Introduction
Differentiated embryo chondrocyte 2 (DEC2; also
known as SHARP-1 or BHLHE41) is a member of the basic
helix-loop-helix-Orange (bHLH-O) family, characterized by a basic
DNA binding domain, a helix-loop-helix (HLH) dimerization domain,
and Orange extended dimerization domain (1,2).
DEC2 is a transcriptional repressor similar to other bHLH-O
proteins such as HES and HESR (3,4).
The DEC2 homodimer directly binds to the E-box sequence (CACGTG)
and represses the transcription of target genes through the
recruitment of a corepressor, histone deacetylase 1 (3,5).
DEC2 also antagonizes the transcriptional activity of other
transcription factors, including BMAL1 and SREBP-1, by direct
protein-protein interactions and/or by competing for binding to the
E-box (5,6). Furthermore, DEC2 modulates various
biochemical processes, including circadian rhythm (5,7–9),
cancer metastasis (10,11), proliferation (12–16) and differentiation (3,4,17,18).
DEC2 is ubiquitously expressed in varying
amounts in both embryonic and adult tissues (1,19).
The expression level of DEC2 is high in developing limbs,
muscle and brain. In our preliminary experiments, DEC2 was
expressed at higher levels in mesenchymal stem cells (MSCs) than in
fibroblasts (data not shown). MSCs have the capacity to
differentiate into a number of types of cells including adipocytes,
chondrocytes, osteoblasts and smooth muscle cells (20). Previous studies have shown that
DEC2 suppresses myogenesis and adipogenesis of MSCs by inhibiting
the transcriptional activities of MyoD and C/EBP, respectively
(3,4,17).
These observations suggest that DEC2 may be involved in maintaining
MSCs in an undifferentiated state. However, whether DEC2 modulates
differentiation of MSCs into osteoblasts or chondrocytes remains
unclear.
In preliminary experiments, the overexpression of
DEC2 in human MSCs suppressed chondrogenic differentiation, whereas
it had a marginal effect on osteogenic differentiation (data not
shown). Therefore, we focused on the role of DEC2 in chondrogenic
differentiation of MSCs. The results revealed that DEC2 is a
negative regulator for the proliferation and differentiation of
chondrocyte lineage-committed MSCs, although it has little effect
on the proliferation of undifferentiated MSCs. We also examined the
effects of DEC2 overexpression on the expressions of fibroblast
growth factors (FGFs) and cell cycle-related genes, as these genes
play important roles in the proliferation and/or differentiation of
chondrogenic cells (21,22).
Materials and methods
Cells
Human iliac bone marrow MSCs were obtained from
RIKEN (Tsukuba, Japan). The cells were maintained in Dulbecco's
modified Eagle's medium (DMEM; Sigma, St. Louis, MO, USA)
supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT,
USA), 1 ng/ml FGF2 (Kaken Pharmaceutical, Tokyo, Japan) and 1%
antibiotic-antimycotic solution (Sigma) at 37°C in 5%
CO2. MSCs harvested at the 4th–6th passage
were used in the experiments.
Chondrogenic differentiation in pellet
cultures
The MSCs were centrifuged at 500 × g for 5 min in
15-ml conical polypropylene tubes to form pellets
(2.5×105 cells/pellet). The pellets were cultured at
37°C in a humidified atmosphere of 5% CO2 for 3 days,
with 0.5 ml chondrogenesis induction medium consisting of α-minimum
essential medium (α-MEM) supplemented with 100 nM dexamethasone, 50
µg/ml ascorbic acid-2-phosphate, 4.5 mg/ml D-glucose (all
from Sigma), 100 µg/ml sodium pyruvate (Wako Pure Chemical,
Osaka, Japan), 1% ITS+ Premix (Thermo Fisher Scientific, Waltham,
MA, USA), and 10 ng/ml TGF-β3 (Peprotech, Rocky Hill, NJ, USA).
Thereafter, the cells were fed every other day with 1 ml fresh
medium.
Low-density monolayer cultures
The MSCs were seeded at a low density (700
cells/cm2) on 6-well plates for RNA isolation or 96-well
plates for the proliferation assay and incubated with DMEM
supplemented with 10% FBS at 37°C in a CO2
incubator.
Construction of an adenoviral expression
vector
The recombinant adenovirus containing Dec2
(Ad-Dec2) was constructed using the Adeno-X Expression System
(Clontech, Mountain View, CA, USA). Full-length mouse Dec2
(mDec2) cDNA (1) was inserted
into the NheI and XbaI sites of the pShuttle vector
to generate the expression cassette under the regulation of the
cytomegalovirus promoter. The expression cassette was ligated to
Adeno-X viral DNA. The adenoviral vector pAd-Dec2 was digested with
PacI and then transfected into 293 cells using Lipofectamine
(Invitrogen, Carlsbad, CA, USA). The resulting adenoviruses were
further propagated in 293 cells and purified using the Adeno-X™
purification kit (Clontech). Viral titers were determined using the
Adeno-X™ rapid titer kit (Clontech). The recombinant adenovirus
containing lacZ (Ad-LacZ) was prepared as a negative control
(23). MSCs were mock-infected or
infected with Ad-Dec2 or Ad-LacZ at the indicated multiplicity of
infection (MOI) for 2 h.
Western blot analysis
MSCs were infected with Ad-Dec2 at the indicated MOI
(control: MOI =0), and then incubated in DMEM supplemented with 10%
FBS. The cells were lysed with Laemmli buffer, and the lysates were
subjected to sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE). The proteins were transferred to a PVDF
membrane (Millipore, Bedford, MA, USA). The membrane was incubated
overnight with rabbit antibodies specific for DEC2 (1:2,000
dilution at 4°C). After washes with TBST buffer (25 mM TBS, 0.05%
Tween-20, pH 7.4), the membrane was incubated for 1 h with alkaline
phosphatase-conjugated anti-rabbit IgG (1:2,000 dilution at room
temperature; Dako, Carpinteria, CA, USA). The antibody-bound bands
were visualized using nitro blue tetrazolium and
5-bromo-4-chloro-3-indolyl phosphate (Nacalai Tesque, Kyoto,
Japan). The anti-DEC2 antibodies were produced by immunizing
synthetic peptide fragment
Cys-Asn-Pro-Glu-Ser-Ser-Gln-Glu-Asp-Ala-Thr-Gln-Pro-Ala. The
obtained antibodies were purified by affinity column
chromatography.
