Instruction
Glioma is the most common malignant tumor of the
brain, and accounts for approximately 30% of central nervous system
tumors and 80% of all malignant brain tumors (1). Despite the remarkable development of
therapies for other types of cancer in recent decades, the
prognosis of patients with advanced glioma remains poor, mainly due
to its resistance to radiotherapy, chemotherapy and adjuvant
therapies (2–5). The deregulation of oncogenes or
tumor suppressors has been implicated in glioma (6). Therefore, the investigation of the
roles of genetic and epigenetic factors may aid in the developmetn
of novel diagnostic and therapeutic strategies for glioma (7).
MicroRNAs (miRNAs or miRs), a class of non-coding
RNAs 18–25 nucleotides in length, which are able to suppress gene
expression by targeting the complementary regions of mRNAs and
inhibiting protein translation (8). By negatively mediating their target
genes, miRNAs act as key regulators in a variety of physiological
and pathological biological processes, including tumorigenesis
(9,10). The deregulation of miRNAs has been
observed in glioma, and is associated with tumor growth, metastasis
and drug resistance (11,12). Therefore, the investigation of the
regulatory mechanisms of miRNAs is important for the treatment of
glioma.
Recently, miR-16 has been implicated in the
development and progression of glioma (13). Malzkorn et al investigated
the expression profiles of 157 miRNAs in 4 patients with primary
WHO grade II gliomas that spontaneously progressed to WHO grade IV
secondary glioblastomas, and found that miR-16 showed increased an
expression upon progression, suggesting that its upregulation may
play a role in the progression of glioma (14). On the contrary, however, the
following studies identified miR-16 as a tumor suppressor in
glioma, by demonstrating that miR-16 exerts inhibitory effects on
growth, migration, invasion, epithelial-mesenchymal transition
(EMT), and angiogenesis in glioma (13,15–17). Moreover, several targets of miR-16
have been identified in glioma, including Zyxin, Bcl-2, matrix
metalloproteinase (MMP)9 and BMI1 proto-oncogene, polycomb ring
finger Bmi-1 (13,15,17). As one miRNA has many targets
(18), whether other targets are
also involved in the miR-16-mediated inhibition of glioma remains
unknown.
Sal-like protein 4 (SALL4) is a zinc finger
transcription factor, and has been identified as a marker for stem
cells, involved in the maintenance of self-renewal in embryonic
stem cells (19). Moreover, SALL4
was recently identified as an important biomarker for several
common human cancers (20,21).
Zhang et al found that SALL4 was significantly upregulated
in glioma, and a high level of SALL4 expression correlated with a
poor outcome (22). A previous
study demonstrated that the upregulation of SALL4, caused by the
low expression of miR-107, inhibited cell apoptosis in glioma
(23). Accordingly, SALL4 acts as
an oncogene in glioma, and may become an important target for the
treatment of glioma. However, evidence of the regulatory mechanisms
of SALL4 expression in glioma is limited.
In the present study, we aimed to investigate the
regulatory mechanisms of miR-16 in glioma growth and metastasis. We
found that miR-16 was significantly downregulated in glioma tissues
compared to normal brain tissues, and that the reduced miR-16
levels were associated with a greater malignancy of glioma. We
further revealed that miR-16 inhibited cell proliferation,
migration and invasion, and EMT in glioma, at least in part by
directly targeting SALL4, which was markedly upregulated in glioma
tissues and inversely correlated with the miR-16 levels.
Materials and methods
Ethics statement and clinical sample
collection
This study was approved by the Ethics Committee of
Central South University, Changsha, China. A total of 23 cases of
glioma specimens and 7 cases of non-tumorous brain tissues were
collected from the Second Xiangya Hospital of Central South
University between January 2012 and March 2013. Non-tumorous brain
tissues were collected by the partial resection of normal brain
tissue in order to reduce increased intracranial pressure in the
treatment of severe head injury. All tissue samples were
immediately snap-frozen in liquid nitrogen and stored at −80°C
until use.
Written informed consent was obtained from all
patients involved in this study. All glioma patients included 14
males and 9 females who ranged in age from 35 to 71 years, with a
mean age of 51.3 years. None of the patients had any radiotherapy
or chemotherapy prior to surgical resection. The glioma specimens
were classified according to the World Health Organization (WHO)
criteria (24). Among these
glioma samples, 5 cases were pilocytic astrocytomas (WHO I), 6 were
diffuse astrocytomas (WHO II), 6 were anaplastic astrocytomas (WHO
III), and 6 were glioblastomas (WHO IV).
