Introduction
Cholangiocarcinoma is a rare, gastrointestinal
cancer which responds poorly to surgery and chemoradiotherapy and
therefore has a poor long-term outcome. Early diagnosis is
difficult due to the anatomical and biological characteristics of
cholangiocarcinoma. Thus, the identification of molecular
mechanisms underlying the carcinogenesis in cholangiocarcinoma is
critical for the development of novel potent drugs and
therapies.
Cyclin-dependent kinase inhibitor 1B
(p27Kip1) is under-expressed and exhibits abnormal
subcellular localization in the cytoplasm of some types of
malignant tumors (1,2). As mutations of p27Kip1
are rare, this has been attributed to the post-transcriptional
regulation of nucleocytoplasmic transport as well as the
nuclear-cytoplasmic distribution of p27Kip1 (3). p27Kip1 is transported
through nuclear pore complexes within the nuclear membrane and is
transferred either into or out of the nuclei by specific carriers,
mediated by nuclear localization signals (4). The aberrant expression and
cytoplasmic detention of p27Kip1 involves an increase in
proteolysis, which correlates with the nuclear export and import of
p27Kip1, and leads to its abnormal localization outside
the nuclei. However, the mechanisms responsible for the
nucleocytoplasmic transport of p27Kip1 remain poorly
understood.
The nuclear export factor, chromosome region
maintenance 1 (CRM-1; also referred to as exportin 1 or Xpo1) plays
a key role in the nuclear export of proteins that have a nuclear
export signal sequence in eukaryotic cells. Previous research has
shown that CRM-1 closely correlates with the nuclear export and
import of the p27Kip1-encoded protein, which in turn is
closely associated with the phosphorylation of Ser10, to produce
phosphorylated (p-)p27Kip1 (Ser10) (5). The nuclear export factor Jab1
facilitates the CRM-1-mediated export of p27Kip1 by
specifically recognizing and binding to p-p27Kip1
(Ser10) (3). However, studies
regarding the association between CRM-1, p27Kip1 and
p-p27Kip (Ser10) in human cholangiocarcinoma are
limited.
In the present study, we examined the expression
patterns of CRM-1 and p27Kip1 proteins, and the
phosphorylation of p27Kip1 (Ser10) in cholangiocarcinoma
tissues compared with that in chronic cholangitis tissues. We aimed
to examine the roles of CRM-1 in the nucleocytoplasmic transport of
p27Kip1 and in the development and progression of human
cholangiocarcinoma.
Materials and methods
Ethics statement
The present study was approved by the Ethics
Committee of Tongji Hospital [Tongji Medical College, Huazhong
University of Science and Technology (HUST), Wuhan, China]. All the
mice in our experiment were housed in the SPF Animal Center at
Tongji Medical College (HUST, Wuhan, China). All mice were
underwent general anesthesia which was induced with isoflurane
inhalation prior to sacrifice (mice were euthanized in a box filled
with CO2 followed by cervical dislocation) in order to
minimize their suffering. Written consent was obtained from all
patients before enrollment. The present study complied with the
principles that govern the use of human tissues as outlined in the
Declaration of Helsinki.
Histological assessment
The extrahepatic cholangiocarcinoma tissue samples
were classified into perihilar (or proximal) and middle or distal
subgroups, according to their anatomic location along the biliary
tree (6,7). Clinical staging was based on the
tumor-node-metastasis (TNM) classification system defined by the
American Joint Committee on Cancer (AJCC) and the Union for
International Cancer Control (UICC). The TNM classification in
extrahepatic cholangiocarcinoma mainly includes perihilar bile duct
carcinoma and distal bile duct carcinoma. The TNM staging and
grading of perihilar bile duct carcinoma has been summarized by
Ganeshan et al (6).
Study population and specimen
collection
The tumor tissue specimens were collected from 53
patients with extrahepatic cholangiocarcinoma and 10 patients with
chronic proliferative cholangitis as a control group. The patients
all underwent surgery at the Department of General Surgery at
Tongji Hospital (Tongji Medical College, HUST, Wuhan, China)
between 2008 and 2010. None of the patients had undergone
preoperative treatment, such as radiotherapy or chemotherapy. The
clinicopathological details of the patients, including gender, age,
tumor diameter, histological subtype, tumor location, tumor grade
and clinical TNM stage were accessed from the hospital database.
The mean and median ages of the cholangiocarcinoma patients were
51.5 and 45.0 years, respectively (range, 33–75 years) and the
male:female ratio was 1.3:1 (30 males; 23 females). The locations
of the lesions in the bile duct were as follows: 16/53 (30.2%)
proximal; 14/53 (26.4%) middle; and 23/53 (43.4%) distal. These
included 24/53 (45.3%) well-differentiated, 12/53 (22.6%)
moderately-differentiated and 17/53 (32.1%) poorly-differentiated
adenocarcinomas. There were 22/53 (41.5%) patients in stages I–II,
and 31/53 (58.5%) patients in stages III–IV.
Antibodies, plasmids and reagents
p27Kip1 (ZA-0557) mouse anti-human
monoclonal antibody, horseradish peroxidase (HRP)-labeled
goat-anti-mouse IgG (ZB-2301), streptavidin-peroxidase (SP)
immunostaining kit and 3,3′-diaminobenzidine (DAB) were purchased
from Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.
(Beijing, China); rabbit anti-CRM-1 (H-300; sc-5595),
p-p27Kip1 (Ser10; sc-12939-R), proliferating cell
nuclear antigen (PCNA; sc-7907) and GAPDH (FL-335) were purchased
from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The
electrochemiluminescence (ECL) reagents were purchased from Pierce
Chemical Co. (Rockford, IL, USA). We constructed two specific short
hairpin RNAs (shRNAs) using a pSicoR vector bought from Addgene
(Cambridge, MA, USA) targeting CRM-1 as previously describd by Wang
et al (8) and the
corresponding shRNA sequences were as follows: shRNA1,
GAAGTACTGACACATTTAA; and shRNA2, GGCTGCTGAACTCTATAGA. The data
shown in Figs. 4,5 and 6
are from shRNA1 (shCRM-1) and the results from shRNA2 targeting
CRM-1 were generally the same as those from shRNA1 (data not
shown).
