Introduction
Macrophages are an important component of innate
immunity, and play a major function in host homeostasis (1). Macrophages are versatile
multifunctional cells, which change their phenotype and functional
capacity depending on various conditions. In response to
micro-environmental signals, macrophages undergo differential
activation, including classical activation characterized by a
pro-inflammatory phenotype (also called M1), and alternative
activation (M2) characterized by polarization associated with an
anti-inflammatory profile (2).
Flexibility and plasticity are key features of mononuclear
phagocytes, and specific cytokine stimuli can induce mononuclear
phagocytes to express specialized and polarized functional
properties (3,4). Classical polarized M1 macrophages
activated by interferon-γ (IFN-γ) or other microbial products such
as LPS, in turn produce high levels of pro-inflammatory cytokines,
such as tumor necrosis factor-α (TNF-α), interleukin (IL)-12,
IL-23, IL-6, IL-1β and a high production of reactive oxygen and
nitrogen intermediates. M1 macrophages contribute as inducer and
effector cells in promotion of Th1 responses, and mediate
resistance against intracellular parasites and tumor cells
(5–9). In contrast, alternatively activated
M2 macrophages induced by IL-4, IL-13 or immune complex (10,11), are characterized by a phenotype of
low production of IL-12 and IL-13, and high levels of IL-10, Arg-1,
YM-1, Fizz1 and Mrc-1 (12).
However, the mechanisms regulating the expression of the
constellation of genes in the response of macrophages to polarizing
conditions remain to be understood.
Circular RNA (circRNAs), formed by the covalent
linkage of the ends of a single RNA molecule, are new players in
the regulation of post-transcriptional gene expression. circRNAs
have been shown to cause loss of microRNA (miRNA) function
accompanied by increased levels of endogenous targets, acting as
miRNA sponges (13,14). Although circRNAs have been
identified for decades now, only recently have studies revealed the
numbers of circRNAs that are endogenous to mammalian cells. Many
circRNAs are abundant, stable, conserved and potentially function
as competing endogenous RNAs (15–17). circRNAs can arise from exons
(exonic circRNA) or introns (intronic circRNA), showing distinct
modes of generation. Evidence of potential functions in the
regulation of gene expression is emerging for both exonic and
intronic circRNAs (15,17–20). However, the role of circRNAs in
gene expression occurring during macrophage polarization remains
unknown.
In the present study, we used a specific circRNA
microarray platform to investigate the changes in the abundance of
circRNAs induced by the activation of primary bone marrow-derived
macrophages (BMDMs) under two distinct polarizing conditions to
expand the spectrum of the activation patterns of M1 and M2
macrophages. Our data revealed that a number of circRNAs were
consistently altered under distinct polarizing conditions.
circRNA-miRNA interaction during macrophage polarization was then
predicted. Thus, the present study is valuable for further
investigation of the precise roles of circRNAs in macrophage
differentiation and polarized activation processes.
Materials and methods
Mice
A total of 40 BALB/c male mice (8 weeks old) were
obtained from the Experimental Animal Center of Qinglongshan
(Nanjing, China), and were housed in pathogen-free mouse colonies.
All animal experiments were performed according to the guidelines
for the Care and Use of Laboratory Animals (Ministry of Health,
China, 1998). All experimental protocols were approved by the
Animal Ethics Committee of Yijishan Hospital (Wuhan, China).
Isolation and cultivation of murine
BMDMs
The mice were sacrificed by cervical dislocation
following anesthesia, and the abdomen and hind legs were sterilized
with 75% ethanol. An incision was made in the midline of the
abdomen, and clipped outward to expose the hind legs. Scissors were
then used to remove all muscle tissue from the bones and the bones
were cut at both ends to free them. The bones were crushed in a
mortar with 5 ml DMEM. The femur and tibia were separated by
cutting at the knee joint. BMDMs were isolated from BALB/c mice by
flushing the femurs with Dulbecco's modified Eagle's medium (DMEM)
(Gibco, Eggenstein, Germany). The cells were collected in 15-ml
tubes and centrifuged for 10 min at 100 × g. The supernatant was
removed, and the pellet was suspended in DMEM containing 20% fetal
bovine serum (FBS) and 20% L929 supernatant. The cells were then
seeded at 1×106 cells/well to 6-well cell-culture plates
(Corning Costar, Corning, NY, USA) at 37°C in 5% CO2.
