Introduction
Histone modifications, including acetylation,
methylation and phosphorylation have been demonstrated to play
critical roles in gene-specific transcriptional regulation, DNA
replication, DNA damage response and repair (1,2).
Acetylation, as the earliest identified histone modification,
involves the transfer of an acetyl group from acetyl-CoA to the
e-amino group of lysine residue by histone acetyltransferases
(HATs) (3).
Histone acetyltransferase binding to origin
recognition complex (ORC) 1 (HBO1), belonging to the MYST family,
was discovered in the context of DNA replication (3). HBO1, as an H4-specific histone
acetylase, can interact with transcription factors (4), mRNA coding regions (5) and ORC (6). Researches have demonstrated that
HBO1 is necessary for licensing and DNA replication (7) and that it is associated with
replication origins specifically during the G1 phase of the cell
cycle (8). Considering the
critical role of HBO1 in DNA replication, it has been demonstrated
that HBO1 mutations can lead to profound negative consequences, and
even to oncogenesis (3). On the
one hand, the HBO1 coding region was identified to be a common
retroviral integration site (9).
As infections with some viruses are considered to lead to
oncogenesis, HBO1 may indirectly contribute to the induction of
cancer (10). On the other hand,
the binding partners of HBO1 are considered to be related to
diseases. The interaction of HBO1 with androgen receptor was
putatively linked to prostate cancer development (11) and the interaction of HBO1 with
nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB)
was connected with tumor suppression (12). Hence, HBO1 is critical in the
development of tumors, although the mechanisms involved remain
unclear.
In the present study, the possible binding sites of
HBO1 on all chromosomes were explored based on chromatin
immunoprecipitation sequencing (ChIP-seq) analysis. To explore the
regulatory mechanisms of HBO1 during tumor development, the genes
that can be regulated putatively by HBO1 were subjected to
functional enrichment analysis. Furthermore, the structure and
inhibitors of HBO1 were systematically explored, which may prove to
be helpful for the discovery of novel anticancer drugs.
Data collection methods
ChIP-seq data of HBO1 in the RKO human
colon cancer cell line
The ChIP-seq data [GSE33007 (13)] were downloaded from the Gene
Expression Omnibus (GEO) (14)
database (http://www.ncbi.nlm.nih.gov/geo/). The data included
two samples: one was obtained in the RKO colon cancer cell line and
the other one was obtained in a normal cell line. The samples were
sequenced on the Illumina Genome Analyzer IIx platform.
Analysis of ChIP-seq data
The reads were mapped back to a reference genome
hg19 from the University of California, Santa Cruz (UCSC) Genome
Browser (15) by using Bowtie
software (16) (version 0.12.9).
The reads were screened out for subsequent analysis with the
criteria of unique alignment position and a mismatch number <2.
The PCR duplicate reads were then removed using SAMtools (17) to keep at most one read per genomic
position. The shifts of 5′ and 3′ are defined as a half of the
average size of the ChIP-seq fragments for the experimental and
control groups using SPP and MaSC software. The peak calling was
carried out using MACS 1.4.0 software (18). The reads with a P-value
>0.00001 were considered as the HBO1 binding sites.
Gene functional analysis
The target genes of HBO1 were identified using the
ChIPpeakAnno (19) package within
R. The functions of those genes were subjected to functional
enrichment analysis based on the Gene Ontology (GO) database
(http://www.geneontology.org/) using the
getEnrichedGO module within the ChIPpeakAnno package (19). The criteria were set to a P-value
<2e-14.
HBO1 structure analysis and inhibitor
screening
The 3D structure of HBO1 (PDB ID: 2pq8) was
downloaded from Protein Data Bank (PDB; http://www.rcsb.org/pdb/home/home.do) and the coenzyme
binding site analysis and structure visualization were conducted
using PyMOL software (20). The
virtual screening of inhibitors was carried out using DOCK 6.6
software (21) following
structure preprocessing with Chimera software (22). The force field was set to
Amber99sb and the binding sites were set to be within 6 Å around
the coenzyme. The Specs compound database was used for the
screening and the top 5 compounds with best binding affinity were
screened out and analyzed.