5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal)
staining
MSCs were cultured for 2 days after infection with
Ad-LacZ at the indicated MOI. The transduction efficiency of MSCs
with the adenovirus was estimated by staining with X-gal (Wako Pure
Chemical). The cells were fixed with phosphate-buffered saline
(PBS) containing 2% formaldehyde and 0.2% glutaraldehyde (both from
Wako Pure Chemical) for 5 min, washed twice with PBS and incubated
in X-gal staining solution overnight at 37°C. After rinsing in
water, images of the cells were captured using an Olympus IX70
microscope and an Olympus DP20 camera (Olympus, Tokyo, Japan).
Proliferation in monolayer culture
Following mock infection or infection with Ad-Dec2
or Ad-LacZ at the indicated MOI, MSCs were seeded at a low density
(700 cells/cm2) on 96-well plates, and incubated with
DMEM supplemented with 10% FBS at 37°C in a CO2
incubator for 4 days. Cell proliferation was estimated with a Cell
Counting kit-8 (CCK-8; Dojindo, Kumamoto, Japan) using WST-8
according to the manufacturer's instructions.
Reverse transcription-quantitative PCR
(RT-qPCR)
Chondrocyte pellets were frozen in liquid nitrogen
and crushed using an SK-Mill (Tokken, Chiba, Japan). Total RNA was
prepared using an RNeasy kit (Qiagen, Hilden, Germany) and reverse
transcribed with ReverTra Ace (Toyobo, Osaka, Japan) according to
the manufacturer's instructions. The synthesized First-Strand cDNA
was used for PCR amplification with specific primers and TaqMan
probe sets (Table I).
Quantitative PCR analyses were performed using an ABI PRISM 7900HT
Sequence Detection System instrument and software (Applied
Biosystems Inc., Foster City, CA, USA) based on the ΔΔCq method
(24). Data were normalized
against 18S rRNA levels. The PCR cycling conditions included an
incubation of 50°C for 2 min and a denaturation of 95°C for 10 min,
which was followed by 40 cycles of 95°C for 15 sec and 60°C for 1
min (25).
 | Table IPrimer and probe sequences used for
RT-qPCR. |
Table I
Primer and probe sequences used for
RT-qPCR.
| Gene | Primer (5′→3′) | Probe (5′→3′) |
|---|
| ACAN | F:
AGTATCATCAGTCCCAGAATCTAGCA
R:GGAATGCAGAGGTGGTTTCAC |
AGACGTCCGCCTATCCTGAAGCTG |
| ALPL | F:
AGACGTCCGCCTATCCTGAAGCTG
R: GCCATACAGGATGGCAGTGA |
CCCCATGCTGAGTGACACAGACAAGAA |
| COL2A1 | F:
GAGACAGCATGACGCCGAG
R: GGCTGCGGATGCTCTCAAT |
ATGCCACACTCAAGTCCCTCAACAACCA |
| COL10A1 | F:
CTAGTATCCTTGAACTTGGTTCATGGA
R: ACTGTGTCTTGGTGTTGGGTAGTG |
CGCTGAACGATACCAAACGCCCAC |
| OSX | F:
ATGAGCTGGAGCGTCATGTG
R: AGGTGGTCGCTTCGGGTAA |
TCACCTGCCTGCTCTGCTCCAAGC |
| hDEC2 | F:
GCATCAGAAGATAATTGCTTTACAGAA
R: TCTCAAACCGGGAGAGGTATTG |
CGTTCCACTCGGGATTTCAAACATGC |
| mDec2 | F:
ATTGCTTTACAGAATGGGGAGCG
R: AAAGCGCGCGAGGTATTGCAAGAC |
CGACTTGGATGCGTTCCACTCGG |
| FGF2 | F:
TTCTTCCTGCGCATCCAC
R: TGCTTGAAGTTGTAGCTTGATGT | Roche Universal
Probe #7 |
| FGF7 | F:
GCAAAGTAAAAGGGACCCAAG
R: TCACTTTCCACCCCTTTGAT | Roche Universal
Probe #59 |
| FGF18 | F:
ATGAACCGCAAAGGCAAG
R: GAACACACACTCCTTGCTGGT | Roche Universal
Probe #60 |
| Cyclin
D1 | F:
GAAGATCGTCGCCACCTG
R: GACCTCCTCCTCGCACTTCT | Roche Universal
Probe #67 |
| p21 | F:
TCACTGTCTTGTACCCTTGTGC
R: GGCGTTTGGAGTGGTAGAAA | Roche Universal
Probe #32 |
| p16INK4 | F:
GTGGACCTGGCTGAGGAG
R: CTTTCAATCGGGGATGTCTG | Roche Universal
Probe #34 |
Toluidine blue staining
In order to estimate the extent of proteoglycan
accumulation in the pellets, the diameter of the pellets was
measured using a ruler, and toluidine blue staining was performed.
The pellets were fixed in 10% buffered formalin, and embedded in
paraffin. Sections of 5 mm were prepared and stained with toluidine
blue (26). The sections were
then examined with an Olympus IX70 microscope and an Olympus DP20
camera.
Quantification of glycosaminoglycan (GAG)
and DNA content
The pellets were washed twice with
phosphate-buffered saline (PBS) and digested with 0.3 mg/ml papain
(Wako Pure Chemical) in 50 mM phosphate buffer, pH 6.5, containing
2 mM EDTA and 2 mM N-acetyl-cysteine (Wako Pure Chemical) at 60°C.
The papain-digested extracts were assayed for GAG and DNA content.