Cell culture and transfection
The human glioma cell lines, U87 and U251, were
obtained from the Chinese Academy of Sciences (Shanghai, China).
The cells were cultured in Dulbecco's modified Eagle's medium
(DMEM) supplemented with 10% fetal bovine serum (FBS) (both from
Life Technologies, Carlsbad, CA, USA) at 37°C in a humidified
incubator containing 5% CO2. Lipofectamine 2000 (Life
Technologies) was used to perform transfection according to the
manufacturer's instructions. Briefly, the U87 and U251 cells were
cultured to 70% confluence, and resuspended in serum-free medium.
Scramble miR (miR-NC), miR-16 mimic, miR-16 inhibitor (all from
Genecopoeia, Rockville, MD, USA), the pc-DNA3.1-SALL4 plasmid
(Amspring, Changsha, China), and Lipofectamine 2000 were diluted in
OPTI-MEM (Life Technologies). The diluted Lipofectamine 2000 was
added to the diluted miR or plasmid, and incubated for 20 min at
room temperature, and then added to the cell suspension. Following
incubation at 37°C, 5% CO2 for 6 h, the transfection
mixture was replaced with DMEM with 10% FBS. The cells were then
cultured for 48 h prior to being used in the following assays.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the tissues and cells
using TRIzol Reagent (Life Technologies) in accordance with the
manufacturer' s instructions. RT-qPCR was used to examine the
relative miR-16 expression using the mirVana™ qRT-PCR microRNA
detection kit (Life Technologies) in accordance with the
manufacturer' s instructions. U6 was used as an internal reference.
The relative mRNA expression of SALL4 was detected by RT-qPCR using
the standard SYBR-Green RT-PCR kit (Takara, Otsu, Japan) in
accordance with the manufacturer's instructions. Glyceraldehyde
3-phosphate dehydrogenase (GAPDH) was used as an internal
reference. For both miRNA and mRNA detection, the reaction
conditions were 95°C for 3 min, followed by 40 cycles of 95°C for
15 sec and 60°C for 30 sec. The specific primers for miR-16 and U6
were purchased from Genecopoeia. The specific primers for SALL4
were as follows: forward, 5′-TAGCCCTGCGTA GCCAGTTA-3′ and reverse,
5′-TCATGCTTAGTCCACT GTCTGT-3′. The specific primers for GAPDH were
as follows: forward, 5′-ACAACTTTGGTATCGTGGAAGG-3′ and reverse,
5′-GCCATCACGCCACAGTTTC-3′. The relative expression level was
quantified using the 2−ΔΔCt method.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
Cell proliferation was examined by MTT assay. The
U87 and U251 cells (2×103) were seeded in a 96-well
plate. Each well was supplemented with 100 µl of fresh serum-free
medium with 0.5 g/l MTT. Following incubation at 37°C, 5%
CO2 for 12, 24, 48 and 72 h, the medium containing MTT
was removed, and 50 μl of DMSO were added to each well. Following
incubation at 37°C, 5% CO2 for 10 min, the absorbance at
570 nm (A570) of each sample was measured using a plate reader
(Tecan Infinite M200; Tecan, Männedorf, Switzerland).
Wound healing assay
Wound healing assay was used to examine the
migratory capacity of the glioma cells. The U87 and U251 cells were
cultured to full confluence. Wounds of approximately 1 mm in width
were created using a plastic scriber. The cells were washed and
then cultured in DMEM containing 10% FBS for 48 h. The cells were
then observed and photographed under a microscope (BX53; Olympus,
Tokyo, Japan).
Transwell assay
Cell invasion was examined using 24-well Transwell
chambers with a layer of Matrigel (Chemicon, Temecula, CA, USA).
For each group, 300 μl of cell suspension, each containing
5,000 cells, was added to the upper chamber. DMEM containing 10%
FBS was added to the lower chamber. Following culture for 24 h,
non-invading cells on the interior of the inserts were removed
using a cotton-tipped swab. Invading cells on the lower surface of
the inserts were stained with 0.1% gentian violet (Sigma, St.
Louis, MO, USA), rinsed with water, and dried in air. The invading
cells were observed under a microscope (BX53; Olympus). The cell
number was counted.