Results
HBO1 binding sites
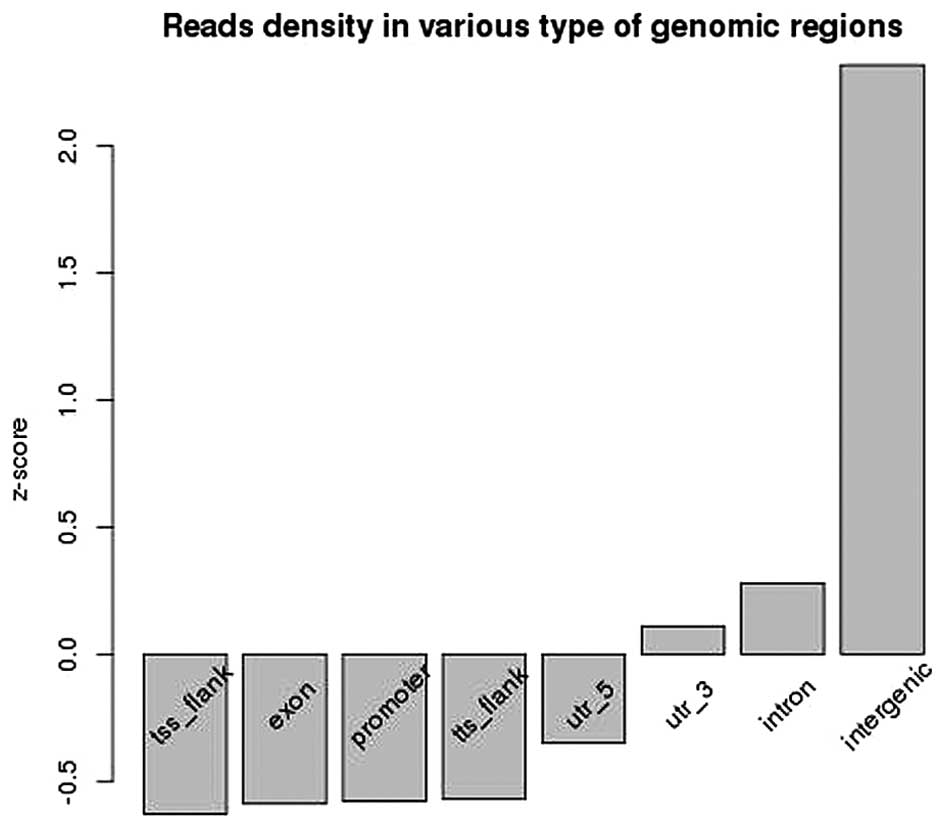
Based on the ChIP-seq analysis, the enriched
positions of HBO1 binding sites were mainly distributed in the
intergenic, intron and 3′-end regions (Fig. 1).
Identification and functional analysis
HBO1 target genes
To explore the regulatory mechanism of HOB1, the
HBO1 target genes were identified and subjected to functional
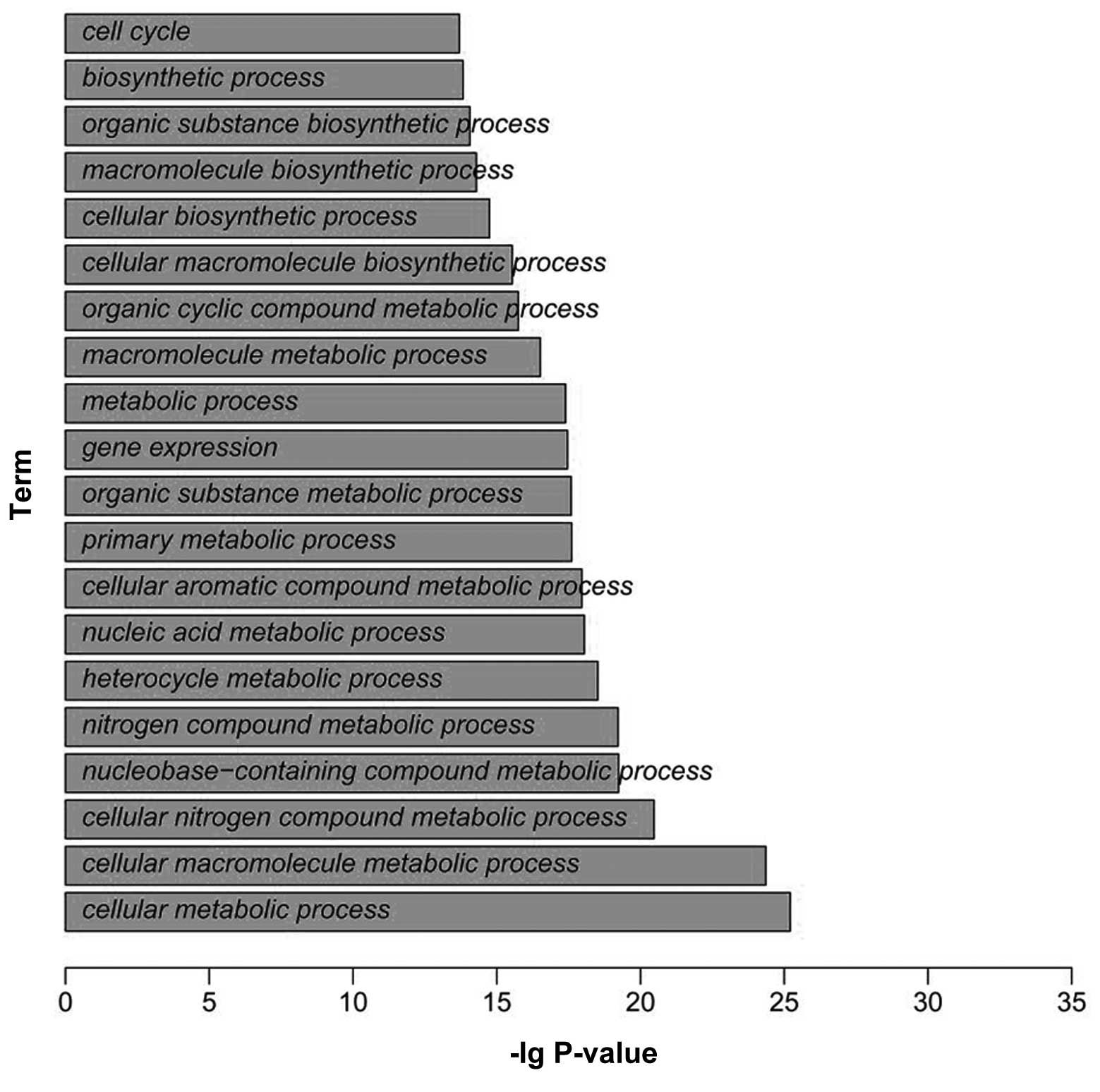
enrichment analysis. A total of 9,467 target genes was identified
around HBO1 binding sites in the RKO cell line. Functional