Sulfated GAG content was quantified using the Blyscan™ Sulfated
Glycosaminoglycan assay kit (Biocolor, Newtownabbey, UK) according
to the manufacturer's instructions. The DNA content of the pellets
was determined using the Quant-iT™ PicoGreen dsDNA assay kit
(Thermo Fisher Scientific) with lambda DNA as a standard.
Statistical analysis
All experiments were performed at least in
triplicate. Data are expressed as the means ± standard deviation
(SD). One-way ANOVA followed by Dunnett's post hoc test was
performed to determine the significance of differences in multiple
comparisons. P<0.05 was considered to indicate a statistically
significant difference.
Results
Endogenous expression of DEC2 during
chondrogenic differentiation of MSCs
To examine the role of DEC2 in chondrocyte
differentiation, we examined the changes in the expression levels
of DEC2 and the main chondrocyte marker genes, namely
aggrecan (ACAN), alkaline phosphatase liver/bone/kidney
(ALPL), type II collagen alpha 1 (COL2A1), type X
collagen alpha 1 (COL10A1) and osterix (OSX) in MSC
pellet cultures after exposure to chondrogenesis induction medium
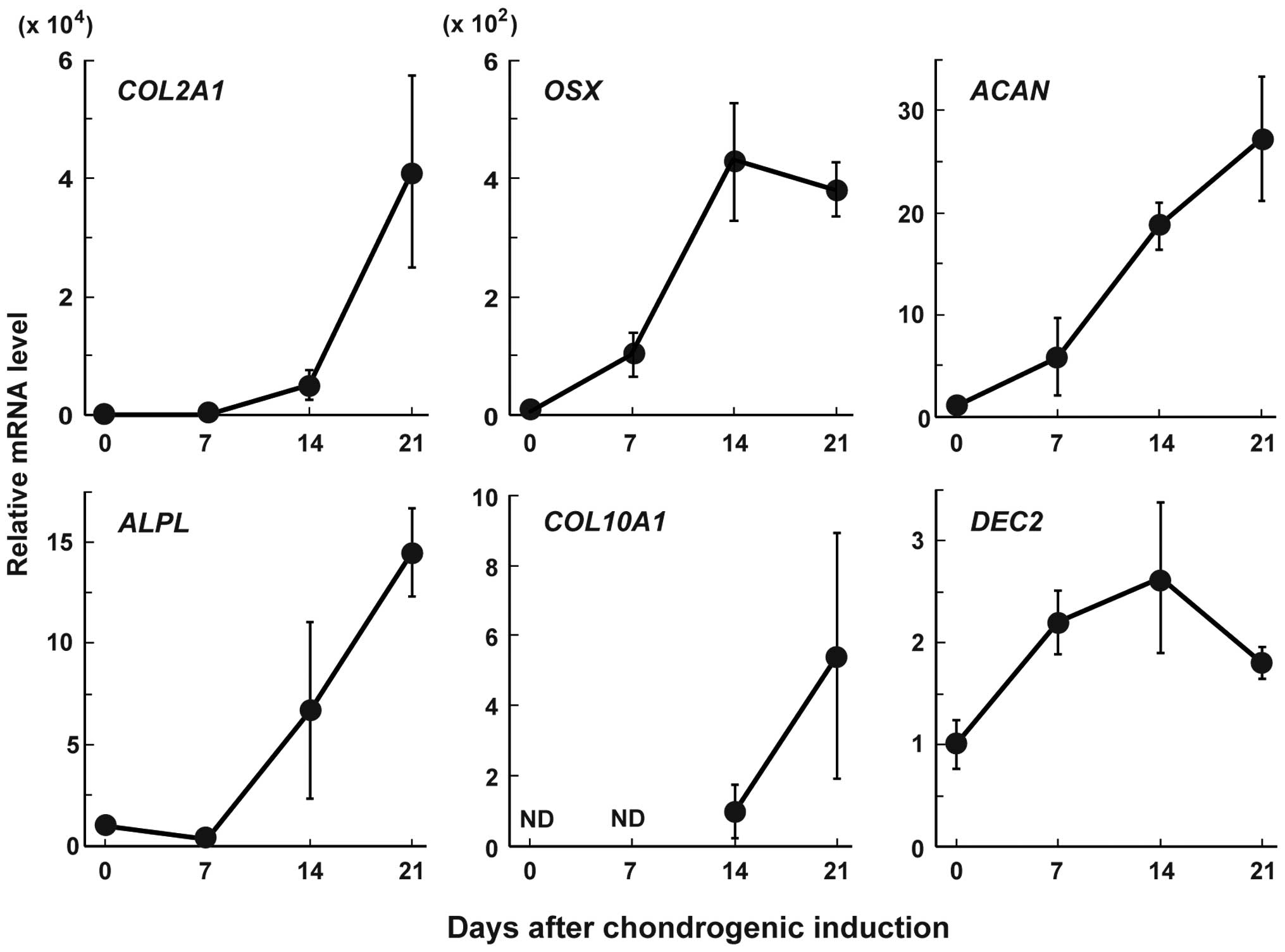
(Fig. 1). The mRNA levels of
COL2A1, OSX and ACAN started to increase on
day 7, and then those of ALPL and COL10A1 started to
increase on day 14. The expression levels of these mRNAs, with the
exception of that of OSX, continued to increase at least
until day 21. DEC2 mRNA expression started to increase on
day 7 and then decreased on day 21.
Effect of DEC2 overexpression on MSC
proliferation in monolayer cultures
MSC monolayer cultures were infected with Ad-Dec2.