Western blot analysis
The cells were lysed in protein lysis buffer (Xinyu
Biotechnology, Shanghai, China). The protein concentration was
determined using the BCA Protein assay kit (Pierce Chemical Co.,
Rockford, IL, USA). Protein was separated with 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred
onto a PVDF membrane (Life Technologies), and then blocked in 5%
non-fat dried milk (Mengniu, Beijing, China) in TBST (Sigma) for 2
h. The PVDF membrane was then incubated with primary antibodies
against SALL4 (rabbit polyclonal; ab29112) or GAPDH (rabbit
polyclonal; ab9485) (Abcam, Cambridge, MA, USA) at 4°C overnight,
and then washed with TBST 4 times. The PVDF membrane was incubated
with mouse anti-rabbit secondary antibody (ab99702; Abcam) for 1 h
at room temperature, and then washed with TBST 3 times. The immune
complexes were then detected using the ECL western blotting kit
(Pierce Chemical Co.) and X-film (Kodak, Tokyo, Japan). ImageJ
software was used to analyze the relative protein expression,
represented as the density ratio versus GAPDH.
Bioinformatics analysis
TargetScan Human 7.0 online software (www.targetscan.org) was used to predict the putative
target of miR-16.
Dual luciferase reporter assay
The wild-type (WT) sequence of the 3′UTR of SALL4
was constructed by PCR and inserted into the pMiR-Report miRNA
Expression Reporter vector (Thermo Fisher Scientific, Carlsbad, CA
USA). The mutant type (MT) sequence of the 3′UTR of SALL4 was
constructed using the Easy Mutagenesis System kit (Promega,
Madison, WI, USA) in accordance with the manufacturer's
instructions, and then inserted into the pMiR-Report miRNA
Expression Reporter vector. The U87 and U251 cells were
co-transfected with the WT SALL4-3′UTR plasmid (200 ng) or the MT
SALL4-3′UTR plasmid (200 ng), and miR-NC (100 nM) or miR-16 mimic
(100 nM), using Lipofectamine 2000. Following co-transfection for
48 h, the dual-luciferase reporter assay system (Promega) was used
to determine the activities of Renilla luciferase and
Firefly luciferase. The Renilla luciferase activity was
normalized to the Firefly luciferase activity.
Statistical analysis
The data in this study are expressed as the means ±
SD. Statistical analysis was performed using SPSS 17.0 (SPSS,
Armonk, NY, USA). The differences between 2 groups were analyzed
using the Student' t-test. The differences among more than 2 groups
were analyzed using ANOVA. A value of P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-16 is downregulated in glioma
To reveal the role of miR-16 in glioma, RT-qPCR was
used to determine its expression levels. The expression levels of
miR-16 were markedly reduced in the glioma tissues compared to the
normal brain tissues (Fig. 1).
Moreover, its levels were markedly lower in the glioma tissues at
stages T2-T4 compared to those at stage T1, suggesting that its
downregulation was associated with the malignant progression of
glioma. Therefore, miR-16 is downregulated in glioma, and the lower
miR-16 levels were associated with its malignant progression.
miR-16 inhibits the proliferation,
migration and invasion of U87 and U251 glioma cells
As miR-16 was found to be downregulated in glioma,
we transfected the U87 and U251 glioma cells with miR-16 mimic in
order to upregulate its expression. Following transfection, the
miR-16 levels were markedly increased compared with those in the
cells transfected with the scramble (control) miR (Fig. 2A and B). MTT assay, wound healing
assay and Transwell assay were further used to examine the
proliferation, migration and invasion of glioma cells,
respectively. We observed that the overexpression of miR-16
significantly suppressed the proliferation, migration and invasion
of the U87 and U251 cells compared to the control cells (Fig. 2C–H). Therefore, miR-16 exerts
inhibitory effects on the proliferation, migration and invasion of
glioma cells.
SALL4, a target gene of miR-16, is
negatively mediated by miR-16 in glioma cells
We then investigated the putative targets of miR-16
in glioma cells. Bioinformatics analysis predicted that SALL4 was a
potential target gene of miR-16, and their targeting relationship
was evolutionarily conserved (Fig. 3A
and B). To verify their targeting relationship, the luciferase
reporter plamids containing the WT or MT of SALL4 3′UTR were
generated (Fig. 3C and D), and
luciferase reporter assay was conducted using the U87 and U251
cells. As demonstrated in Fig. 3E and
F, the luciferase activity was significantly decreased in the
U87 and U251 cells co-transfected with the WT SALL4 3′UTR plamid
and miR-16 mimic, but was unaltered in the U87 and U251 cells
co-transfected with the MT SALL4 3′UTR plamid and miR-16 mimic,
when compared to the control group, respectively, indicating that
SALL4 is a direct target gene of miR-16.