Immunostaining
The tissue specimens were fixed in 10% formalin and
embedded in paraffin prior to sectioning into 3 µm slices
and stained with hematoxylin and eosin (H&E). The sections were
pathologically diagnosed by two independent, double-blind
pathologists (Z. Hongbo and D. Hao). The tissue sections were
deparaffinized, washed in phosphate-buffered saline (PBS) and
prepared for immunohistochemistry according to the SP protocol.
Briefly, antigen retrieval was performed using a microwave, and
endogenous peroxidase activity was blocked with 0.3%
H2O2 and non-specific reactions were blocked
with normal goat serum. The samples were incubated with the primary
antibody overnight at 4°C, followed by incubation with biotinylated
secondary antibody and HRP-conjugated streptavidin. For the control
group, the primary antibody was replaced with PBS; for the negative
control group, the primary antibody was replaced with normal goat
serum. Immunohistochemical (IHC) staining of the sections was
followed by DAB staining and counterstaining with hematoxylin. The
slides were dehydrated, and mounted.
A minimum of 1×103 cells were counted
from 10 randomly selected fields (magnification, x400) for each
section under a light microscope (1X71 inverted microscope; Olympus
Corp., Tokyo, Japan). The proteins appearing brown or tan in the
nuclei, the cytoplasm or both were defined as positive. The mean
percentage of positive cells was calculated for each patient. The
tissue samples were defined as having nuclear or cytoplasmic
expression of a protein if >25% of the cells showed positive
staining in the nuclei or cytoplasm, respectively.
Isolation of cytoplasmic and nuclear
proteins
The cytoplasmic and nuclear proteins were separated
using the ProteoJET Cytoplasmic and Nuclear Protein Extraction kit
(Fermentas International, Inc., Burlington, ON, Canada) according
to the manufacturer's instructions. Briefly, the fresh tissue
samples were rinsed with ice-cold PBS and blotted dry. The tissues
were then gently homogenized in PBS with protease inhibitors. The
homogenate was centrifuged in a microcentrifuge at 250 × g for 5
min at 4°C. The supernatant was discarded and 500 µl of cell
lysis buffer containing protease inhibitors and dithiothreitol
(DTT) was added to 100 mg tissue, mixed gently by vortexing, and
set on ice for 10 min. The cytoplasmic fraction was separated from
the nuclei by centrifugation at 500 × g for 7 min at 4°C. The
nuclear pellet was set on ice and the supernatant was centrifuged
at 20,000 × g for 15 min at 4°C to separate the cytoplasmic protein
extract. This was then transferred to a new tube and either used
immediately or stored at −70°C. The nuclear pellet was washed in
500 µl nuclei washing buffer containing protease inhibitors
and DTT by vortexing briefly, and then set on ice for 2 min. The
suspension was centrifuged at 500 × g for 7 min at 4°C, and the
supernatant was carefully removed. This procedure was repeated 1–2
times. The volume of the nuclear pellet was estimated and 10
volumes of ice-cold nuclei storage buffer containing protease
inhibitors and DTT were added. Any clumps of nuclei were broken up
by gentle pipetting. The suspension was either used immediately or
stored at −70°C. Following centrifugation at 20,000 × g for 5 min,
150 µl ice-cold nuclei storage buffer with protease
inhibitors and DTT was added to the nuclear pellet. Any clumps were
broken up as before. The nuclei were lysed by adding 1/10 volume of
nuclei lysis reagent to the suspension, vortexed briefly and shaken
on a rotating bed (900–1,200 rpm) for 15 min at 4°C. The resulting
nuclear lysate was cleared by centrifugation at 20,000 × g for 5
min at 4°C, and the supernatant containing the nuclear protein
extract was transferred to a new tube and either used immediately
or stored at −70°C for subsequent analysis.
Western blot analysis
Equivalent amounts of nuclear or cytoplasmic
proteins were separated by SDS polyacrylamide gel electrophoresis.
The proteins were electrolytically transferred to a PVDF membrane
and blocked in TBST containing 20% non-fat milk for 2 h at room
temperature. The membrane was incubated at room temperature for 1 h
with primary antibodies against p27Kip1 (1:2,000); CRM-1
(1:1,000); and p-p27Kip1 (Ser10) (1:1,000). After
further incubation overnight at 4°C, the membrane was incubated
with HRP-conjugated secondary antibodies (1:1,000) for 2 h at room
temperature. The protein bands were developed by ECL and exposed to
X-ray film. The intensities of the protein bands were quantified
using a grayscale scanner (GeneGnome XRQ; Syngene Corp., Cambridge,
UK). PCNA (1:2,000) and GAPDH (1:5,000) were used as loading
controls.
Cell viability assay
The cholangiocarcinoma cell line QBC939 (kindly
donated by Professor Shu-Guang Wang, Hepatobiliary Department of
Xinan Hospital, Third Military Medical University, Chongqing,
China) was infected with lentivirus encoding shCRM-1 or the vector
(psicoR plasmid) after lentivirus packaging for approximately 24 h.
Following treatment, the above two groups together with untreated
QBC939 cells (blank group) were transferred to 96-well plates at a
density of 1,000 cells/well. Cell viability was evaluated using a
Cell Counting Kit-8 (CCK-8) assay (Promoter, Wuhan, China) and a
5-ethynyl-2′-deoxyuridine (EdU) assay (Guangzhou RiboBio Co., Ltd.,
Guangzhou, China) according to the manufacturer's instructions.
Colony formation assay
Exponentially proliferating QBC939 cells treated as
described above were seeded at a density of approximately 1,000
cells/well in a 6-well plate and the media was replaced every 3
days. The colonies were counted and evaluated after 10 days by
staining with 0.05% crystal violet solution (C8470; Solarbio Corp.,
Beijing, China) for 15 min.
Cell cycle analysis
Exponentially proliferating QBC939 cells stably
transfected with either lenti-vector or lenti-shCRM-1 were counted
after digestion with EDTA-free trypsin and rinsed with PBS followed
by fixation in 70% precooled ethanol at 4°C overnight. After
incubation with RNase A (Sigma, St. Louis, MO, USA) at 37°C for 30
min, the cells were stained with propidium iodide (PI; KeyGen
Biotech, Nanjing, China) at 4°C for 30 min. The cells were then
analyzed by flow cytometry (BD Biosciences, Franklin Lakes, NJ,
USA) to measure the proportion of cells in the G1, S and G2/M
stages.