After 7 days in culture, the medium was removed and the cells were
cultured in RPMI-1640 medium supplemented with 10% FBS for an
additional 24 h. Macrophage polarization was obtained by adding 100
ng/ml LPS plus 20 ng/ml IFN-γ (for M1 polarization), or 20 ng/ml
IL-4 (for M2 polarization) in DMEM containing 10% FBS and
incubating for 48 h.
FACS analysis
BMDMs were harvested and stained with
FITC-anti-F4/80 monoclonal antibody (Cat. no. 11-4801-82;
eBioscience, Inc., San Diego, CA, USA). Their F4/80 surface
expression was assessed by fluorescence-activated cell sorting
(FACS) analysis and data were analyzed using CellQuest software (BD
Biosciences, Franklin Lakes, NJ, USA).
circRNA microarray analysis
BMDMs were cultured with polarizing stimuli for 48
h; total RNA was extracted by TRIzol reagent (Invitrogen, Carlsbad,
CA, USA) and quantified using the NanoDrop ND-1000. Sample
preparation and microarray hybridization were performed based on
the Arraystar's standard protocols. Briefly, total RNA from each
sample was treated with RNase R (Epicentre Inc., Madison, WI, USA)
to remove linear RNAs. Then, each sample was amplified and
transcribed into fluorescent cRNA utilizing random primers
according to Arraystar's Super RNA Labeling protocol (Arraystar
Inc., Rockville, MD, USA). The labeled cRNAs were purified using
the RNeasy Mini kit (Qiagen, Hilden, Germany). The concentration
and specific activity of the labeled cRNAs (pmol Cy3/µg cRNA) were
measured by NanoDrop ND-1000. Next, 1 µg of each labeled
cRNA was fragmented by adding 5 µl 10X blocking agent and 1
µl of 25X fragmentation buffer, and then heating the mixture
at 60°C for 30 min. Finally, 25 µl 2X hybridization buffer
was added to dilute the labeled cRNA. Subsequently, 50 µl of
hybridization solution was dispensed into the gasket slide and
assembled to the circRNA expression microarray slide. The slides
were incubated for 17 h at 65°C in an Agilent hybridization oven.
After washing, the slides were scanned by the Axon GenePix 4000B
microarray scanner. Scanned images were then imported into GenePix
Pro 6.0 software (Axon, Foster City, CA, USA) for grid alignment
and data extraction. Quantile normalization and subsequent data
processing were performed using the R software package.
Differentially expressed circRNAs with statistical significance
between the two groups were identified through volcano plot
filtering and fold-change filtering. Hierarchical clustering was
performed to show the distinguishable circRNA expression pattern
among samples. All the experimental results were saved as Microsoft
Excel files.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA was isolated from the BMDMs as described above.
For linear RNA expression analysis, total RNA was reverse
transcribed using the SuperScript III First-Strand synthesis system
(Life Technologies, Carlsbad, CA, USA) with oligo(dT) according to
the manufacturer's instructions. Quantitative PCR (qPCR) was
performed using QuantiTect SYBR®-Green PCR kits (Qiagen)
and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an
internal control utilizing a Bio-Rad CFX96 real-time PCR system.
The reactions were incubated in 96-well plates at 95°C for 5 min,
followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min,
followed by a dissociation curve. For circRNA analysis, total RNA
was treated with RNase R for enzymatic digestion to remove linear
RNAs, and SuperScript III reverse transcriptase using random
primers (Life Technologies, Carlsbad, CA, USA) according to the
manufacturer's instructions. Primers assaying for circular products
were designed to cross the back-splice junction site. qPCR was
carried out on a BioRad CFX96 cycler using QuantiTect
SYBR®-Green PCR kits (Qiagen). All the PCR reactions
were run in triplicate. A complete list of primers used in this
study is listed in Table I.
 | Table IPrimer sequences used in real-time PCR
are listed in the 5′-3′ direction. |
Table I
Primer sequences used in real-time PCR
are listed in the 5′-3′ direction.