As miRNAs inhibit the expression of their target
genes, we then examined the effects of miR-16 overexpression on the
expression of SALL4 in glioma cells. We found that the
overexpression of miR-16 did not affect the mRNA expression of
SALL4 (Fig. 3G), but
significantly inhibited the protein expression of SALL4 in the U87
and U251 cells (Fig. 3H).
Therefore, miR-16 can inhibit the expression of SALL4 at the
post-transcriptional level in glioma cells by directly targeting
the 3′UTR of SALL4 mRNA.
SALL4 is involved in the miR-16-mediated
inhibition of the proliferation, migration and invasion of glioma
cells
As SALL4 has been reported to be upregulated in
glioma and to be associated with its malignant progression
(22), we speculated that SALL4
may be involved in the miR-16-mediated proliferation, migration and
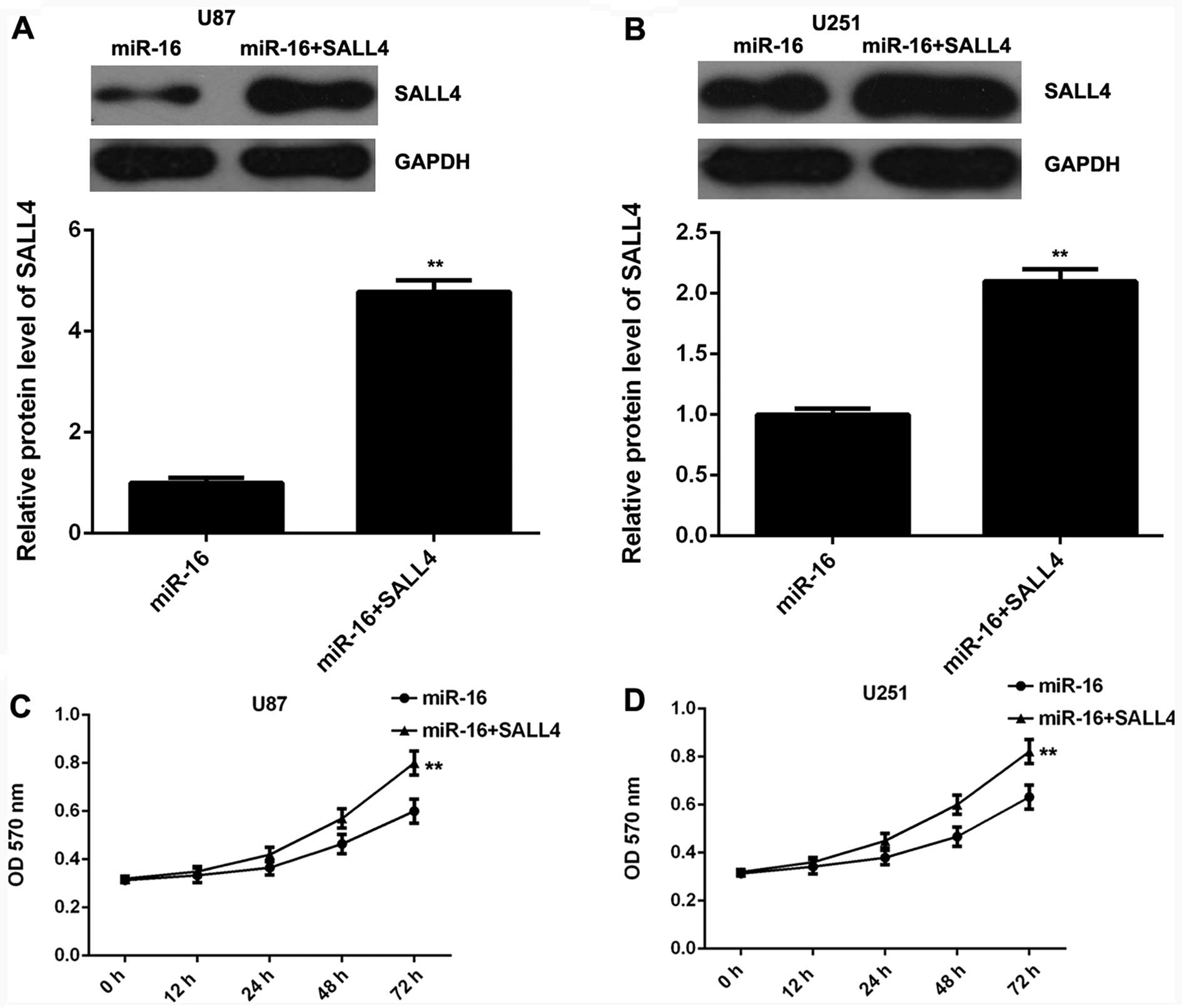
invasion of glioma cells. miR-16-overexpressing glioma cells were
transfected with the pc-DNA3.1-SALL4 plasmid to restore SALL4
expression. Following transfection, western blot analysis was
conducted to examine the protein levels of SALL4 in each group. The
SALL4 protein levels were markedly higher in the U87 and U251 cells
co-trasnfected with the miR-16 mimic and SALL4 overexpression
plasmid, when compared to the U87 and U251 cells transfected only
with the miR-16 mimic (Fig. 4A and
B). Subsequently, MTT assay, wound healing assay and Transwell
assay were performed to examine the proliferation, migration and
invasion of glioma cells in each group, respectively. Our data
demonstrated that the proliferation, migration and invasion of thye
U87 and U251 cells were significantly enhanced following
co-transfection with themiR-16 mimic and SALL4 plasmid, when
compared to the cells transfected only with the miR-16 mimic
(Fig. 4C–H), indicating that the
overexpression of SALL4 reversed the suppressive effects of miR-16
overexpression on glioma cell proliferation, migration and
invasion. Based on these data, we suggest that miR-16 inhibits the
proliferation, migration and invasion of glioma cells, at least
partly by directly targeting SALL4.