enrichment analysis revealed that the genes were mainly enriched in
the functions of cell cycle, biosynthetic process, organic
substance biosynthetic process and macromolecule biosynthetic
process (Fig. 2).
HBO1 structure analysis
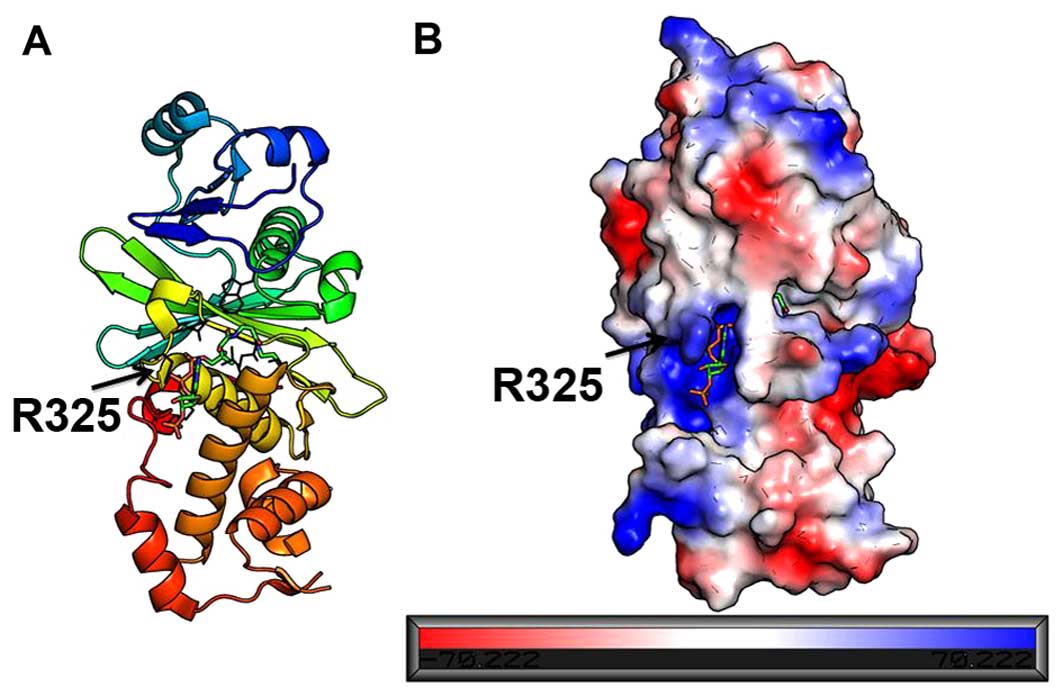
HBO1 combined with an acetyl-CoA belongs to the HATs
family. The surface of HBO1 was covered by charged residues and the
acetyl-CoA was located at a cavity which was full of positively
charged residues (Fig. 3). The
entrance of the cavity was guarded by a positively charged side
chain of R325, promoting the binding of negatively charged
compounds.