| Genes | Primer (5′-3′) |
|---|
| Nos2 |
ATCTTTGCCACCAAGATGGCCTGGTTCCTGTGCTGTGCTACAGTTCCG |
| TNF-α |
CCAGTGTGGGAAGCTGTCTTAAGCAAAAGAGGAGGCAACA |
| IL-12 |
GATGTCACCTGCCCAACTGTGGTTTGATGATGTCCCTGA |
| Arg-1 |
TGACTGAAGTAGACAAGCTGGGGATCGACATCAAAGCTCAGGTGAATCGG |
| YM-1 |
ATGAAGCATTGAATGGTCTGAAAGTGAATATCTGACGGTTCTGAGGAG |
| Fizz1 |
AGGTCAAGGAACTTCTTGCCAATCCAAGCACACCCAGTAGCAGTCATCCC |
| Mrc-1 |
GGAGTGGCAGGTGGCTTATTGGACATTTGGGTTCAGGAG |
| circRNA-003780 |
AGTGCCTCAGGTTTCTGGATTCTGTCTTCCTTTCTTGC |
| circRNA-010056 |
TCACCAGGAGAATCCCAGTCGAACTCTAAAATCAGGCT |
| circRNA-010231 |
TTGAGGCGAATGGCTGAGGCGGGAGGCTTGAATGTC |
| circRNA-003424 |
GAAAGCAGCACAGAAATCAGACCGTGTACTTGCAGACA |
| circRNA-013630 |
TAGGTGCTCTAATGTGAGTCAAAGTTTGGATGAGGGAT |
| circRNA-001489 |
GGAAAGGGTCTTAGCAAGCCCTAAATTACAGCAGGTA |
| circRNA-018127 |
AAGACGCTGGCATCCAAACCTCGGCTGGCTTCAACA |
Statistical analysis
Data are presented as the means ± SEM. Statistical
analysis of the data was performed with the two-tailed independent
Student's t-test or ANOVA analysis using GraphPad Prism (version
4.0) statistical program. A value of P<0.05 was considered to
indicate a statistically significant difference.
Results
Identification of ex vivo polarized M1
and M2 macrophages
To identify alterations in the circRNA expression
profile during macrophage polarization, we first prepared M1 and M2
phenotype macrophages in vitro. BMDMs were isolated from
BALB/c mice and stimulated with LPS and IFN-γ (for M1 polarization)
or IL-4 (for M2 polarization). FACS analysis showed that the purity
of isolated BMDMs was ~96% by assessing surface F4/80 expression
(Fig. 1A). TNF-α, Nos2 and IL-12
were used as markers for the M1 phenotype, and other four markers,
Arg-1, YM-1, Fizz1 and Mrc-1, were used as markers for the M2
phenotype. BMDMs stimulated with LPS and IFN-γ had significantly
higher TNF-α, Nos2 and IL-12 levels, while the levels of Arg1,
YM-1, Fizz1 and Mrc-1 were upregulated in IL-4-treated BMDMs