SALL4 is upregulated in glioma tissues
and its expression inversely correlates with the miR-16 levels
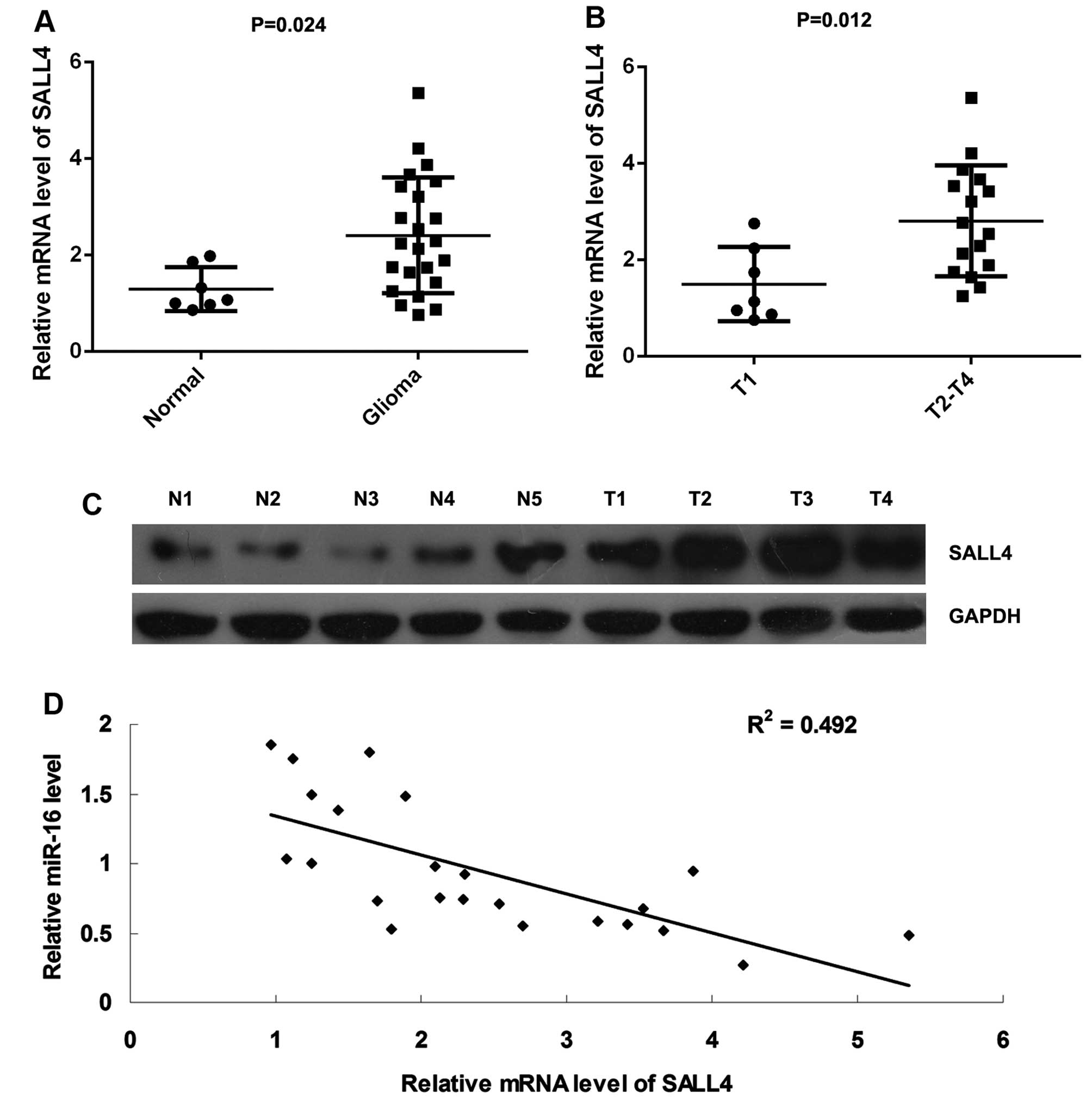
Finally, we conducted RT-qPCR to examine the mRNA
levels of SALL4 in glioma tissues and normal brain tissues. We
found that SALL4 was significantly upregulated in thye glioma
tissues compared with the normal brain tissues (Fig. 5A). Moreover, its mRNA levels were
markedly higher in the glioma samples at stages T2-T4, when
compared with those at T1 stage, suggesting that its upregulation
was associated with the malignant progression of glioma (Fig. 5B). Moreover, the results of
western blot analysis further demonstrated that the protein
expression of SALL4 was increased in the glioma tissues compared
with the normal brain tissues (Fig.
5C). In addition, we found that the mRNA levels of SALL4
inversely correlated with the miR-16 levels in glioma tissues
(Fig. 5D), suggesting that the
downregulation of miR-16 may be an important cause for the
upregulation of SALL4 in glioma.
Discussion
Recently, miR-16 has been demonstrated to play a
role in the inhibition of the growth and metastasis of glioma
(13,15–17). However, the underlying mechanisms
remain to be fully elucidated. In this study, we found that the
miR-16 levels were markedly decreased in glioma tissues compared to
normal brain tissues, and its downregulation was associated with
the malignant progression of the disease. Further in vitro
experiments revealed that the overexpression of miR-16 suppressed
the proliferation, migration and invasion of glioma cells by
directly targeting SALL4, which was significantly upregulated and
inversely correlated with the miR-16 levels in glioma tissues.
The central tumor suppressor, p53, has been found to
enhance the post-transcriptional maturation of miR-16 with
growth-suppressive function in response to DNA damage (25). Moreover, miR-16 has been
demonstrated to be deregulated and to play a role in a variety of
human cancers. Amaral et al reported that miR-16 was
significantly downregulated in ACTH-secreting pituitary tumors when
compared to normal pituitary tissues (26). Furthermore, miR-16 has been shown
to be deleted or downregulated in the majority of chronic
lymphocytic leukemia cases, and to induce the apoptosis of leukemic
cells by directly targeting Bcl-2 (27). Recently, miR-16 was found to act
as a tumor suppressor in glioma (13,15–17). For instance, Li et al found