Xenograft tumor growth assay
The in vivo tumorigenicity of
cholangiocarcinoma cells was determined by subcutaneously injecting
1×106 cancer cells per mouse into the left axillary
fossa of the nude BALB/cA-nu mice (purchased from HFK Bioscience
Corp., Beijing, China). The 3 groups of nude mice (5 mice/group)
were injected with either shCRM-1, the vector or blank cells. Tumor
volume was measured with calipers at the same site of injection
every 3 days by two trained laboratory staff at different times on
the same day starting from the 15th day using the formula,
V=0.5ab2, in which 'a' stands for the longer axis and
'b' stands for the shorter axis of the tumor. The animals were
sacrificed and the tumors were weighed 60 days after injection. The
nude mice were sacrificed and cared for according to the NIH Animal
Care and Use Committee guidelines of the Experimental Animal Center
of Tongji Medical School (HUST, Wuhan, China).
Statistical analysis
Statistical analyses were performed using SPSS
software v. 20.0 (SPSS, Inc., Chicago, IL, USA). Fisher's exact
test was used to determine significant differences between groups
of data, the Chi-square (χ2) test was used to compare
protein expression percentages and Spearman's rank correlation test
was used to compare pairs of variables. All values are presented as
the means ± SEM unless otherwise indicated and the t-test was
adopted to examine the difference between the CRM-1 knockdown
group, blank group and vector group. P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression patterns of CRM-1,
p27Kip1 and p-p27Kip1 (Ser10) proteins in
cholangiocarcinoma tissues
In order to examine the role of CRM-1,
p27Kip1 and p-p27Kip1 (Ser10) in the
development of cholangiocarcinoma, we analyzed 53
cholangiocarcinoma tissue samples and 10 control chronic
cholangitis samples using immunohistochemistry. The protein
expression of CRM-1 and p-p27Kip1 (Ser10) was
significantly higher in the cholangiocarcinoma tissues compared
with that in the control tissues (71.7 vs. 0.0% and 60.4 vs. 20.0%,
respectively; P<0.05; Table
I). By contrast, p27Kip1 expression was
significantly lower in the cholangiocarcinoma tissues compared with
that in the control tissues (37.7 vs. 80.0%; P<0.05; Table I). These results were supported by
correlation analyses which showed that there was a negative
correlation between the expression of CRM-1 and
p-p27Kip1 (Ser10) proteins and the expression of
p27Kip1 (correlation coefficients, r=−0.461 and
r=−0.484, respectively; P<0.01; Table II); whereas CRM-1 expression
positively correlated with p-p27Kip1 (Ser10)
expression.
 | Table IImmunohistochemical analysis of
p27Kip1, CRM-1 and p-p27Kip1 (Ser10) protein
expression in cholangiocarcinoma (n=53) and chronic proliferative
cholangitis tissue samples (n=10). |
Table I
Immunohistochemical analysis of
p27Kip1, CRM-1 and p-p27Kip1 (Ser10) protein
expression in cholangiocarcinoma (n=53) and chronic proliferative
cholangitis tissue samples (n=10).
| Tissue samples | p27Kip1
n (%)
| P-value | CRM-1 n (%)
| P-value |
p-p27Kip1 (Ser10) n (%)
| P-value |
|---|
| + | − | + | − | + | − |
|---|
| CCA (n=53) | 20 (37.7) | 33 (62.3) | 0.034 | 38 (71.7) | 15 (28.3) | 0.013 | 32 (60.4) | 21 (39.6) | 0.035 |
| CPC (n=10) | 8 (80.0) | 2 (20.0) | | 0 (0) | 10 (100) | | 2 (20.0) | 8 (80.0) | |
 | Table IICorrelations between the expression
of p27Kip1 and CRM-1 and p-p27Kip1 (Ser10)
expression in cholangiocarcinoma tissue samples (n=53). |
Table II
Correlations between the expression
of p27Kip1 and CRM-1 and p-p27Kip1 (Ser10)
expression in cholangiocarcinoma tissue samples (n=53).
| CRM-1 n
| r-value |
p-p27Kip1 (Ser10) n
| r-value |
|---|
| + | − | + | − |
|---|
|
p27Kip1 | | | | | | |
| + | 9 | 11 | −0.461a | 6 | 14 | −0.484a |
| − | 29 | 4 | | 26 | 7 | |
Correlations between p27Kip1,
CRM-1, p-p27Kip1 (Ser10) protein expression and
clinicopathological features in cholangiocarcinoma
The correlations between the expression of CRM-1,
p27Kip1 and p-p27Kip1 (Ser10) proteins and
clinicopathological features in cholangiocarcinoma tissues are
summarized in Table III. The
results revealed that the expression of all three proteins
significantly correlated with the tumor grade and clinical stage in
cholangiocarcinoma (P<0.05; Table III). By contrast, there were no
significant correlations between protein expression and patient
age, gender, tumor size or site (P>0.05; Table III). The expression of CRM-1 and
p-p27Kip1 (Ser10) positively correlated with the degree
of malignancy and clinical stage whereas the expression of
p27Kip1 decreased with the degree of malignancy and
clinical stage.
 | Table IIICorrelations between the expression
of p27Kip1, CRM-1 and p-p27Kip1 (Ser10)
proteins and clinicopathological features in cholangiocarcinoma
tissue samples (n=53). |
Table III
Correlations between the expression
of p27Kip1, CRM-1 and p-p27Kip1 (Ser10)
proteins and clinicopathological features in cholangiocarcinoma
tissue samples (n=53).