(Fig. 1B). These data confirmed
that the polarization used in this study resulted in distinct
macrophage phenotypes.
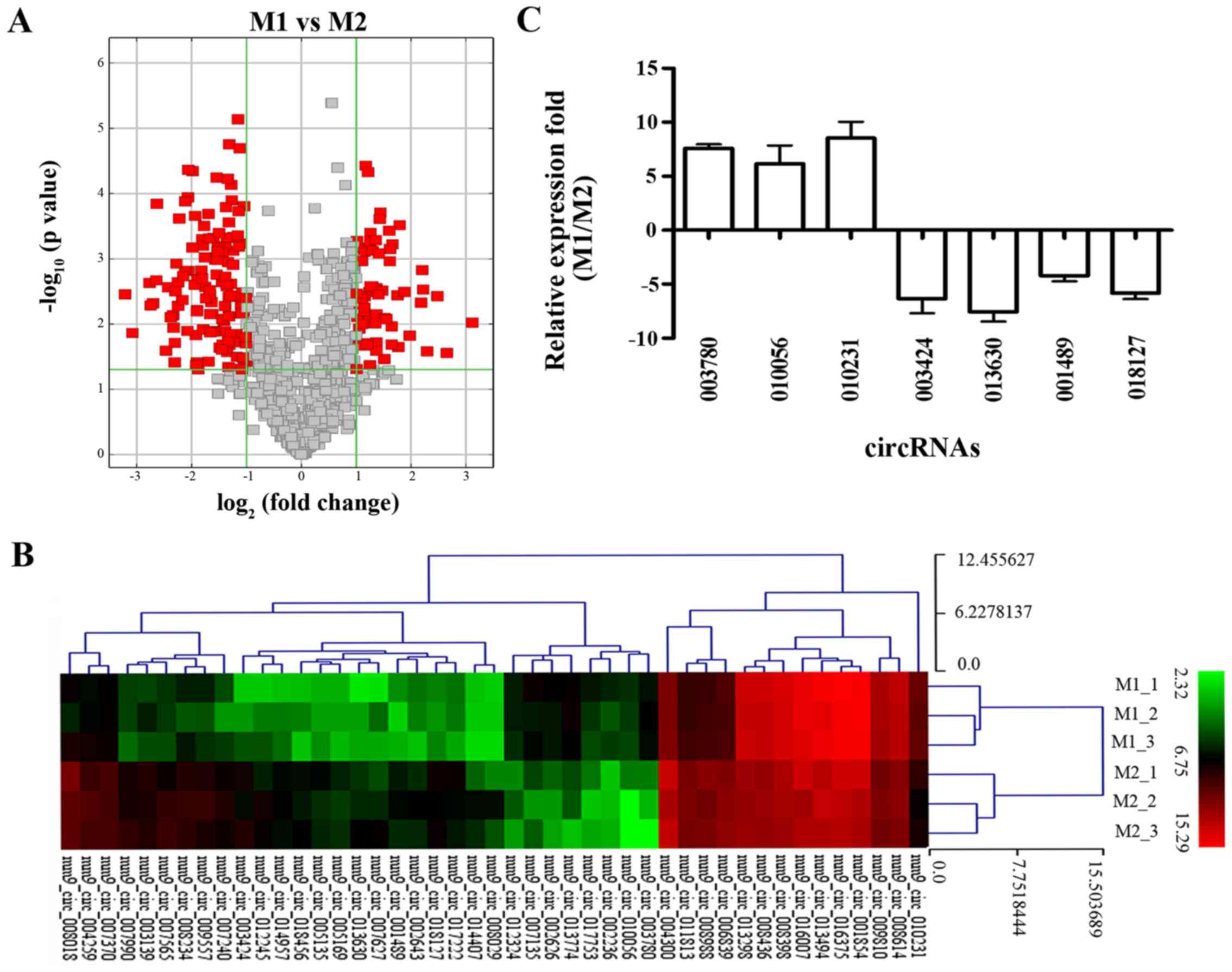
Analysis of the circRNA microarray
results
To screen for circRNAs that were differentially
expressed between the M1 and M2 macrophages, we determined the
circRNA expression profiles with a mouse circRNA microarray, and
the circRNA expression patterns for M1 and M2 were compared. We
found that 189 circRNAs were differentially expressed through a
combination of statistical significance (fold-change >2;
P<0.05). Among these, 62 circRNAs were upregulated and 127
circRNAs were downregulated in M1 compared with that noted in the
M2 macrophages (Table II). The
expression ratios (log2 scale) of the circRNAs between
M1 and M2 are shown as volcano plots at different P-values and
fold-change (Fig. 2A) and heat
maps (Fig. 2B).
 | Table IIThe number of differentially expressed
circRNAs in the polarized macrophages (M1 vs. M2, expression fold
>2). |
Table II
The number of differentially expressed
circRNAs in the polarized macrophages (M1 vs. M2, expression fold
>2).
| Regulation | Expression fold
>2 | Expression fold
>4 |
|---|
| Upregulation | 62 | 7 |
| Downregulation | 127 | 27 |
RT-qPCR validation of the differentially
expressed circRNAs
To verify the microarray results, we selected 7
differentially expressed exonic circRNAs (fold-change >4;
P<0.05), including 3 upregulated circRNAs and 4 downregulated
circRNAs as having the highest fold-change among the differentially
expressed circRNAs in M1 compared to M2 by the microarray results,
and validated their expression levels by RT-qPCR analysis. The
results showed that 3 circRNAs (circRNA-003780, circRNA-010056 and
circRNA-010231) were overexpressed, while 4 circRNAs
(circRNA-003424, circRNA-013630, circRNA-001489 and circRNA-018127)
were underexpressed in M1 compared with M2. The data from RT-qPCR
were consistent with the microarray analysis (Fig. 2C).