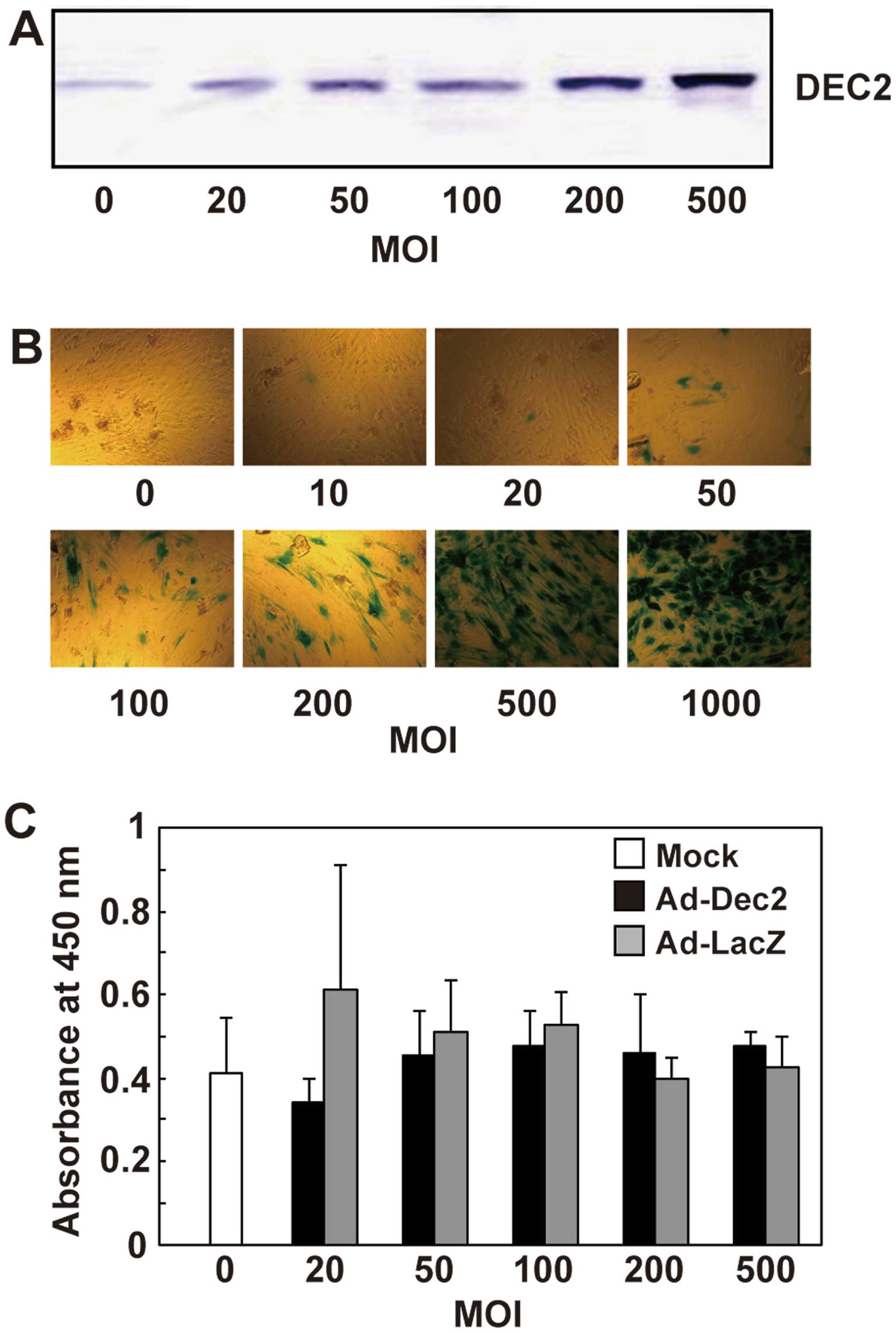
The DEC2 protein levels successfully increased in an MOI-dependent
manner up to an MOI of 500 in MSCs infected with Ad-Dec2 (Fig. 2A), and a high level of DEC2
expression in MSCs was maintained until at least 7 days after
infection with Ad-Dec2 (data not shown). In similar cultures, the
transduction efficiency of the adenovirus was determined with
Ad-LacZ. It increased the intensity of β-galactosidase staining in
an MOI-dependent manner, and >90% of the cells showed positive
staining for β-galactosidase at an MOI of 500 (Fig. 2B), suggesting that this method
allows the overexpression of DEC2 in most MSCs.
To examine the effect of DEC2 overexpression on cell
proliferation, MSCs were infected with either Ad-Dec2 or Ad-LacZ at
various MOI levels and incubated at a low density with DMEM
supplemented with 10% FBS. On day 4, there was no significant
difference with regard to proliferation between Ad-Dec2- and
Ad-LacZ-infected cells, and infection with Ad-Dec2 or Ad-LacZ did
not show cytotoxicity up to an MOI of 500 (Fig. 2C).