that miR-16 inhibited the expression of Zyxin, and suppressed the
proliferation, migration and invasion of high-invasive glioma cells
(13). Wang et al reported
that miR-16 suppressed the invasion, adhesion, cell cycle
progression, production of interleukin (IL)-6, IL-8 and
transforming growth factor-β, and EMT-related gene expression,
including vimentin, β-catenin and E-cadherin in U87 and U251 glioma
cells (16). Yang et al
showed that miR-16 suppressed glioma cell growth and invasion,
while it induced cell apoptosis in vitro and in vivo
by inhibiting NF-κB1, MMP9 and Bcl-2 (17). In the present study, we found that
miR-16 was downregulated in glioma tissues compared to normal brain
tissues, and its downregulation was associated with the tumor
grade, TMN stage and vascular invasion of glioma. These data
suggest that miR-16 may serve as a diagnostic marker for
glioma.
Moreover, we identified SALL4 as a direct target
gene of miR-16 by using luciferase reporter assay, and found that
the overexpression of miR-16 led to a significant decrease in the
protein levels of SALL4 in glioma U87 and U251 cells. SALL4, a stem
cell-related transcription factor, has recently been demonstrated
to play an oncogenic role in many common human cancers, such as
hepatocellular carcinoma, endometrial cancer, lung cancer,
colorectal cancer, esophageal squamous cell carcinoma and breast
cancer, by enhancing tumor cell survival, growth, metastasis,
angiogenesis and drug resistance (28–33). Recently, Zhang et al
reported that the expression of SALL4 was significantly increased
in glioma specimens compared to normal brain tissues, and that its
upregulation tightly correlated with a higher pathological grade,
as well as with a poor prognosis (22). Moreover, the knockdown of SALL4
effectively suppressed the proliferation of U251 cells (22). Accordingly, we hypothesized that
the tumor suppressive role of miR-16 may be mediated through the
inhibition of SALL4. To verify this hypothesis,
miR-16-overexpressing glioma cells were further transfected with a
pc-DNA3.1-SALL4 plasmid to restore its protein levels. Our data
indicated that the overexpression of SALL4 reversed the inhibitory
effects of miR-16 on the proliferation, migration and invasion of
U87 and U251 cells. Therefore, we suggest that miR-16 inhibits the
malignant phenotypes of glioma cells by directly targeting SALL4.