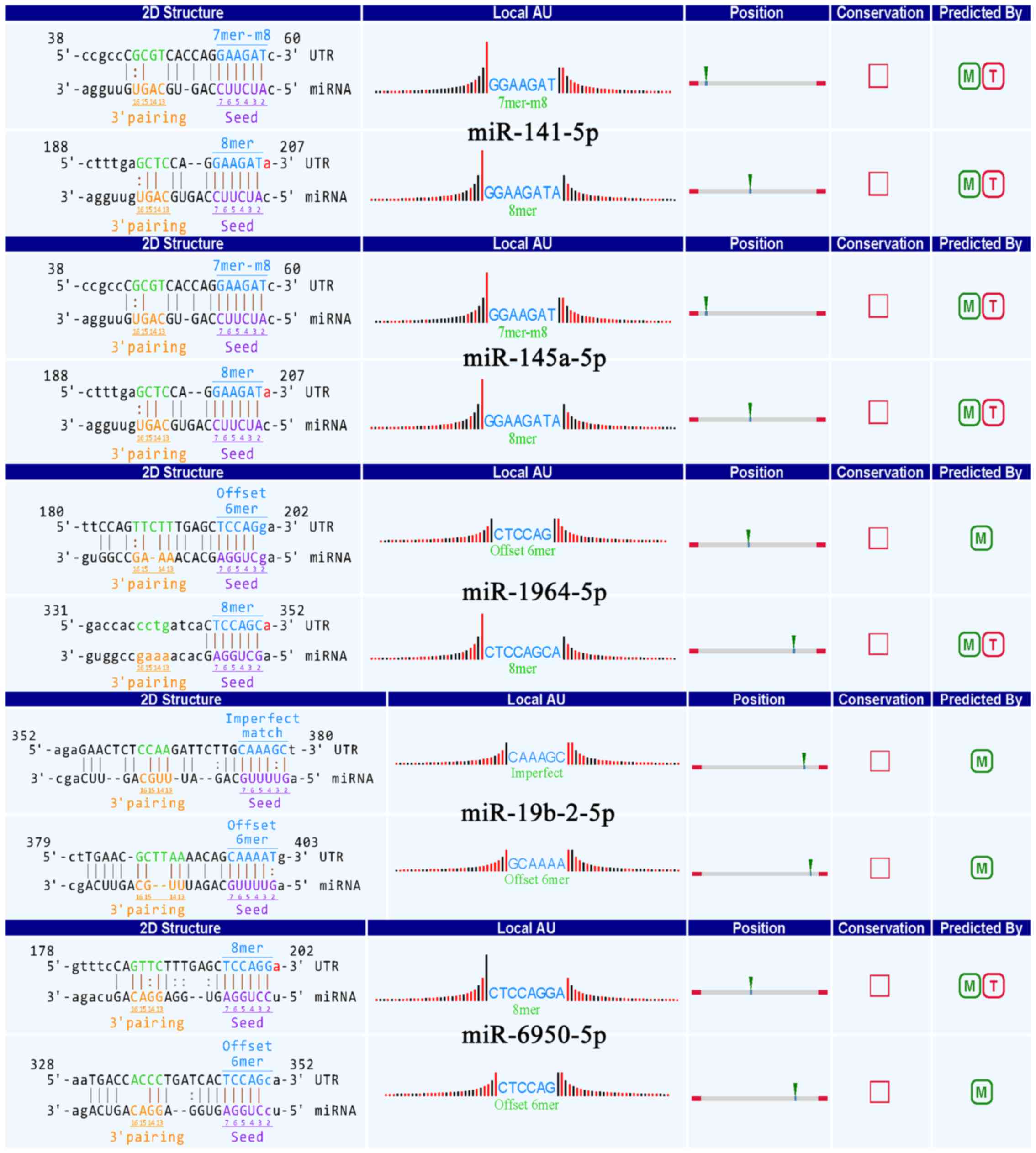
Annotation for circRNA/microRNA
interaction
To further facilitate the implication of our
research study, we used the Arraystar's home-made miRNA target
prediction software based on TargetScan (21) and miRanda (22) to predict circRNA/microRNA
interaction. We selected 29 differentially expressed exonic circRNA
with the highest fold-change (fold-change >4; P<0.05) to
predict their microRNA response elements (MREs), including 7
upregulated exonic circRNAs and 22 downregulated circRNAs. Five
MREs with good mirSVR scores for each circRNA are shown (Table III). Furthermore, the
overexpressed circRNA-010231 (fold-change, 5.56; P<0.05) in M1
compared to M2, showed detailed annotation for interaction with
various miRNAs (miR-141-5p, miR-145a-5p, miR-1964-5p, miR-19b-2-5p
and miR-6950-5p) (Fig. 3).