HBO1 inhibitor screening
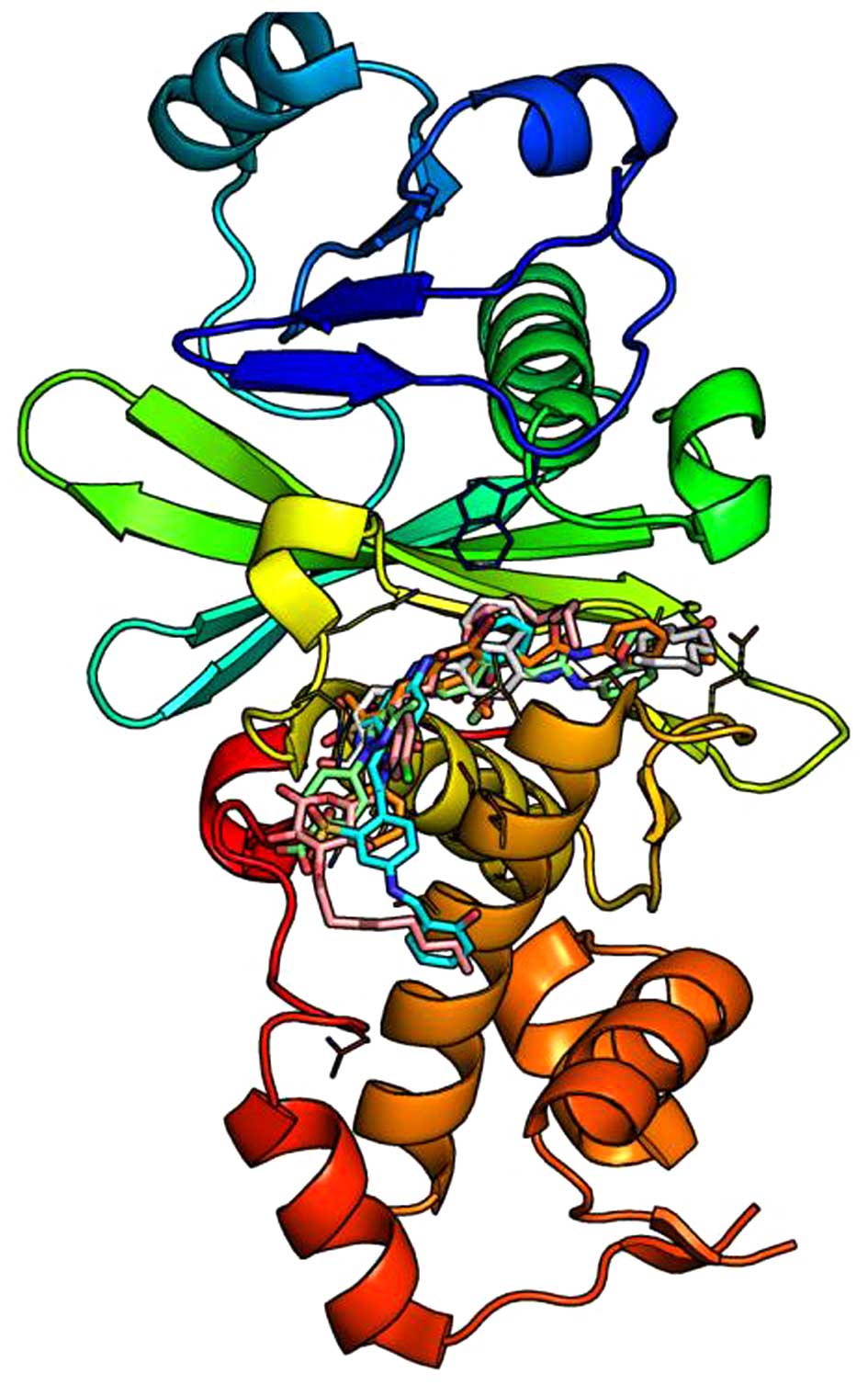
Based on the HBO1 3D structure and acetyl-CoA