| Variable | Total | p27Kip1
n (%) | P-value | CRM-1 n (%) | P-value |
p-p27Kip1 (Ser10) n (%) | P-value |
|---|
| Gender | 53 | | 0.779 | | 0.380 | | 1.000 |
| Male | 30 | 12 (40.0) | | 20 (66.7) | | 18 (60.0) | |
| Female | 23 | 8 (34.8) | | 18 (78.3) | | 14 (60.9) | |
| Age (years) | 53 | | 1.000 | | 0.761 | | 1.000 |
| <60 | 28 | 11 (39.3) | | 21 (75.0) | | 17 (64.7) | |
| ≥60 | 25 | 9 (36.0) | | 17 (68.0) | | 15 (60.0) | |
| Size (cm) | 53 | | 0.773 | | 0.546 | | 0.397 |
| <2 | 21 | 7 (33.3) | | 14 (66.7) | | 11 (52.4) | |
| ≥2 | 32 | 13 (40.6) | | 24 (75.0) | | 21 (65.6) | |
| Location | 53 | | 0.769 | | 0.929 | | 0.938 |
| Proximal | 16 | 5 (31.2) | | 11 (68.75) | | 9 (56.3) | |
| Middle | 14 | 5 (35.7) | | 10 (71.43) | | 9 (64.3) | |
| Distal | 23 | 10 (43.5) | | 17 (73.91) | | 14 (60.9) | |
| Grade | 53 | | 0.022 | | 0.048 | | 0.003 |
| Well | 24 | 14 (58.3) | | 13 (54.17) | | 10 (41.67) | |
| Moderate | 12 | 3 (25.0) | | 10 (83.33) | | 8 (66.67) | |
| Poor | 17 | 3 (17.6) | | 15 (88.24) | | 14 (82.35) | |
| Stage | 53 | | 0.010 | | 0.003 | | 0.023 |
| I–II | 22 | 13 (59.09) | | 12 (54.55) | | 9 (40.91) | |
| III–IV | 31 | 7 (22.58) | | 26 (83.87) | | 23 (74.19) | |
Associations between the subcellular
localization of CRM-1, p27Kip1 and p-p27Kip1
(Ser10) with clinicopathological features in
cholangiocarcinoma
The subcellular distributions of CRM-1,
p27Kip1 and p-p27Kip1 (Ser10) proteins in
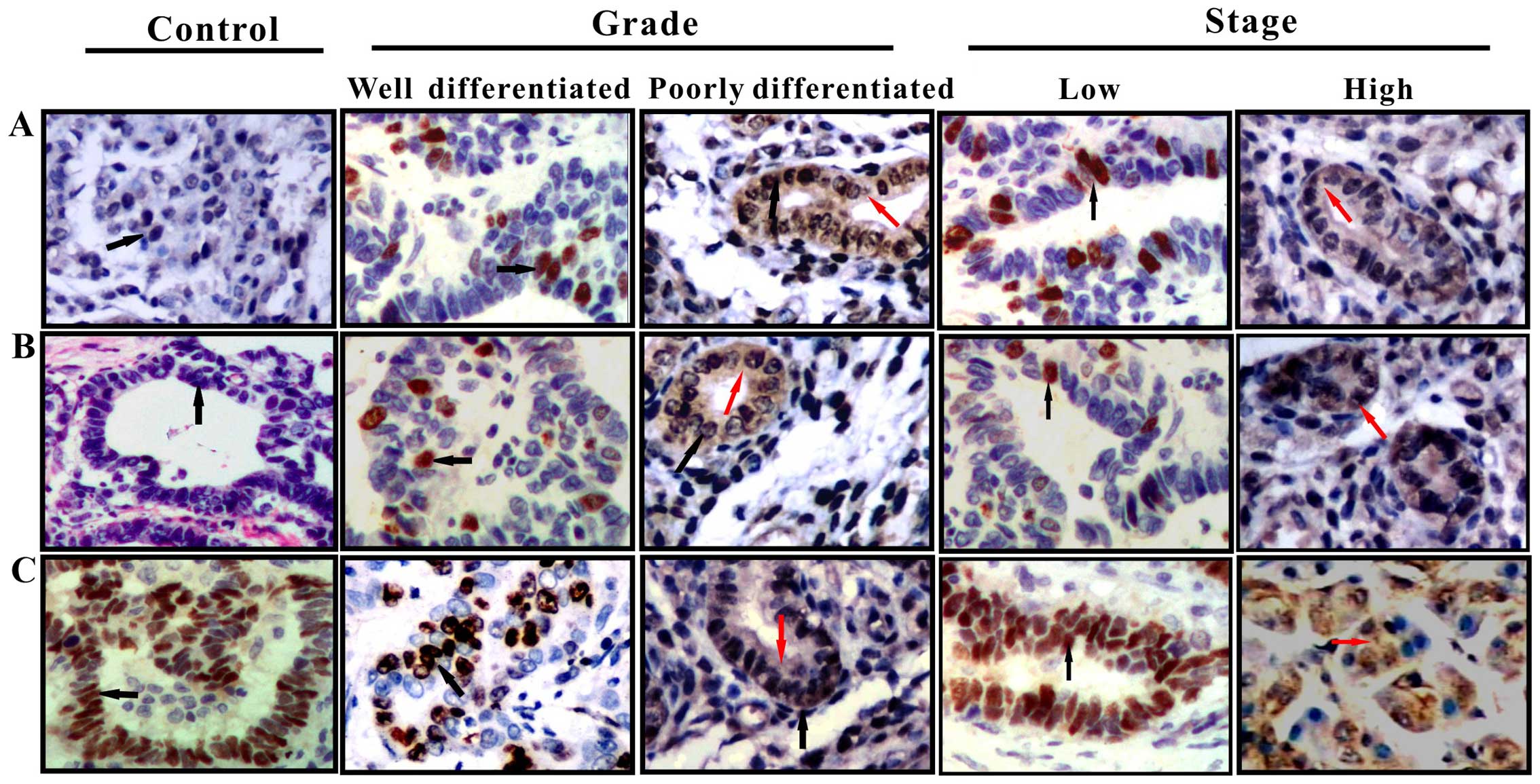
cholangiocarcinoma tissues were examined by IHC analysis (Fig. 1). The staining patterns revealed
that p27Kip1 was principally localized in the nuclei in
chronic cholangitis tissue samples and was primarily localized in
the nuclei, with little cytoplasmic staining in the
cholangiocarcinoma tissues of low malignant potential
(well-differentiated and TNM stage I–II); and was diffusely
distributed in the nuclei and cytoplasm in the tissues of highly
malignant tumors (poorly-differentiated and TNM stage III–IV).
Evaluation of the IHC staining signals showed that the expression
of cytoplasmic p27Kip1 increased from 12.5% (3/24) in
well-differentiated tumors to 17.6% (3/17) in poorly-differentiated
tumors; whereas the corresponding expression of nuclear
p27Kip1 decreased from 45.8% (11/24) to 0.0% (0/17).
These changes were significant (P<0.05; Table IV). A similar pattern was
observed between the subcellular expression levels of
p27Kip1 and the clinical stage of
cholangiocarcinoma.
 | Table IVCorrelations between the subcellular
localizations of p27Kip1, CRM-1 and p-p27Kip1
(Ser10) proteins and the malignancy of tumors in patients with
cholangiocarcinoma (n=53) (Fig.