Moreover, the binding sites of the conserved miRNAs are represented
(Fig. 3).
 | Table IIIAnnotation for differentially
expressed circRNAs/miRNA interaction. |
Table III
Annotation for differentially
expressed circRNAs/miRNA interaction.
| circRNAs | FC (abs) | Regulation | circRNA_type | Gene symbol | MRE1 | MRE2 | MRE3 | MRE4 | MRE5 |
|---|
| circRNA_003780 | 86.475.919 | Up | Exonic | Cdyl | miR-7083-5p | miR-6902-5p | miR-185-3p | miR-3085-3p | miR-3064-5p |
| circRNA_010056 | 62.674.297 | Up | Exonic | Ube3a | miR-138-5p | miR-6981-5p | miR-3079-5p | miR-181b-2-3p | miR-541-5p |
| circRNA_010231 | 55.637.673 | Up | Exonic | Fut8 | miR-6950-5p | miR-19b-2-5p | miR-141-5p | miR-1964-5p | miR-145a-3p |
| circRNA_013774 | 48.818.098 | Up | Exonic | Erbb2ip | miR-21b | miR-6399 | miR-32-3p | miR-450b-3p | miR-450a-1-3p |
| circRNA_001854 | 4.660.326 | Up | Exonic | Atrnl1 | miR-376c-3p | miR-7673-5p | miR-7065-3p | miR-691 | miR-7221-3p |
| circRNA_002236 | 45.968.504 | Up | Exonic | St3gal5 | miR-7669-3p | miR-5621-5p | miR-1943-5p | miR-7659-5p | miR-7662-3p |
| circRNA_002626 | 45.184.143 | Up | Exonic | Raf1 | miR-3552 | miR-1964-5p | miR-297a-5p | miR-505-5p | miR-1941-5p |
| circRNA_003424 | 8.459.959 | Down | Exonic | Dgkb | miR-7220-3p | miR-133c | miR-7657-3p | miR-873a-5p | miR-6905-5p |
| circRNA_013630 | 67.069.108 | Down | Exonic | Thada | miR-7234-5p | miR-324-3p | miR-33-3p | miR-541-5p | miR-7119-5p |
| circRNA_001489 | 62.485.413 | Down | Exonic | Rnf2 | miR-6400 | miR-1933-5p | miR-374c-5p | miR-7667-5p | miR-7031-5p |
| circRNA_018127 | 6.203.339 | Down | Exonic | Ipo11 | miR-493-3p | miR-29b-1-5p | miR-383-3p | miR-7087-3p | miR-3064-5p |
| circRNA_012245 | 55.332.692 | Down | Exonic | Abca8b | miR-7013-5p | miR-742-3p | miR-6981-5p | miR-717 | miR-7051-3p |
| circRNA_007990 | 53.930.793 | Down | exonic | Tead1 | miR-5627-3p | miR-145a-3p | miR-188-3p | miR-6962-3p | miR-6241 |
| circRNA_007565 | 52.050.665 | Down | Antisense | Nufip2 | miR-7680-5p | miR-301a-3p | miR-301b-3p | miR-6341 | miR-145b |
| circRNA_014957 | 50.518.057 | Down | Exonic | Trim37 | miR-677-3p | miR-148a-5p | miR-7672-5p | miR-3065-5p | miR-880-3p |
| circRNA_007240 | 50.199.436 | Down | Exonic | Bre | miR-6985-5p | miR-6365 | miR-6340 | miR-497a-5p | miR-16-5p |
| circRNA_007627 | 49.545.711 | Down | Exonic | Zfat | miR-6998-3p | miR-377-3p | miR-6337 | miR-497a-5p | miR-1906 |
| circRNA_008029 | 49.454.406 | Down | Exonic | Gphn | miR-6418-5p | miR-7067-5p | miR-1933-5p | miR-145a-3p | miR-23a-5p |
| circRNA_017222 | 49.041.543 | Down | Exonic | Tmem26 | miR-148a-5p | miR-148b-5p | miR-6414 | miR-3068-5p | miR-499-3p |
| circRNA_005135 | 48.433.047 | Down | Exonic | Zcchc11 | miR-1904 | miR-7116-3p | miR-146a-3p | miR-6951-3p | miR-7235-3p |
| circRNA_008234 | 47.541.014 | Down | Antisense | Fasn | miR-361-3p | miR-504-5p | miR-1291 | miR-3090-3p | miR-212-5p |
| circRNA_004259 | 47.454.669 | Down | Exonic | Mga | miR-7092-3p | miR-7007-5p | miR-298-5p | miR-7682-3p | miR-7090-5p |
| circRNA_005169 | 46.792.326 | Down | Exonic | Lrp1b | miR-9769-5p | miR-6896-3p | miR-320-5p | miR-7118-3p | miR-6901-3p |
| circRNA_009557 | 4.491.621 | Down | Exonic | Nrg3 | miR-22-5p | miR-532-5p | miR-1291 | miR-7578 | miR-1912-3p |
| circRNA_004300 | 44.410.193 | Down | Antisense | Malat1 | miR-1960 | miR-6715-3p | miR-495-3p | miR-667-5p | miR-3078-3p |
| circRNA_003139 | 43.402.291 | Down | Exonic | Dtnb | miR-667-5p | miR-3066-5p | miR-6938-3p | miR-496b | miR-6929-5p |
| circRNA_002643 | 42.171.155 | Down | Exonic | Kpna4 | miR-107-5p | miR-103-1-5p | miR-103-2-5p | miR-6359 | miR-7060-5p |