Effects of DEC2 overexpression on
extracellular matrix formation during chondrogenesis
MSCs were infected with either Ad-Dec2 or Ad-LacZ at
an MOI of 500 and then exposed to chondrogenesis induction medium
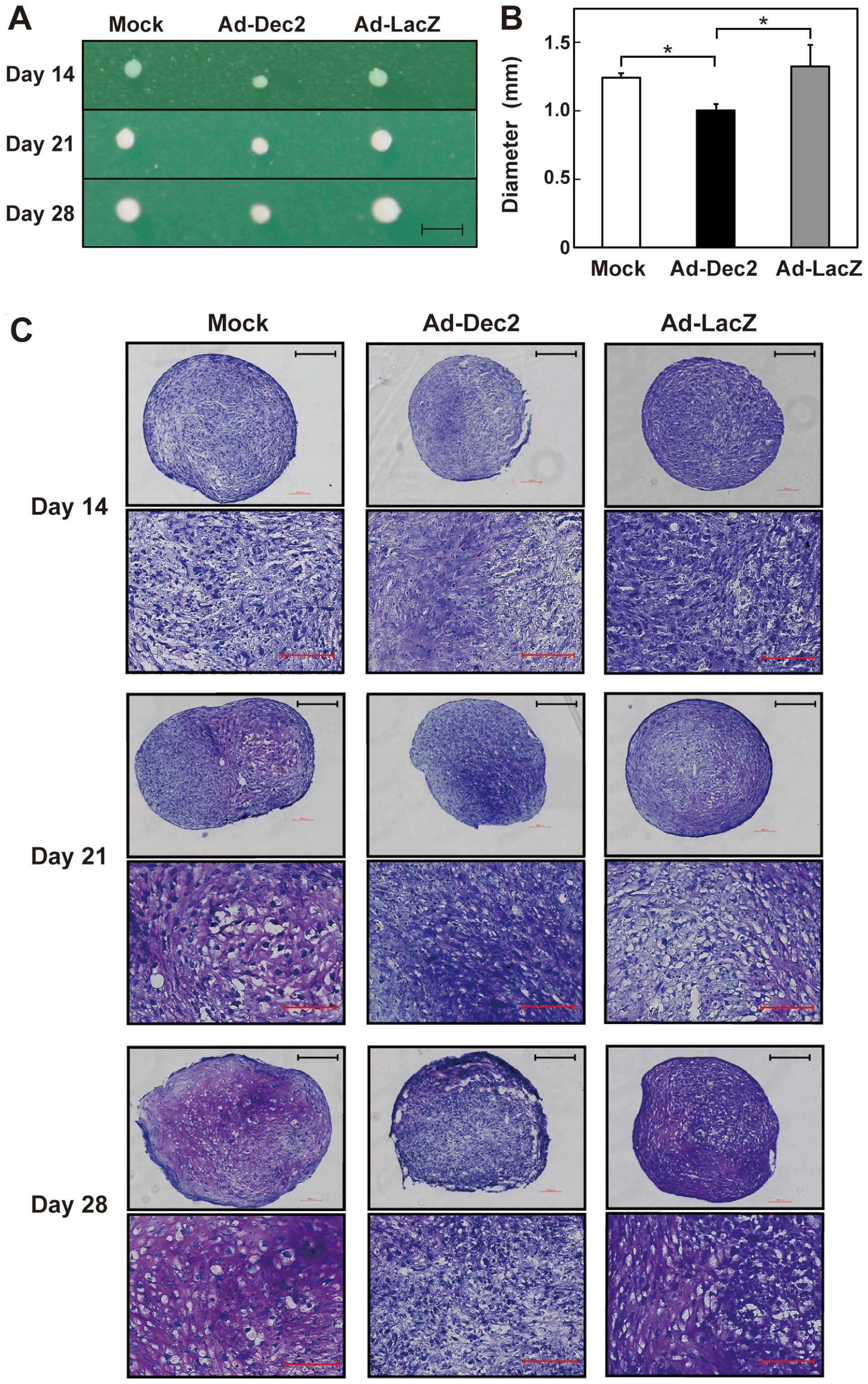
in pellet cultures. The size of these pellets gradually increased
after the onset of differentiation (Fig. 3A). However, pellets of
Ad-Dec2-infected cells were smaller than those of Ad-LacZ- or
mock-infected cells. Further, the diameter of Ad-Dec2-infected cell
pellets was significantly smaller than that of Ad-LacZ- or
mock-infected cell pellets on day 28 (Fig. 3B). There were no differences
between the diameters of Ad-LacZ- and mock-infected cell
pellets.
To estimate the extent of proteoglycan accumulation
in the pellets, we performed toluidine blue staining. On days 21
and 28, proteoglycan deposition (characterized by a red-purple
color) was observed at high levels in the Ad-LacZ- and
mock-infected pellets; however, it was scarcely detected in the
Ad-Dec2-infected pellets (Fig.
3C). Microscopic analysis also indicated the accumulation of
abundant proteoglycans and typical lacunae structures around
chondrocytes in both the Ad-LacZ- and mock-infected pellets on days
21 and 28, whereas the proteoglycan accumulation and lacunae
structures were scarcely observed in the Ad-Dec2-infected
pellets.
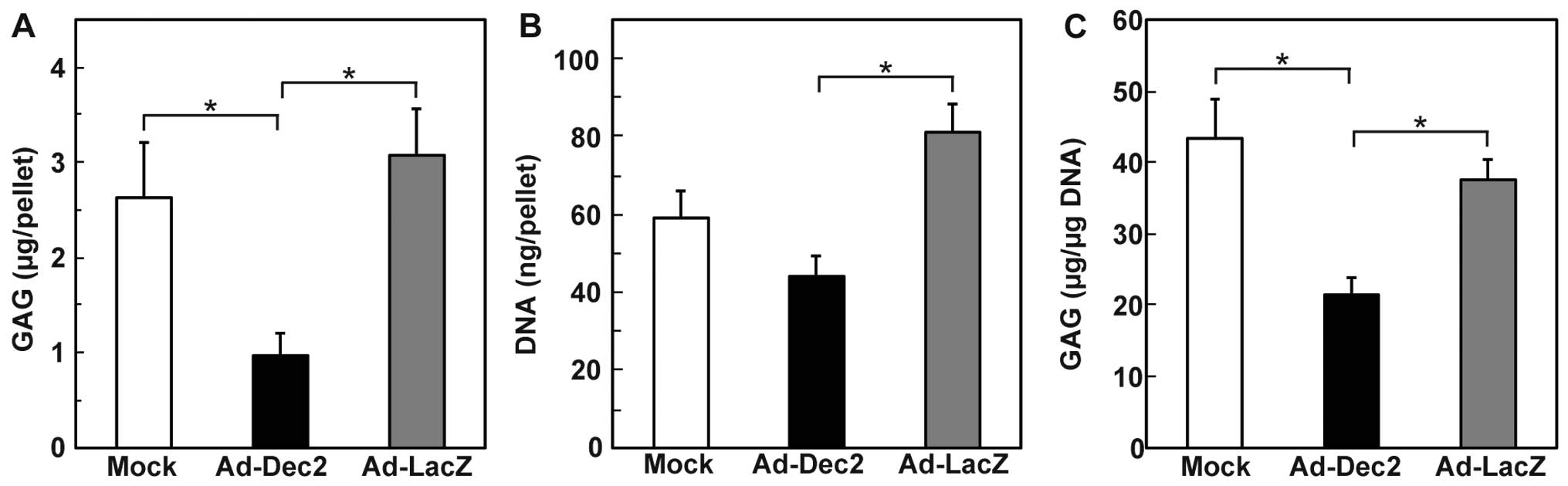
We subsequently quantified the total GAG content per
pellet (Fig. 4A), along with the
GAG content per microgram DNA on day 21 (Fig. 4B and C). The total GAG content and
GAG/DNA of the Ad-Dec2-infected pellets were lower than that in the
Ad-LacZ- and mock-infected pellets (Fig. 4A and C). In addition, a
significant decrease in the DNA content per pellet was observed in
the Ad-Dec2-infected pellets in comparison with the
Ad-LacZ-infected pellets, indicating the suppression of cell growth
by DEC2 (Fig. 4B). By contrast,
there were no significant differences between the Ad-LacZ- and
mock-infected pellets with respect to the GAG content, DNA content
and GAG/DNA.