binding site, the inhibitors of HBO1 were screened out from the
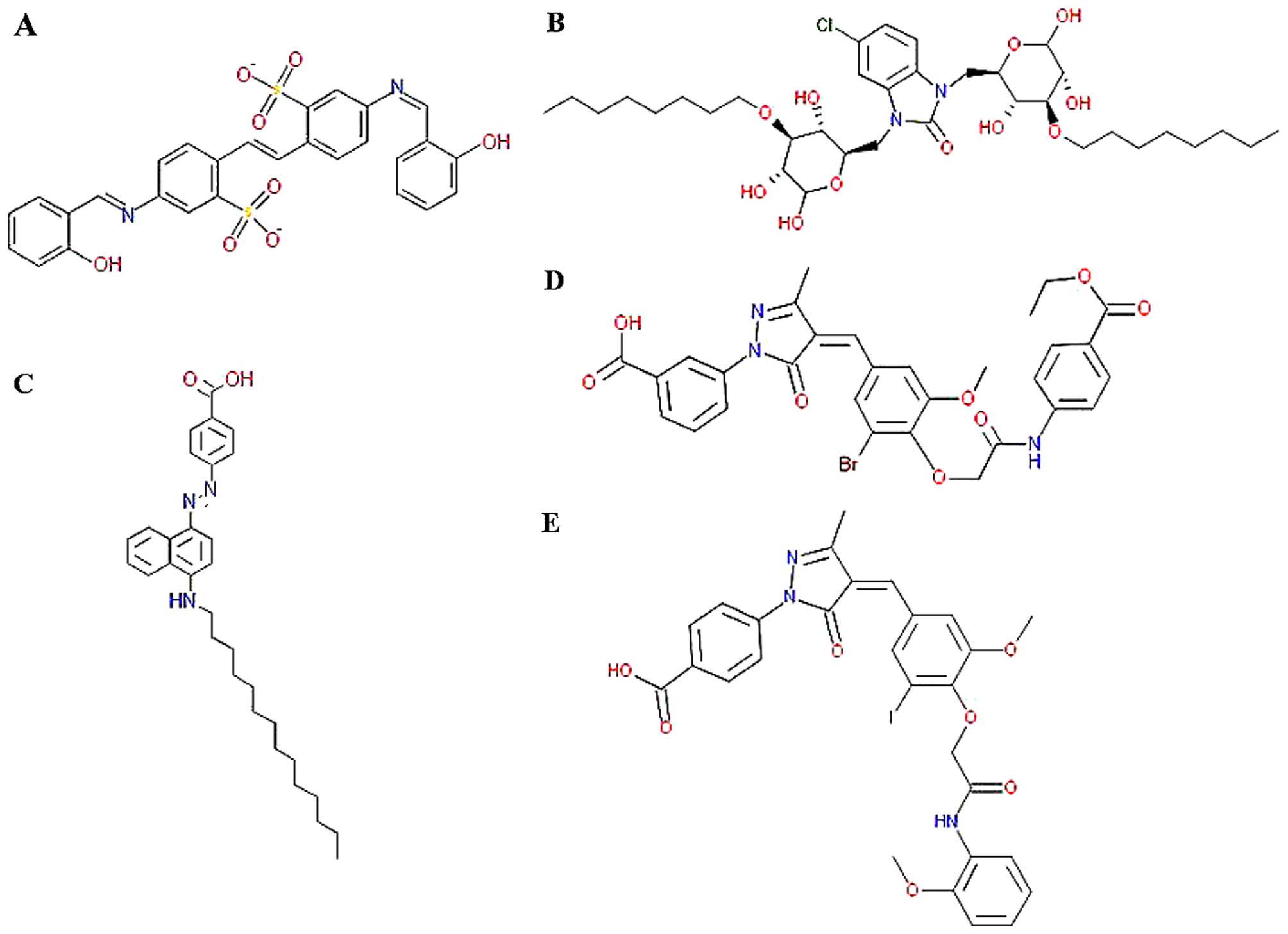
Specs database. As shown in Fig.