We further found that SALL4 was upregulated in glioma, consistent
with previous findings (22), and
that its upregulation inversely correlated with the downregulation
of miR-16 in glioma tissues. These data further suggest that
downregulation of miR-16 may contribute to the upregulation of
SALL4 in glioma. In future studies, we aim to further investigate
the role of miR-16 and SALL4 in glioma in vivo, and to
explore the downstream factors of the miR-16/SALL4 axis in
glioma.
In addition, another miRNA (miR-107) was found to
play a role in glioma by targeting SALL4. He et al reported
that miR-107 was significantly downregulated in glioma, and
suppressed glioma cell growth by directly targeting SALL4, leading
to the activation of FADD/caspase-8/caspase-3/7 signaling pathway
of cell apoptosis (23).
Therefore, our study expands the understanding of the functions of
miRNAs in glioma.
In conclusion, this study demonstrates that miR-16
plays a suppressive role in regulating cell proliferation,
migration and invasion, and EMT in glioma, at least in part by
directly targeting SALL4. Therefore, we suggest that the
miR-16/SALL4 axis may serve as a potential therapeutic target for
the treatment of glioma.
References
|
1
|
Goodenberger ML and Jenkins RB: Genetics
of adult glioma. Cancer Genet. 205:613–621. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stewart LA: Chemotherapy in adult
high-grade glioma: A systematic review and meta-analysis of
individual patient data from 12 randomised trials. Lancet.
359:1011–1018. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhu VF, Yang J, Lebrun DG and Li M:
Understanding the role of cytokines in Glioblastoma Multiforme
pathogenesis. Cancer Lett. 316:139–150. 2012. View Article : Google Scholar
|
|
4
|
Sathornsumetee S, Reardon DA, Desjardins
A, Quinn JA, Vredenburgh JJ and Rich JN: Molecularly targeted
therapy for malignant glioma. Cancer. 110:13–24. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pulkkanen KJ and Yla-Herttuala S: Gene
therapy for malignant glioma: Current clinical status. Mol Ther.
12:585–598. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Marumoto T and Saya H: Molecular biology
of glioma. Adv Exp Med Biol. 746:2–11. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen C and Wang G: Mechanisms of
hepatocellular carcinoma and challenges and opportunities for
molecular targeted therapy. World J Hepatol. 7:1964–1970. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tessitore A, Cicciarelli G, Del Vecchio F,
Gaggiano A, Verzella D, Fischietti M, Vecchiotti D, Capece D,
Zazzeroni F and Alesse E: MicroRNAs in the DNA Damage/Repair
Network and Cancer. Int J Genomics. 2014:8202482014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brower JV, Clark PA, Lyon W and Kuo JS:
MicroRNAs in cancer: Glioblastoma and glioblastoma cancer stem
cells. Neurochem Int. 77:68–77. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Auffinger B, Thaci B, Ahmed A, Ulasov I
and Lesniak MS: MicroRNA targeting as a therapeutic strategy
against glioma. Curr Mol Med. 13:535–542. 2013. View Article : Google Scholar
|
|
13
|
Li X, Ling N, Bai Y, Dong W, Hui GZ, Liu
D, Zhao J and Hu J: MiR-16-1 plays a role in reducing migration and
invasion of glioma cells. Anat Rec (Hoboken). 296:427–432. 2013.
View Article : Google Scholar
|
|
14
|
Malzkorn B, Wolter M, Liesenberg F,
Grzendowski M, Stühler K, Meyer HE and Reifenberger G:
Identification and functional characterization of microRNAs
involved in the malignant progression of gliomas. Brain Pathol.
20:539–550. 2010. View Article : Google Scholar
|
|
15
|
Chen F, Chen L, He H, Huang W, Zhang R, Li
P, Meng Y and Jiang X: Up-regulation of microRNA-16 in glioblastoma
inhibits the function of endothelial cells and tumor angiogenesis
by targeting Bmi-1. Anticancer Agents Med Chem. 16:609–620. 2016.