| circRNA_007370 | 41.685.939 | Down | Exonic | Neo1 | miR-8092 | miR-3098-5p | miR-7007-5p | miR-670-3p | miR-367-3p |
| circRNA_008988 | 41.658.594 | Down | Exonic | Unc13b | miR-5623-3p | miR-1903 | miR-7652-3p | miR-7688-3p | miR-326-3p |
Discussion
Mammalian macrophages are induced to diverse
phenotypes in response to different external stimuli. We and other
researchers have reported that a subset of miRNA expression changes
was repeatedly found to be involved in macrophage polarization
(5,6,9,12,23–25).
circRNAs, as miRNA sponges, are stable transcripts
expressed from diverse genomic locations, and have been recently
identified as important players in the regulation of cellular miRNA
abundance and thus are a major component in the miRNA-mediated
post-transcriptional regulatory network. Available studies suggest
that interactions between circRNAs and miRNAs indicate that
circRNAs are potentially associated with many disease, cell
processes and gene expression (13,26).
The present study aimed to identify the expression
patterns of circRNAs in response to stimuli polarizing two distinct
patterns of macrophage activation (M1 and M2). We performed an
assay using a circRNA microarray to profile the expression of
circRNAs. We demonstrated that the expression of 189 circRNAs was
significantly different in the M1 compared with that found in the
M2 macrophages. Among these, 62 circRNAs were upregulated, while
127 circRNAs were downregulated. Based on the microarray analysis,
high levels of circRNA-003780, circRNA-010056 and circRNA-010231 in
M1 cells and circRNA-003424, circRNA-013630, circRNA-001489 and
circRNA-018127 in M2 cells with fold-change >5 were selected and
validated by RT-qPCR to confirm the results of the microarray
analysis. Although, there were some discrepancies in the results of
the microarray and the RT-qPCR analyses, the microarray provided a
rapid method for identifying a large number of differentially
expressed circRNAs in M1 macrophages which could then be confirmed
by RT-qPCR.
Recent evidence suggests that circRNAs play a
crucial role in fine-tuning the level of miRNA-mediated regulation
of gene expression by sequestering miRNAs. The interaction of
circRNAs with disease-associated miRNAs indicates that circRNAs are
important for disease regulation (26). For example, the circRNA ciRS-7
contains multiple tandem miRNA-7 binding sites, thereby acting as
an endogenous miRNA 'sponge' to adsorb, and hence quench normal
miRNA-7 function (27). Thus, we
annotated the circRNA/miRNA interaction for the differentially
expressed circRNAs and performed a detailed annotation for
representing the binding sites of circRNA-010231 and the conserved
MREs with good scores with miR-6950-5p, miR-19-2-5p, miR-141-5p,
miR-1964-5p and miR-145a-3p, respectively. However, few
circRNA/miRNA interactions have been experimentally validated.
Therefore, the functional effect of their interaction will be the
focus of our future research.
In conclusion, the present study investigated the
global expression patterns of circRNAs in macrophage activation and
contributes to the understanding of the role of circRNAs in
macrophage polarization under different stimulating conditions. The
illustration from our database is an important starting point for
understanding the role of circRNAs in macrophage function and their
potential effect on disease pathology.
Acknowledgments
The present study was supported by grants from the
National Natural Science Foundation of China (Nos. 81300172,
81301497 and 81472017) and the Natural Science Foundation of Anhui
Province (Nos. 1308085QH137 and 1408085QH148).