4, a total of 5 compounds with best binding affinity in the
cavity were screened out: i)
5-[(2-hydroxybenzylidene)amino]-2-(2-{4-[(2-hydroxybenzylidene)amino]-2-sulfonatophenyl}vinyl)benzenesulfonate,
ii)
3-[4-(3-bromo-4-{2-[4-(ethoxycarbonyl)anilino]-2-oxoethoxy}-5-methoxybenzylidene)-3-methyl-5-oxo-4,5-dihydro-1H-pyrazol-1-yl]benzoic
acid, iii)
4-(4-{3-iodo-5-methoxy-4-[2-(2-methoxyanilino)-2-oxoethoxy]benzylidene}-3-methyl-5-oxo-4,5-dihydro-1H-pyrazol-1-yl)benzoic
acid, iv)
5-chloro-1,3-bis{[3,5,6-trihy-droxy-4-(octyloxy)tetrahydro-2H-pyran-2-yl]methyl}-1,3-dihydro-2H-benzimidazol-2-one
and v) 4-{[4-(tetradecylamino)-1-naphthyl]diazenyl}benzoic acid.
The structures of the 5 compounds are depicted in Fig. 5 and the corresponding Specs IDs
are AO-861/15351016, AO-763/14815006, AA-516/33241036,
AK-968/13031190 and AK-968/13031237, respectively.
Discussion
HBO1 has been demonstrated play critical roles in
genome expression and maintenance (3). In this study, we explored the
possible binding sites of HBO1 on all chromosomes. By focusing our
efforts on the functions of HBO1, the genes that can be regulated
by HBO1 were identified and subjected to functional analysis.
Furthermore, the coenzyme binding site and surface charge
distribution of HBO1 were explored and the potential inhibitors
were screened out.
Based on the results of ChIP-seq analysis, HBO1
mainly binds upstream of gene transcription start sites and
putatively enhances gene expression. Experiments have indicated
that a strong enrichment of HOB1 near the homeobox A9 (HOXA9) and
homeobox A10 (HOXA10) genes and the expression of HOXA10 is found
to be downregulated upon the knockdown of HBO1 (13). ChIP analysis has also revealed
that HBO1 is highly enriched throughout the coding regions of a
large number of human genes, from the middle of the genes to the
transcription end sites or centered around the transcription start
sites (5). The critical role of
HBO1 in gene transcriptional regulation implies that HBO1 probably
participates in the regulation of tumor development.
The target genes of HBO1 were significantly related
to the cell cycle process, based on functionalo enrichment
analysis. The association of HBO1 with various inhibitor of growth
(ING) tumor suppressor proteins suggests that it plays critical
roles in gene regulation and DNA replication through histone H3 and
H4 acetylation (13). Genomic
analysis has indicated that the HBO1 and ING complexes are mainly
involved in the regulation of the p53 pathways (13). Histone acetylase defective mutant
experiments have also demonstrated that HBO1 acetylase activity is
necessary for replication licensing. HOB1 can regulate the
expression of cell division cycle 27 (CDC27), cyclin-dependent
kinase inhibitor 2D (CDKN2D) and cell division cycle 20 (CDC20),
which can participate in cell cycle progression. The protein
encoded by CDC27 is a component of anaphase-promoting complex (APC)
(23). Experiments have shown
that the interaction between this protein and mitotic checkpoint
proteins, such as mitotic arrest deficient 2 (Mad2), p55
cell-division cycle protein 20 (p55CDC) and BUB1 mitotic checkpoint
serine/threonine kinase B (BUBR1) are involved in controlling the
timing of mitosis (24). The
protein encoded by CDKN2D can suppress the activation of the CDK
kinases and regulate cell cycle G1 progression by forming a stable
complex with CDK4 or CDK6 (25).