1). |
Table IV
Correlations between the subcellular
localizations of p27Kip1, CRM-1 and p-p27Kip1
(Ser10) proteins and the malignancy of tumors in patients with
cholangiocarcinoma (n=53) (Fig.
1).
| Variable | Total | p27Kip1
(positive)
| P-value | CRM-1 (positive)
| P-value |
p-p27Kip1 (Ser10) (positive)
| P-value |
|---|
| N | C | N | C | N | C |
|---|
| Grade | 53 | | | 0.018 | | | 0.851 | | | 0.583 |
| Well | 24 | 11 | 3 | | 7 | 1 | | 8 | 2 | |
| Moderate | 12 | 1 | 2 | | 5 | 2 | | 8 | 1 | |
| Poor | 17 | 0 | 3 | | 10 | 3 | | 9 | 4 | |
| Stage | 53 | | | 0.017 | | | 1.000 | | | 1.000 |
| I–II | 22 | 10 | 3 | | 6 | 1 | | 11 | 2 | |
| III–IV | 31 | 1 | 6 | | 17 | 4 | | 15 | 4 | |
These results suggested that p27Kip1 may
be transported from the nucleus to the cytoplasm, and that this is
associated with increasing malignancy in cholangiocarcinoma. This
further suggested that the nuclear export of p27Kip1 may
contribute to the progression and malignant transformation of
cholangiocarcinoma.
By contrast, the IHC staining patterns revealed that
the levels of nuclear CRM-1 and p-p27Kip1 (Ser10)
proteins were low in the chronic cholangitis tissue samples;
however, the levels increased with increasing malignancy and
clinical stage in the cholangiocarcinoma tissue samples. There was
no significant change in their cytoplasmic levels (P>0.05;
Table IV).
These results demonstrated that the cytoplasmic
distribution of p27Kip1 was consistent with the
cytoplasmic expression of CRM-1 and p-p27Kip1 (Ser10)
(Fig. 3), suggesting that the
involvement of p27Kip1 nuclear export mediated by CRM-1
in the development and progression of cholangiocarcinoma. It
suggested that CRM-1 and p-p27Kip1 (Ser10) may be
implicated in the nuclear export of p27Kip1 during the
development and progression of cholangiocarcinoma.
Furthermore, semi-quantitative western blot analysis
and grayscale analysis were performed in order to confirm the
results of IHC staining by evaluating the protein concentrations of
p27Kip1, CRM-1 and p-p27Kip1 (Ser10)
according to their subcellular localizations in the
cholangiocarcinoma tissues (Tables
V and VI). The results of
the grayscale analysis revealed that the nuclear levels of CRM-1
protein as well as the phosphorylation of p27Kip1
(Ser10) increased with increasing malignancy and clinical stage in
cholangiocarcinoma; whereas the opposite effect was observed for
p27Kip1 (Table V;
Fig. 2). However, there was
little change in the cytoplasmic levels of CRM-1 protein and the
phosphorylation of p27Kip1 (Ser10) with malignancy or
clinical stage, and only a small increase in the cytoplasmic
expression of p27Kip1 (Table VI; Fig. 3). These results indicated that the
subcellular distribution of p27Kip1 protein was closely
associated with its phosphorylation and the expression of CRM-1.
These findings support the hypothesis that the nuclear export of
p27Kip1 may be implicated in the progression and
malignancy of cholangiocarcinoma.
 | Table VAssociations between nuclear
expression of p27Kip1, CRM-1 and p-p27Kip1
(Ser10) proteins with different tumor grades and clinical stages in
53 cholangiocarcinoma tissues samples relative to 10 chronic
cholangitis control samples (Fig.
2). |
Table V
Associations between nuclear
expression of p27Kip1, CRM-1 and p-p27Kip1
(Ser10) proteins with different tumor grades and clinical stages in
53 cholangiocarcinoma tissues samples relative to 10 chronic
cholangitis control samples (Fig.
2).
| Protein | Control | Grade
(differentiation)
| Stage
|
|---|
| Well | Moderate | Poor | I–II | III–IV |
|---|
|
p27Kip1 | 1.0 | 0.91±0.17 | 0.5±0.11 | 0.35±0.19 | 0.84±0.15 | 0.47±0.22 |
| CRM-1 | 1.0 | 1.69±0.31 | 1.83±0.25 | 2.45±0.34 | 1.57±0.19 | 1.98±0.27 |
|
p-p27Kip1 (Ser10) | 1.0 | 1.24±0.19 | 1.43±0.33 | 1.76±0.27 | 1.42±0.32 | 1.58±0.24 |
 | Table VIAssociations between the cytoplasmic
expression of p27Kip1, CRM-1 and p-p27Kip1
(Ser10) proteins with different grades and clinical stages in 53
cholangiocarcinoma tissue samples relative to 10 chronic
cholangitis control samples (Fig.
3). |
Table VI
Associations between the cytoplasmic
expression of p27Kip1, CRM-1 and p-p27Kip1
(Ser10) proteins with different grades and clinical stages in 53
cholangiocarcinoma tissue samples relative to 10 chronic
cholangitis control samples (Fig.
3).