Effects of DEC2 overexpression on the
expression of chondrocyte-related genes
We examined the effects of DEC2 overexpression on
the expression of chondrocyte-related genes in the pellet cultures.
DEC2 overexpression suppressed the expression of ACAN and
COL10A1, but not the expression of OSX and
ALPL, on day 28 (Fig. 5A).
The inhibitory effect of DEC2 on ACAN and COL10A1
expression was not observed on day 7, suggesting that DEC2 may
indirectly suppress the expression of the two chondrocyte genes. We
also examined the expression levels of Dec2 in
DEC2-overexpressing MSC pellets. The mDec2 mRNA level was
~80-fold higher than the endogenous human expression on day 7, and
the high-level expression of mDec2 decreased to less than
one tenth on day 28 (Fig. 5A),
suggesting that the infection with Ad-Dec2 affects the chondrogenic
potential of MSCs in the early stage.
Because FGFs play an important role in the control
of proliferation and differentiation of chondrocytes (21,22,27), we also examined whether DEC2
affected the expression of some FGFs. DEC2 overexpression strongly
suppressed the expression of FGF18, but not that of
FGF2 and FGF7, on days 1 and 7 (Fig. 5B).
Effects of DEC2 overexpression on the
expression of cell cycle-related genes
The regulation of cell cycle progression is closely
associated with cell proliferation and differentiation (28). Therefore, we examined whether DEC2
overexpression in MSCs altered the expression levels of the cell
cycle-related genes, namely cyclin D1, p21 and p16INK4. Cyclin D1
acts as a positive regulator of cell cycle progression, whereas p21
and p16INK4 negatively regulate the cell cycle by inhibiting
cyclin-dependent kinases (CDKs) (28). The mRNA expression of p16INK4 was
upregulated on day 1 by DEC2 overexpression in the MSC pellets
exposed to chondrogenesis induction medium but not in the monolayer
cultures exposed to growth medium (Fig. 5C and D). In the pellets, DEC2
overexpression also increased the average level of p21 and
decreased cyclin D1 expression levels, although the observed
differences were not statistically significant (Fig. 5C). These findings suggest that
DEC2 suppresses cell cycle progression of the mesenchymal cells in
pellet cultures exposed to the differentiation induction medium by
regulating the expression of cell cycle-related genes in a
stage-dependent manner.
Discussion
In this study, we demonstrated that DEC2 acts as a
negative regulator of chondrogenic differentiation. The forced
expression of DEC2 resulted in the inhibition of both cell
proliferation and GAG accumulation during chondrogenesis in MSC
pellet cultures, whereas in repeated experiments, it did not
suppress the proliferation of MSC monolayer cultures. These
findings indicate that DEC2 inhibits the proliferation of
chondrocyte lineage-committed MSCs, but not undifferentiated MSCs.
In pellet cultures of MSCs, the expression of DEC2 began to
increase earlier than that of chondrocyte markers, such as
ALPL and COL10A1, and then decreased in the late
differentiation stage, suggesting that DEC2 inhibits chondrogenic
differentiation of MSCs in premature stage, and may delay the
maturation of chondrocytes.
DEC2 suppresses the proliferation of various types
of cells, including human lung cancer cell lines (A549, NCI-H520
and NCI-H596 cells), human epidermoid carcinoma cells (HEp3), human
mammary epithelial cells (HMECs), and NIH3T3 cells (12–15), although DEC2 has little effect on
the proliferation of HepG2 cells (13). DEC2-induced cell growth arrest is
reportedly associated with decreased cyclin D1 expression in
C2C12 and NCI-H520 cells as well as HMECs (4,13,14). Consistent with these findings, the
DEC2-mediated inhibition of proliferation in MSC pellet cultures
was associated with a decrease in the average cyclin D1
levels and an increase in the average p16INK4 and p21
levels. However, DEC2 exerted minimal effects on MSC proliferation,
and the expression of p16INK4 in the undifferentiated state
in low-density monolayer cultures. Taken together, these findings
suggest that the growth inhibitory effect of DEC2 depends upon cell
type, cell density, and/or differentiation stage. However, we have
not examined the effect of DEC2 on the proliferation and
differentiation of other tissue-derived MSCs, including adipose
tissue-derived MSCs and umbilical cord-derived MSCs as bone
marrow-derived MSCs have been most widely used in studies of
chondrogenesis. This issue warrants further investigation in the
future using the other tissues-derived MSCs.
FGF signaling has been implicated in the regulation
of endochondral bone growth (21,22). Even though FGF3 receptor (FGF3R)
activation suppresses the terminal differentiation of growth plate
chondrocytes, FGF18, a ligand of FGF3R, may exert a positive effect
on the proliferation and differentiation of immature committed
chondrocytes (29–32). FGF18 knockout
(FGF18−/−) mice exhibited decreased chondrocyte
proliferation activity and a delay in the initiation of chondrocyte
hypertrophy at the embryonic stage (29). We found that DEC2 overexpression
strongly represses the expression of FGF18. Although the role of
FGF18 in cartilage development remains controversial, the signaling
may be associated with DEC2-mediated suppression of proliferation
and/or differentiation of chondrocyte-lineage committed MSCs.