References
|
1
|
Gordon S and Martinez FO: Alternative
activation of macrophages: Mechanism and functions. Immunity.
32:593–604. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mosser DM: The many faces of macrophage
activation. J Leukoc Biol. 73:209–212. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Biswas SK and Mantovani A: Macrophage
plasticity and interaction with lymphocyte subsets: Cancer as a
paradigm. Nat Immunol. 11:889–896. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mantovani A, Sozzani S, Locati M, Allavena
P and Sica A: Macrophage polarization: Tumor-associated macrophages
as a paradigm for polarized M2 mononuclear phagocytes. Trends
Immunol. 23:549–555. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen T, Huang Z, Wang L, Wang Y, Wu F,
Meng S and Wang C: MicroRNA-125a-5p partly regulates the
inflammatory response, lipid uptake, and ORP9 expression in
oxLDL-stimulated monocyte/macrophages. Cardiovasc Res. 83:131–139.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cheng Y, Kuang W, Hao Y, Zhang D, Lei M,
Du L, Jiao H, Zhang X and Wang F: Downregulation of
miR-27a* and miR-532-5p and upregulation of miR-146a and
miR-155 in LPS-induced RAW264.7 macrophage cells. Inflammation.
35:1308–1313. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Coley W, Van Duyne R, Carpio L, Guendel I,
Kehn-Hall K, Chevalier S, Narayanan A, Luu T, Lee N, Klase Z, et
al: Absence of DICER in monocytes and its regulation by HIV-1. J
Biol Chem. 285:31930–31943. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dalton DK, Pitts-Meek S, Keshav S, Figari
IS, Bradley A and Stewart TA: Multiple defects of immune cell
function in mice with disrupted interferon-gamma genes. Science.
259:1739–1742. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Forrest AR, Kanamori-Katayama M, Tomaru Y,
Lassmann T, Ninomiya N, Takahashi Y, de Hoon MJ, Kubosaki A, Kaiho
A, Suzuki M, et al: Induction of microRNAs, mir-155, mir-222,
mir-424 and mir-503, promotes monocytic differentiation through
combinatorial regulation. Leukemia. 24:460–466. 2010. View Article : Google Scholar
|
|
10
|
Goerdt S and Orfanos CE: Other functions,
other genes: Alternative activation of antigen-presenting cells.
Immunity. 10:137–142. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gordon S: Alternative activation of
macrophages. Nat Rev Immunol. 3:23–35. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cai X, Yin Y, Li N, Zhu D, Zhang J, Zhang
CY and Zen K: Re-polarization of tumor-associated macrophages to
pro-inflammatory M1 macrophages by microRNA-155. J Mol Cell Biol.
4:341–343. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ebert MS, Neilson JR and Sharp PA:
MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian
cells. Nat Methods. 4:721–726. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
16
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lasda E and Parker R: Circular RNAs:
Diversity of form and function. RNA. 20:1829–1842. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5:R12003. View Article : Google Scholar
|
|
22
|
Pasquinelli AE: MicroRNAs and their
targets: Recognition, regulation and an emerging reciprocal
relationship. Nat Rev Genet. 13:271–282. 2012.PubMed/NCBI
|
|
23
|
Zhang Y, Zhang M, Zhong M, Suo Q and Lv K:
Expression profiles of miRNAs in polarized macrophages. Int J Mol
Med. 31:797–802. 2013.PubMed/NCBI
|
|
24
|
Rückerl D, Jenkins SJ, Laqtom NN,
Gallagher IJ, Sutherland TE, Duncan S, Buck AH and Allen JE:
Induction of IL-4Rα-dependent microRNAs identifies PI3K/Akt
signaling as essential for IL-4-driven murine macrophage
proliferation in vivo. Blood. 120:2307–2316. 2012. View Article : Google Scholar
|
|
25
|
Chaudhuri AA, So AY, Sinha N, Gibson WS,
Taganov KD, O'Connell RM and Baltimore D: MicroRNA-125b potentiates
macrophage activation. J Immunol. 187:5062–5068. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ghosal S, Das S, Sen R, Basak P and
Chakrabarti J: Circ2Traits: A comprehensive database for circular
RNA potentially associated with disease and traits. Front Genet.
4:2832013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lukiw WJ: Circular RNA (circRNA) in
Alzheimer's disease (AD). Front Genet. 4:3072013. View Article : Google Scholar
|