View Article : Google Scholar
|
|
16
|
Wang Q, Li X, Zhu Y and Yang P:
MicroRNA-16 suppresses epithelial-mesenchymal transition related
gene expression in human glioma. Mol Med Rep. 10:3310–3314.
2014.PubMed/NCBI
|
|
17
|
Yang TQ, Lu XJ, Wu TF, Ding DD, Zhao ZH,
Chen GL, Xie XS, Li B, Wei YX, Guo LC, et al: MicroRNA-16 inhibits
glioma cell growth and invasion through suppression of Bcl-2 and
the nuclear factor-κB1/MMP9 signaling pathway. Cancer Sci.
105:265–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ambros V: microRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar
|
|
19
|
Chen X, Vega VB and Ng HH: Transcriptional
regulatory networks in embryonic stem cells. Cold Spring Harb Symp
Quant Biol. 73:203–209. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang X, Yuan X, Zhu W, Qian H and Xu W:
SALL4: An emerging cancer biomarker and target. Cancer Lett.
357:55–62. 2015. View Article : Google Scholar
|
|
21
|
Oishi N, Yamashita T and Kaneko S:
Molecular biology of liver cancer stem cells. Liver Cancer.
3:71–84. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang L, Yan Y, Jiang Y, Cui Y, Zou Y,
Qian J, Luo C, Lu Y and Wu X: The expression of SALL4 in patients
with gliomas: High level of SALL4 expression is correlated with
poor outcome. J Neurooncol. 121:261–268. 2015. View Article : Google Scholar
|
|
23
|
He J, Zhang W, Zhou Q, Zhao T, Song Y,
Chai L and Li Y: Low-expression of microRNA-107 inhibits cell
apoptosis in glioma by upregulation of SALL4. Int J Biochem Cell
Biol. 45:1962–1973. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cunliffe CH, Fischer I, Parag Y and Fowkes
ME: State-of-the-art pathology: new WHO classification,
implications, and new developments. Neuroimaging Clin N Am.
20:259–271. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Suzuki HI, Yamagata K, Sugimoto K, Iwamoto
T, Kato S and Miyazono K: Modulation of microRNA processing by p53.
Nature. 460:529–533. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Amaral FC, Torres N, Saggioro F, Neder L,
Machado HR, Silva WA Jr, Moreira AC and Castro M: MicroRNAs
differentially expressed in ACTH-secreting pituitary tumors. J Clin
Endocrinol Metab. 94:320–323. 2009. View Article : Google Scholar
|
|
27
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting Bcl-2. Proc
Natl Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar
|
|
28
|
Oikawa T, Kamiya A, Zeniya M, Chikada H,
Hyuck AD, Yamazaki Y, Wauthier E, Tajiri H, Miller LD, Wang XW, et
al: Sal-like protein 4 (SALL4), a stem cell biomarker in liver
cancers. Hepatology. 57:1469–1483. 2013. View Article : Google Scholar
|
|
29
|
Li A, Jiao Y, Yong KJ, Wang F, Gao C, Yan
B, Srivastava S, Lim GS, Tang P, Yang H, et al: SALL4 is a new
target in endometrial cancer. Oncogene. 34:63–72. 2015. View Article : Google Scholar
|
|
30
|
Kobayashi D, Kuribayashi K, Tanaka M and
Watanabe N: Overexpression of SALL4 in lung cancer and its
importance in cell proliferation. Oncol Rep. 26:965–970.
2011.PubMed/NCBI
|
|
31
|
Ardalan Khales S, Abbaszadegan MR,
Abdollahi A, Raeisossadati R, Tousi MF and Forghanifard MM: SALL4
as a new biomarker for early colorectal cancers. J Cancer Res Clin
Oncol. 141:229–235. 2015. View Article : Google Scholar
|
|
32
|
Forghanifard MM, Ardalan Khales S,
Javdani-Mallak A, Rad A, Farshchian M and Abbaszadegan MR: Stemness
state regulators SALL4 and SOX2 are involved in progression and
invasiveness of esophageal squamous cell carcinoma. Med Oncol.
31:9222014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Itou J, Matsumoto Y, Yoshikawa K and Toi
M: Sal-like 4 (SALL4) suppresses CDH1 expression and maintains cell
dispersion in basal-like breast cancer. FEBS Lett. 587:3115–3121.
2013. View Article : Google Scholar : PubMed/NCBI
|