang HH, Lin HH, Shih YL, Chu
HC, Chang WK and Hsieh TY: Restriction fragment length polymorphism
effectively identifies exon 1 mutation of UGT1A1 gene in patients
with Gilbert's syndrome. Liver Int. 35:2050–2056. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng L, Ge Q, Xiao P, Sun B, Ke X, Bai Y
and Lu Z: Association study between BDNF gene polymorphisms and
autism by three-dimensional gel-based microarray. Int J Mol Sci.
10:2487–2500. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tang J and Xiao P: Polymerizing
immobilization of acrylamide-modified nucleic acids and its
application. Biosens Bioelectron. 24:1817–1824. 2009. View Article : Google Scholar
|