Similarly, CDC20 was demonstrated to be a regulatory protein at
multiple cell cycle points by interacting with several other
proteins (26). CDC20 can bind to
and promote the ubiquitin ligase activity of anaphase-promoting
complex/cyclosome (APC/C) and enable the degradation of securing
and cyclin B, thus promoting the onset of anaphase and mitotic exit
(27). All these data demonstrate
the importance of HBO1 in controlling cell proliferation and in the
regulation of cell cycle.
Structure analysis indicated that the acetyl-CoA
combined with HBO1 in the cavity was surrounded with positively
charged residues. The acetyl-CoA binds in a bent conformation which
facilitates an extensive set of protein interactions. In addition,
the positively charged side chain of R325 in the cavity entrance
promotes the binding of negatively charged substrates. The
potential inhibitors can combine with HBO1 perfectly in the cavity.
The inhibitors possess various polar groups, such as carbonyl
oxygen, hydroxyl oxygen and heterocyclic nitrogen, which can
mediate interactions between HBO1 and inhibitors by protein
backbone hydrogen bonds or protein side chain van der Waals
interactions. Hence, the inhibition of HBO1 can interrupt the cell
cycle or cell proliferation and putatively suppress tumor
development.
In conclusion, in this study, the location and
target gene function alanalysis of HBO1 indicated that HBO1 plays
critical roles in the regulation of gene expression and the cell
cycle. Furthermore, structure analysis and inhibitor identification
provided the HBO1 regulatory mechanisms, which may prove to be
helpful for the inhibition of tumor development.
References
|
1
|
Karlić R, Chung H-R, Lasserre J,
Vlahoviček K and Vingron M: Histone modification levels are
predictive for gene expression. Proc Natl Acad Sci USA.
107:2926–2931. 2010. View Article : Google Scholar
|
|
2
|
Henikoff S and Shilatifard A: Histone
modification: Cause or cog? Trends Genet. 27:389–396. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Avvakumov N and Côté J: The MYST family of
histone acetyltransferases and their intimate links to cancer.
Oncogene. 26:5395–5407. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Georgiakaki M, Chabbert-Buffet N, Dasen B,
Meduri G, Wenk S, Rajhi L, Amazit L, Chauchereau A, Burger CW, Blok
LJ, et al: Ligand-controlled interaction of histone
acetyltransferase binding to ORC-1 (HBO1) with the N-terminal
transactivating domain of progesterone receptor induces steroid
receptor coactivator 1-dependent coactivation of transcription. Mol
Endocrinol. 20:2122–2140. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Saksouk N, Avvakumov N, Champagne KS, Hung
T, Doyon Y, Cayrou C, Paquet E, Ullah M, Landry AJ, Côté V, et al:
HBO1 HAT complexes target chromatin throughout gene coding regions
via multiple PHD finger interactions with histone H3 tail. Mol
Cell. 33:257–265. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Burke TW, Cook JG, Asano M and Nevins JR:
Replication factors MCM2 and ORC1 interact with the histone
acetyltransferase HBO1. J Biol Chem. 276:15397–15408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Doyon Y, Cayrou C, Ullah M, Landry AJ,
Côté V, Selleck W, Lane WS, Tan S, Yang XJ and Côté J: ING tumor
suppressor proteins are critical regulators of chromatin
acetylation required for genome expression and perpetuation. Mol
Cell. 21:51–64. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Miotto B and Struhl K: HBO1 histone
acetylase is a coactivator of the replication licensing factor
Cdt1. Genes Dev. 22:2633–2638. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Suzuki T, Shen H, Akagi K, Morse HC,
Malley JD, Naiman DQ, Jenkins NA and Copeland NG: New genes
involved in cancer identified by retroviral tagging. Nat Genet.