FGF18 is also involved in cartilage angiogenesis
through the upregulation of vascular endothelial growth factor
(VEGF) (29). By contrast, DEC2
represses the expression of Vegf, resulting in the
reciprocal expression of Dec2 and Vegf in mouse rib
cartilage (33). Therefore, DEC2
and FGF18 appear to exert opposite effects on vasculogenesis as
well as chondrogenesis.
DEC1 and DEC2 are structurally similar, but they
often have different functions. For example, DEC1 has pro-apoptotic
effects, whereas DEC2 has anti-apoptotic effects on TNF-α-induced
apoptosis in MCF-7 cells (34).
Furthermore, DEC2, but not DEC1, suppresses the expression of
Vegf through binding with HIF1α in NIH3T3 cells (33). Previous findings have shown that
the overexpression of DEC1 promotes chondrogenic differentiation of
rabbit MSCs and mouse ATDC5 cells (23). Therefore, DEC1 and DEC2 seem to
have opposite actions on chondrogenesis. Besides direct DNA
binding, DEC1 and DEC2 modulate gene expression through
protein-protein interactions with common and distinct transcription
factors (3,5,6,8,10,17,33). Identifying the partners of DEC2
will clarify how DEC2 regulates the chondrogenic differentiation of
MSCs.
In conclusion, we have shown that DEC2 is involved
in the suppression of proliferation and differentiation of
chondrocyte lineage-committed MSCs. DEC2 inhibits the proliferation
of MSCs by regulating the expression of cell cycle-related genes
under chondrogenic conditions. The decreased proliferation of MSCs
may decrease their subsequent differentiation into chondrocytes. An
alternative possibility is that DEC2 suppresses chondrogenic
differentiation by downregulating FGF18 independently of MSC
proliferation. Besides inhibition of chondrocyte differentiation,
DEC2 suppresses the differentiation of MSCs into adipocytes
(17) and myoblasts (3,4),
suggesting that DEC2 may be implicated in maintaining MSCs in the
undifferentiated state.
Acknowledgments
The present study was supported by Grants-in-Aid for
Challenging Exploratory Research (no. 24659876) and Scientific
Research (C) (no. 22592067) from the Ministry of Education,
Culture, Sports, Science and Technology of Japan. We would like to
thank Dr Eiso Hiyama at the Natural Science Center for Basic
Research and Development, Hiroshima University for the use of their
facilities.
Abbreviations:
|
MSCs
|
mesenchymal stem cells
|
|
DEC2
|
differentiated embryo chondrocyte
2
|
|
FGF
|
fibroblast growth factor
|
|
GAG
|
glycosaminoglycan
|
|
ACAN
|
aggrecan
|
|
ALPL
|
alkaline phosphatase
liver/bone/kidney
|
|
COL2A1
|
type II collagen alpha 1
|
|
mCOL10A1
|
type X collagen alpha 1
|
|
OSX
|
osterix
|
References
|
1
|
Fujimoto K, Shen M, Noshiro M, Matsubara
K, Shingu S, Honda K, Yoshida E, Suardita K, Matsuda Y and Kato Y:
Molecular cloning and characterization of DEC2, a new member of
basic helix-loop-helix proteins. Biochem Biophys Res Commun.
280:164–171. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sun H, Ghaffari S and Taneja R:
bHLH-Orange Transcription Factors in Development and Cancer. Transl
Oncogenomics. 2:107–120. 2007.PubMed/NCBI
|
|
3
|
Fujimoto K, Hamaguchi H, Hashiba T,
Nakamura T, Kawamoto T, Sato F, Noshiro M, Bhawal UK, Suardita K
and Kato Y: Transcriptional repression by the basic
helix-loop-helix protein Dec2: Multiple mechanisms through E-box
elements. Int J Mol Med. 19:925–932. 2007.PubMed/NCBI
|
|
4
|
Azmi S, Ozog A and Taneja R: Sharp-1/DEC2
inhibits skeletal muscle differentiation through repression of
myogenic transcription factors. J Biol Chem. 279:52643–52652. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Honma S, Kawamoto T, Takagi Y, Fujimoto K,
Sato F, Noshiro M, Kato Y and Honma K: Dec1 and Dec2 are regulators
of the mammalian molecular clock. Nature. 419:841–844. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Choi SM, Cho HJ, Cho H, Kim KH, Kim JB and
Park H: Stra13/DEC1 and DEC2 inhibit sterol regulatory element
binding protein-1c in a hypoxia-inducible factor-dependent
mechanism. Nucleic Acids Res. 36:6372–6385. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hamaguchi H, Fujimoto K, Kawamoto T,
Noshiro M, Maemura K, Takeda N, Nagai R, Furukawa M, Honma S, Honma
K, et al: Expression of the gene for Dec2, a basic helix-loop-helix
transcription factor, is regulated by a molecular clock system.
Biochem J. 382:43–50. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kato Y, Kawamoto T, Fujimoto K and Noshiro
M: DEC1/STRA13/SHARP2 and DEC2/SHARP1 coordinate physiological
processes, including circadian rhythms in response to environmental
stimuli. Curr Top Dev Biol. 110:339–372. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rossner MJ, Oster H, Wichert SP, Reinecke
L, Wehr MC, Reinecke J, Eichele G, Taneja R and Nave KA: Disturbed
clockwork resetting in Sharp-1 and Sharp-2 single and double mutant
mice. PLoS One. 3:e27622008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Montagner M, Enzo E, Forcato M, Zanconato
F, Parenti A, Rampazzo E, Basso G, Leo G, Rosato A, Bicciato S, et
al: SHARP1 suppresses breast cancer metastasis by promoting
degradation of hypoxia-inducible factors. Nature. 487:380–384.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sato F, Kawamura H, Wu Y, Sato H, Jin D,
Bhawal UK, Kawamoto T, Fujimoto K, Noshiro M, Seino H, et al: The
basic helix-loop-helix transcription factor DEC2 inhibits
TGF-β-induced tumor progression in human pancreatic cancer BxPC-3
cells. Int J Mol Med. 30:495–501. 2012.PubMed/NCBI
|
|
12
|
Adam AP, George A, Schewe D, Bragado P,
Iglesias BV, Ranganathan AC, Kourtidis A, Conklin DS and
Aguirre-Ghiso JA: Computational identification of a
p38SAPK-regulated transcription factor network required for tumor
cell quiescence. Cancer Res. 69:5664–5672. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Falvella FS, Colombo F, Spinola M,
Campiglio M, Pastorino U and Dragani TA: BHLHB3: a candidate tumor
suppressor in lung cancer. Oncogene. 27:3761–3764. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Y, Shen Q, Kim HT, Bissonnette RP,
Lamph WW, Yan B and Brown PH: The rexinoid bexarotene represses
cyclin D1 transcription by inducing the DEC2 transcriptional
repressor. Breast Cancer Res Treat. 128:667–677. 2011. View Article : Google Scholar
|
|
15
|
Liu JJ, Chung TK, Li J and Taneja R:
Sharp-1 modulates the cellular response to DNA damage. FEBS Lett.