| Protein | Control | Grade
(differentiation)
| Stage
|
|---|
| Well | Moderate | Poor | I–II | III–IV |
|---|
|
p27Kip1 | 1.00 | 1.19±0.17 | 1.59±0.20 | 1.83±0.27 | 1.34±0.19 | 1.62±0.22 |
| CRM-1 | 1.00 | 1.29±0.11 | 1.13±0.05 | 1.49±0.16 | 1.25±0.17 | 1.39±0.26 |
|
p-p27Kip1(Ser10) | 1.00 | 1.30±0.15 | 1.13±0.07 | 1.41±0.27 | 1.26±0.12 | 1.43±0.18 |
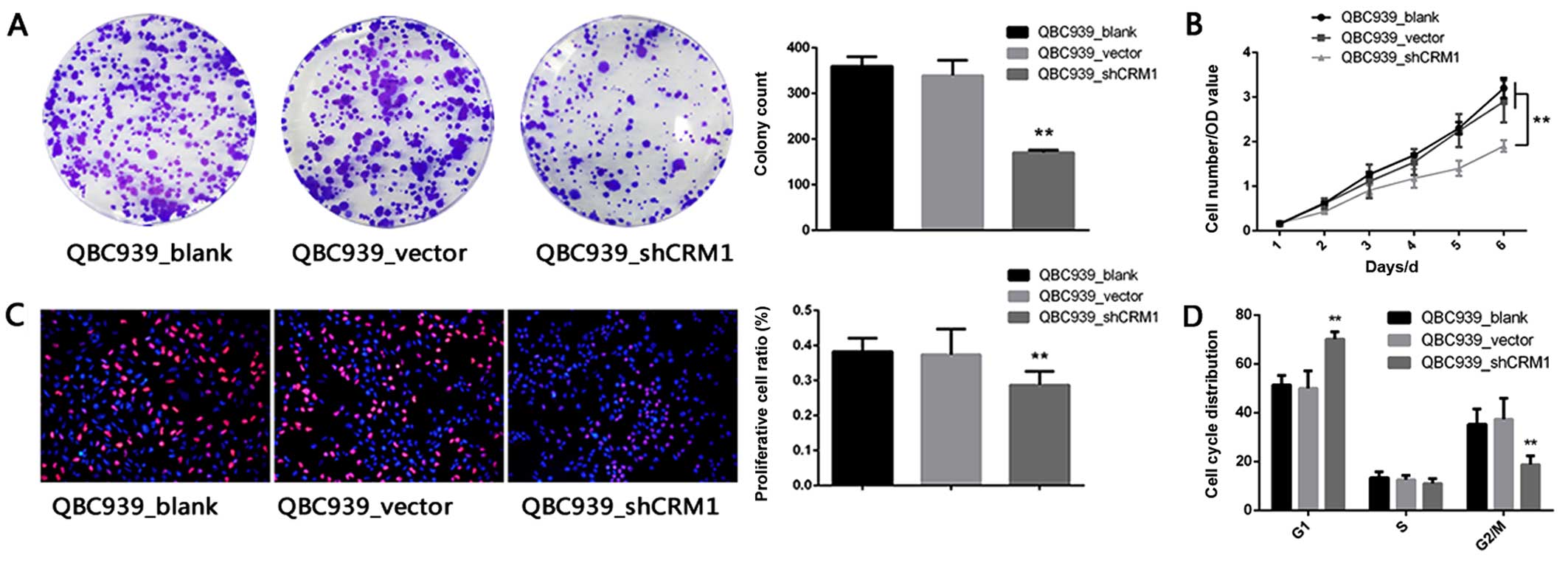
Downregulation of CRM-1 inhibits the
colony forming ability and reduces the viability of the
cholangiocarcinoma cell line QBC939 both in vitro and in vivo
To explore the causal relationship between the
downregulation of CRM-1 and the nuclear export of
p27Kip1 and carcinogenesis in cholangiocarcinoma, a
classical cholangiocarcinoma cell line QBC939 was selected for
CRM-1 knockdown. Notably, the colony formation assay demonstrated
that the colony forming ability of the QBC939 cells was
significantly inhibited by the knockdown of CRM-1 as compared with
that in the blank and vector groups (Fig. 4A). Furthermore, the knockdown of
CRM-1 by shRNA reduced the viability of QBC939 cells (Fig. 4B and C).
Additionally, performing the cell cycle analysis of
these cells using the PI staining method, revealed marked G1 arrest
in the QBC939-shCRM-1 group as compared with that in the blank and
control groups (Fig. 4D).
Furthermore, in order to examine the precise role of
CRM-1 in a xenograft mouse model, in vivo experiments were
undertaken to evaluate the procarcinogenic effect of CRM-1 in
cholangiocarcinoma. As shown in Fig.
5A, the CRM-1 knockdown group exhibited markedly smaller tumor
volumes as compared with those in the blank and vector groups. The
xenograft tumors were weighed and the CRM-1 knockdown group clearly
produced lighter (Fig. 5B) and
smaller tumors (Fig. 5C).
Downregulation of CRM-1 enhances nuclear
p27Kip1 and decreases cytoplasmic p27Kip1
simultaneously
In order to elucidate the underlying mechanism
responsible for the pro-carcinogenic effect of CRM-1 in
cholangiocarcinoma, nuclear and cytoplasmic proteins were
separately extracted from the QBC939 cells treated with shCRM-1 as
well as the blank and the vector groups. Notably, as shown in
Fig. 6, CRM-1 expression was
closely associated with the subcellular location of
p27Kip1. As CRM-1 was downregulated in Fig. 6A, cytoplasmic p27Kip1
was also decreased whereas nuclear p27Kip1 was increased
in the QBC939 cells. Thus, it is quite probable that CRM-1 may
induce the progression of cholangiocarcinoma by regulating the
subcellular distribution of p27Kip1. However, further
experiments examining point mutations in the phosphorylation sites
of p27Kip1 are warranted.
Discussion
p27Kip1 is a newly discovered tumor
suppressor gene that is involved in the negative regulation of cell
cycle progression at the G1/S checkpoint. The aberrant expression
or localization of p27Kip1 weakens its inhibitory effect
on the cell cycle, leading to uncontrolled cell growth and
carcinogenesis (8). The
downregulation and abnormal subcellular localization of
p27Kip1 have been reported in a number of types of tumor
including nasopharyngeal carcinoma (9). Consistent with these previous
findings, we found that p27Kip1 expression was decreased
in the cholangiocarcinoma tissues compared with that in the chronic
cholangitis tissues, suggesting that the aberrant expression of
p27Kip1 may be associated with the development and
progression of cholangiocarcinoma. Immunohistochemistry and western
blot analysis showed that the protein expression of
p27Kip1 increased sequentially from well- to
poorly-differentiated tumors, and from stage I–II cases of
cholangiocarcinoma to stage III–IV cases. In addition, the
cytoplasmic staining patterns indicated that p27Kip1 was
primarily located in the cytoplasm of tissues from
poorly-differentiated tumors and those from patients with advanced
stage disease. These findings suggested that that there may be an
association between the aberrant expression of p27Kip1
and the malignancy and clinical stage of cholangiocarcinoma, and
that p27Kip1 is transported from the nucleus to the
cytoplasm during tumor progression in cholangiocarcinoma. Similar
observations have been reported in other types of cancer;
p27Kip1 expression was decreased in liver cancer tissues
compared with that in para-carcinomatous tissues and normal liver
tissues, and the cytoplasmic localization of p27Kip1
closely correlated with malignant progression, clinical stage and
invasion in liver cancer (10).