32:166–174. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Iizuka M, Takahashi Y, Mizzen CA, Cook RG,
Fujita M, Allis CD, Frierson HF Jr, Fukusato T and Smith MM:
Histone acetyltransferase Hbo1: Catalytic activity, cellular
abundance, and links to primary cancers. Gene. 436:108–114. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sharma M, Zarnegar M, Li X, Lim B and Sun
Z: Androgen receptor interacts with a novel MYST protein, HBO1. J
Biol Chem. 275:35200–35208. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Contzler R, Regamey A, Favre B, Roger T,
Hohl D and Huber M: Histone acetyltransferase HBO1 inhibits
NF-kappaB activity by coactivator sequestration. Biochem Biophys
Res Commun. 350:208–213. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Avvakumov N, Lalonde M-E, Saksouk N,
Paquet E, Glass KC, Landry AJ, Doyon Y, Cayrou C, Robitaille GA,
Richard DE, et al: Conserved molecular interactions within the HBO1
acetyltransferase complexes regulate cell proliferation. Mol Cell
Biol. 32:689–703. 2012. View Article : Google Scholar :
|
|
14
|
Barrett T, Troup DB, Wilhite SE, Ledoux P,
Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M,
Marshall KA, et al: NCBI GEO: Archive for high-throughput
functional genomic data. Nucleic Acids Res. 37(Database):
D885–D890. 2009. View Article : Google Scholar :
|
|
15
|
Fujita PA, Rhead B, Zweig AS, Hinrichs AS,
Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A,
et al: The UCSC Genome Browser database: Update 2011. Nucleic Acids
Res. 39(Database): D876–D882. 2011. View Article : Google Scholar :
|
|
16
|
Langmead B, Trapnell C, Pop M and Salzberg
SL: Ultrafast and memory-efficient alignment of short DNA sequences
to the human genome. Genome Biol. 10:R252009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li H, Handsaker B, Wysoker A, Fennell T,
Ruan J, Homer N, Marth G, Abecasis G and Durbin R: 1000 Genome
Project Data Processing Subgroup: The sequence alignment/map format
and SAMtools. Bioinformatics. 25:2078–2079. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Y, Liu T, Meyer CA, Eeckhoute J,
Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W and
Liu XS: Model-based analysis of ChIP-Seq (MACS). Genome Biol.
9:R1372008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin
SM, Lapointe DS and Green MR: ChIPpeakAnno: a bioconductor package
to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics.
11:2372010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
DeLano WL: The PyMOL Molecular Graphics
System DeLano Scientific. San Carlos, CA, USA: 2002, http://www.pymol.org.
|
|
21
|
Lang PT, Brozell SR, Mukherjee S,
Pettersen EF, Meng EC, Thomas V, Rizzo RC, Case DA, James TL and
Kuntz ID: DOCK 6: combining techniques to model RNA-small molecule
complexes. RNA. 15:1219–1230. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pettersen EF, Goddard TD, Huang CC, Couch
GS, Greenblatt DM, Meng EC and Ferrin TE: UCSF Chimera - a
visualization system for exploratory research and analysis. J
Comput Chem. 25:1605–1612. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Topper LM, Campbell MS, Tugendreich S,
Daum JR, Burke DJ, Hieter P and Gorbsky GJ: The dephosphorylated
form of the anaphase-promoting complex protein Cdc27/Apc3
concentrates on kinetochores and chromosome arms in mitosis. Cell
Cycle. 1:282–292. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Singh N, Wiltshire TD, Thompson JR, Mer G
and Couch FJ: Molecular basis for the association of microcephalin
(MCPH1) protein with the cell division cycle protein 27 (Cdc27)
subunit of the anaphase-promoting complex. J Biol Chem.
287:2854–2862. 2012. View Article : Google Scholar :
|
|
25
|
Carcagno AL, Marazita MC, Ogara MF, Ceruti
JM, Sonzogni SV, Scassa ME, Giono LE and Cánepa ET: E2F1-mediated
upregulation of p19INK4d determines its periodic expression during
cell cycle and regulates cellular proliferation. PLoS One.
6:e219382011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hadjihannas MV, Bernkopf DB, Brückner M
and Behrens J: Cell cycle control of Wnt/β-catenin signalling by
conductin/axin2 through CDC20. EMBO Rep. 13:347–354. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu H: Cdc20: A WD40 activator for a cell
cycle degradation machine. Mol Cell. 27:3–16. 2007. View Article : Google Scholar : PubMed/NCBI
|