584:619–624. 2010. View Article : Google Scholar :
|
|
16
|
Wu Y, Sato H, Suzuki T, Yoshizawa T,
Morohashi S, Seino H, Kawamoto T, Fujimoto K, Kato Y and Kijima H:
Involvement of c-Myc in the proliferation of MCF-7 human breast
cancer cells induced by bHLH transcription factor DEC2. Int J Mol
Med. 35:815–820. 2015.
|
|
17
|
Gulbagci NT, Li L, Ling B, Gopinadhan S,
Walsh M, Rossner M, Nave KA and Taneja R: SHARP1/DEC2 inhibits
adipogenic differentiation by regulating the activity of C/EBP.
EMBO Rep. 10:79–86. 2009. View Article : Google Scholar :
|
|
18
|
Yang XO, Angkasekwinai P, Zhu J, Peng J,
Liu Z, Nurieva R, Liu X, Chung Y, Chang SH, Sun B and Dong C:
Requirement for the basic helix-loop-helix transcription factor
Dec2 in initial TH2 lineage commitment. Nat Immunol. 10:1260–1266.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Azmi S and Taneja R: Embryonic expression
of mSharp-1/mDEC2, which encodes a basic helix-loop-helix
transcription factor. Mech Dev. 114:181–185. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pittenger MF, Mackay AM, Beck SC, Jaiswal
RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and
Marshak DR: Multilineage potential of adult human mesenchymal stem
cells. Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Horton WA and Degnin CR: FGFs in
endochondral skeletal development. Trends Endocrinol Metab.
20:341–348. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ellman MB, Yan D, Ahmadinia K, Chen D, An
HS and Im HJ: Fibroblast growth factor control of cartilage
homeostasis. J Cell Biochem. 114:735–742. 2013. View Article : Google Scholar :
|
|
23
|
Shen M, Yoshida E, Yan W, Kawamoto T,
Suardita K, Koyano Y, Fujimoto K, Noshiro M and Kato Y: Basic
helix-loop-helix protein DEC1 promotes chondrocyte differentiation
at the early and terminal stages. J Biol Chem. 277:50112–50120.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(− Delta DeltaC(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Fujii S, Fujimoto K, Goto N, Kanawa M,
Kawamoto T, Pan H, Srivatanakul P, Rakdang W, Pornprasitwech J,
Saskianti T, et al: Characteristic expression of, and in dental
pulp cells. Biomed Rep. 3:566–572. 2015.PubMed/NCBI
|
|
26
|
Suardita K, Fujimoto K, Oda R, Shimazu A,
Miyazaki K, Kawamoto T and Kato Y: Effects of overexpression of
membrane-bound transferrin-like protein (MTf) on chondrogenic
differentiation in Vitro. J Biol Chem. 277:48579–48586. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Correa D, Somoza RA, Lin P, Greenberg S,
Rom E, Duesler L, Welter JF, Yayon A and Caplan AI: Sequential
exposure to fibroblast growth factors (FGF) 2, 9 and 18 enhances
hMSC chondrogenic differentiation. Osteoarthritis and
cartilage/OARS. Osteoarthritis Res Soc. 23:443–453. 2015.
View Article : Google Scholar
|
|
28
|
Budirahardja Y and Gönczy P: Coupling the
cell cycle to development. Development. 136:2861–2872. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu Z, Lavine KJ, Hung IH and Ornitz DM:
FGF18 is required for early chondrocyte proliferation, hypertrophy
and vascular invasion of the growth plate. Dev Biol. 302:80–91.
2007. View Article : Google Scholar
|
|
30
|
Liu Z, Xu J, Colvin JS and Ornitz DM:
Coordination of chondrogenesis and osteogenesis by fibroblast
growth factor 18. Genes Dev. 16:859–869. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yamaoka H, Nishizawa S, Asawa Y, Fujihara
Y, Ogasawara T, Yamaoka K, Nagata S, Takato T and Hoshi K:
Involvement of fibroblast growth factor 18 in dedifferentiation of
cultured human chondrocytes. Cell Prolif. 43:67–76. 2010.
View Article : Google Scholar
|
|
32
|
Mori Y, Saito T, Chang SH, Kobayashi H,
Ladel CH, Guehring H, Chung UI and Kawaguchi H: Identification of
fibroblast growth factor-18 as a molecule to protect adult
articular cartilage by gene expression profiling. J Biol Chem.
289:10192–10200. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sato F, Bhawal UK, Kawamoto T, Fujimoto K,
Imaizumi T, Imanaka T, Kondo J, Koyanagi S, Noshiro M, Yoshida H,
et al: Basic-helix-loop-helix (bHLH) transcription factor DEC2
negatively regulates vascular endothelial growth factor expression.
Genes Cells. 13:131–144. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu Y, Sato F, Bhawal UK, Kawamoto T,
Fujimoto K, Noshiro M, Morohashi S, Kato Y and Kijima H: Basic
helix-loop-helix transcription factors DEC1 and DEC2 regulate the
paclitaxel-induced apoptotic pathway of MCF-7 human breast cancer
cells. Int J Mol Med. 27:491–495. 2011.PubMed/NCBI
|