In colorectal cancer, the cytoplasmic expression of
p27Kip1 was found to be significantly higher in tumor
cells compared with that in normal mucosal cells (11). The cytoplasmic localization of
p27Kip1 has also been reported to be significantly
associated with poor prognosis in ovarian cancer (12). Taken together, these findings
suggest that abnormal subcellular localization and increased
cytoplasmic expression of p27Kip1 may play an important
role in tumor development. However, the mechanisms that regulate
p27Kip1 expression and intracellular transport remain
poorly understood.
The subcellular localization of a protein is closely
associated with its function. A recent proposal for a
p27Kip1-regulated subcellular regionalization mechanism
suggested that the abnormal cytoplasmic localization of
p27Kip1 may separate p27Kip1 from its nuclear
effectors leading to its deactivation (13). The majority of studies on the
abnormal subcellular localization of p27Kip1 have
focused on ubiquitin-degradation pathways. These include a
cytoplasmic pathway which is dependent on nuclear translocation
through the conjugation of ubiquitin with cytoplasmic ubiquitin
ligase KPC (17), and a nuclear
pathway dependent on ubiquitin ligase SCF-Skp2 (14). The cytoplasmic pathway is mediated
by p-p27Kip1 (Ser10) (15), whereas the nuclear pathway is
mediated by p-p27Kip1 (Thr187). A previous study by our
group has confirmed that the nuclear ubiquitin-degradation pathway
involving SCF-Skp2 and p-p27Kip1 (Thr187) was enhanced
in cholangiocarcinoma tissues and cells (16). This may be associated with the
observed decrease in the nuclear expression of p27Kip1.
However, the precise details regarding the association between
decreasing nuclear expression and increasing cytoplasmic expression
remain unclear. Further investigations in vitro may confirm
whether the two pathways co-exist in cholangiocarcinoma. In
addition, the association between the translocation of
p27Kip1 and the cytoplasmic ubiquitin-degradation
pathway involving KPC remains to be determined.
CRM-1 mediates the nuclear export of
p27Kip1 by binding to p27Kip1 through a
nuclear export signal in a leptomycin B-sensitive manner (17). The nuclear export of
p27Kip1 correlates with the phosphorylation of Ser10,
and reports have shown that CRM-1 specifically recognizes and binds
to Ser10 phosphorylated p27Kip1, thereby promoting its
nuclear export (18). Mutation at
this site has been shown to cause p27Kip1 to lose its
nuclear export capability (19),
indicating that Ser10 phosphorylation may play a critical role in
the subcellular distribution and functional status of
p27Kip1. We found that CRM-1 and p-p27Kip1
(Ser10) were highly expressed in the cholangiocarcinoma tissues,
and their nuclear expression levels increased with increasing
malignancy compared with those in the control samples (P<0.05).
This is consistent with previous findings in neuroglioma which
demonstrated that high expression levels of CRM-1 and
p27Kip1 closely correlated with malignancy, and that
high CRM-1 expression was associated with a poor prognosis
(20,21). Our results demonstrated that the
nuclear expression of CRM-1 and p-p27Kip1 (Ser10)
inversely correlated with nuclear expression of p27Kip1
and that CRM-1 expression positively correlated with that of
p-p27Kip1 (Ser10) in the cholangiocarcinoma tissues,
which was consistent with the role of CRM-1 in the nuclear export
of p27Kip1 in these tumors. Although the nuclear
expression of CRM-1 and p27Kip1 (Ser10) were found to be
associated with the degree of malignancy and clinical stage in
cholangiocarcinoma, we observed little change in the cytoplasmic
levels of these proteins.
The aberrant expression of p27Kip1 may
result in abnormal cytoplasmic proteolysis coupled with nuclear
translocation in cholangiocarcinoma. Our findings suggest that
CRM-1 may play a role in the nuclear and cytoplasmic distribution
of p27Kip1 by recognizing and binding to
p-p27Kip1 (Ser10) and thereby regulating the nuclear
export of p27Kip1.
Previous studies have demonstrated that specific
agents may restore p27Kip1 expression in cells including
cholangiocarcinoma cells and trophoblast cells (22–25). The control of protein localization
by inhibiting the translocation of p27Kip1 has also been
applied in cancer therapies (26).
Taken together, these findings indicate that changes
in the expression levels and subcellular distributions of CRM-1,
p27Kip1 and p-p27Kip1 (Ser10) proteins may be
a potential predictor and indicator of malignancy in
cholangiocarcinoma. Although further investigations are warranted
in order to determine the precise details of how these proteins
interact in cholangiocarcinoma, our findings suggest that these
proteins may prove to be promising novel targets in the diagnosis,
treatment and prognosis of cholangiocarcinoma.
In conclusion, the knockdown of CRM-1 may lead to
decreases in p27Kip1 levels in the cytoplasm, thereby
inhibiting malignant transformation in cholangiocarcinoma. These
findings have revealed potentially potent targets for the
diagnosis, and prognosis of cholangiocarcinoma which may also serve
as novel therapies for the treatment of patients with
cholangiocarcinoma.
Acknowledgments
We thank all the donors who participated in this
program and all our coworkers who contributed to this study. The
present study was supported by the Natural Science Foundation of
Hubei Province of China (project no. 2013CKB020).
References
|
1
|
Duncan TJ, Al-Attar A, Rolland P, Harper
S, Spendlove I and Durrant LG: Cytoplasmic p27 expression is an
independent prognostic factor in ovarian cancer. Int J Gynecol
Pathol. 29:8–18. 2010. View Article : Google Scholar
|
|
2
|
Sgambato A, Camerini A, Genovese G, De
Luca F, Viacava P, Migaldi M, Boninsegna A, Cecchi M, Sepich CA,
Rossi G, et al: Loss of nuclear p27(Kip1) and
α-dystroglycan is a frequent event and is a strong predictor of
poor outcome in renal cell carcinoma. Cancer Sci. 101:2080–2086.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tomoda K, Kubota Y and Kato J: Degradation
of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by
Jab1. Nature. 398:160–165. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Smitherman M, Lee K, Swanger J, Kapur R
and Clurman BE: Characterization and targeted disruption of murine
Nup50, a p27(Kip1)-interacting component of the nuclear pore
complex. Mol Cell Biol. 20:5631–5642. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Connor MK, Kotchetkov R, Cariou S, Resch
A, Lupetti R, Beniston RG, Melchior F, Hengst L and Slingerland JM:
CRM1/Ran-mediated nuclear export of p27(Kip1) involves a nuclear
export signal and links p27 export and proteolysis. Mol Biol Cell.
14:201–213. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ganeshan D, Moron FE and Szklaruk J:
Extrahepatic biliary cancer: new staging classification. World J
Radiol. 4:345–352. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jang JY, Kim SW, Park DJ, Ahn YJ, Yoon YS,
Choi MG, Suh KS, Lee KU and Park YH: Actual long-term outcome of
extrahepatic bile duct cancer after surgical resection. Ann Surg.
241:77–84. 2005.
|
|
8
|
Wang Y, Wang Y, Xiang J, Ji F, Deng Y,
Tang C, Yang S, Xi Q, Liu R and Di W: Knockdown of CRM1 inhibits
the nuclear export of p27(Kip1) phosphorylated at serine 10 and
plays a role in the pathogenesis of epithelial ovarian cancer.
Cancer Lett. 343:6–13. 2014. View Article : Google Scholar
|
|
9
|
Pan Y, Zhang Q, Tian L, Wang X, Fan X,
Zhang H, Claret FX and Yang H: Jab1/CSN5 negatively regulates p27
and plays a role in the pathogenesis of nasopharyngeal carcinoma.
Cancer Res. 72:1890–1900. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dai L, Liu Y, Liu J, Wen X, Xu Z, Wang Z,
Sun H, Tang S, Maguire AR, Quan J, et al: A novel
cyclinE/cyclinA-CDK inhibitor targets p27(Kip1) degradation, cell
cycle progression and cell survival: implications in cancer
therapy. Cancer Lett. 333:103–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ciaparrone M, Yamamoto H, Yao Y, Sgambato
A, Cattoretti G, Tomita N, Monden T, Rotterdam H and Weinstein IB:
Localization and expression of p27KIP1 in multistage colorectal
carcinogenesis. Cancer Res. 58:114–122. 1998.PubMed/NCBI
|
|
12
|
Masciullo V, Ferrandina G, Pucci B,
Fanfani F, Lovergine S, Palazzo J, Zannoni G, Mancuso S, Scambia G
and Giordano A: p27Kip1 expression is associated with clinical
outcome in advanced epithelial ovarian cancer: multivariate
analysis. Clin Cancer Res. 6:4816–4822. 2000.
|
|
13
|
Ibañez IL, Bracalente C, Notcovich C,
Tropper I, Molinari BL, Policastro LL and Durán H: Phosphorylation
and subcellular localization of p27Kip1 regulated by hydrogen
peroxide modulation in cancer cells. PLoS One. 7:e445022012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tsvetkov LM, Yeh KH, Lee SJ, Sun H and
Zhang H: p27(Kip1) ubiquitination and degradation is regulated by
the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr
Biol. 9:661–664. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kazi A, Carie A, Blaskovich MA, Bucher C,
Thai V, Moulder S, Peng H, Carrico D, Pusateri E, Pledger WJ, et
al: Blockade of protein geranylgeranylation inhibits Cdk2-dependent
p27Kip1 phosphorylation on Thr187 and accumulates p27Kip1 in the
nucleus: implications for breast cancer therapy. Mol Cell Biol.
29:2254–2263. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Luo J, Chen YJ, Wang WY and Zou SQ: Effect
of mutant p27(kipl) gene on human cholangiocarcinoma cell line,
QBC(939). World J Gastroenterol. 14:5344–5348. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kamura T, Hara T, Matsumoto M, Ishida N,
Okumura F, Hatakeyama S, Yoshida M, Nakayama K and Nakayama KI:
Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1)
at G1 phase. Nat Cell Biol. 6:1229–1235. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kotoshiba S and Nakayama K: The
degradation of p27 and cancer. Nihon Rinsho. 63:2047–2056. 2005.In
Japanese. PubMed/NCBI
|
|
19
|
Lee JG, Song JS, Smith RE and Kay EP:
Human corneal endothelial cells employ phosphorylation of p27(Kip1)
at both Ser10 and Thr187 sites for FGF-2-mediated cell
proliferation via PI3-kinase. Invest Ophthalmol Vis Sci.
52:8216–8223. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rodier G, Montagnoli A, Di Marcotullio L,
Coulombe P, Draetta GF, Pagano M and Meloche S: p27 cytoplasmic
localization is regulated by phosphorylation on Ser10 and is not a
prerequisite for its proteolysis. EMBO J. 20:6672–6682. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shen A, Wang Y, Zhao Y, Zou L, Sun L and
Cheng C: Expression of CRM1 in human gliomas and its significance
in p27 expression and clinical prognosis. Neurosurgery. 65:153–160.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He W, Wang B, Zhuang Y, Shao D, Sun K and
Chen J: Berberine inhibits growth and induces G1 arrest and
apoptosis in human cholangiocarcinoma QBC939 cells. J Pharmacol
Sci. 119:341–348. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huether A, Höpfner M, Baradari V, Schuppan
D and Scherübl H: Sorafenib alone or as combination therapy for
growth control of cholangiocarcinoma. Biochem Pharmacol.
73:1308–1317. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kotake Y, Nakayama K, Ishida N and
Nakayama KI: Role of serine 10 phosphorylation in p27 stabilization
revealed by analysis of p27 knock-in mice harboring a serine 10
mutation. J Biol Chem. 280:1095–1102. 2005. View Article : Google Scholar
|
|
25
|
Nadeem L, Brkic J, Chen YF, Bui T, Munir S
and Peng C: Cytoplasmic mislocalization of p27 and CDK2 mediates
the anti-migratory and anti-proliferative effects of Nodal in human
trophoblast cells. J Cell Sci. 126:445–453. 2013. View Article : Google Scholar
|
|
26
|
Shiraso S, Katayose Y, Yamamoto K, Mizuma
M, Yabuuchi S, Oda A, Rikiyama T, Onogawa T, Yoshida H, Hayashi H,
et al: Overexpression of adenovirus-mediated p27Kip1
lacking the Jab1-binding region enhances cytotoxicity and inhibits
xenografted human cholangiocarcinoma growth. Anticancer Res.
29:2015–2024. 2009.PubMed/NCBI
|