Introduction
Breast cancer (BC) is one of the most common cancers
and the leading cause of morbidity among all female malignant
tumors, despite improvements in screening and adjuvant systemic
treatment (1). It has been widely
acknowledged that BC is a highly heterogeneous disease, with
different biological behaviors for the same stage of BC among
different patients (2).
Therefore, personalized therapy based on the different tumor
molecular classifications has become the main research field in BC
treatment (3). To date,
personalized therapy of BC is still guided by various traditional
prognostic and predictive parameters, including estrogen receptor
(ER) and progesterone receptor (PR) status, and human epidermal
growth factor receptor 2 (HER2) amplification (4,5).
Therefore, there is an urgent requirement to identify new
prognostic and predictive biomarkers that can be used to optimize
treatments and predict clinical outcomes among BC patients.
Previous studies have indicated that serine
proteases play an important role in cancer invasion and metastasis.
Proteases contribute to degradation of the basement membrane and
extracellular matrix (ECM) (6,7),
which allows tumor cells to invade the surrounding tissue and
nearby blood vessels. In addition, proteases may be involved in all
stages of the development and progression of cancer, including
proliferation, survival, migration, invasion, angiogenesis and
metastasis (8). Type II
transmembrane serine proteases (TTSPs) are a new subfamily of
serine proteases that participate in the regulation of cellular
signaling events at the plasma membrane and in the ECM (9,10).
Many of the TTSPs have been found to be dysregulated in malignant
tumors, implicating their possible roles in tumorigenesis and/or
progression (11).
Transmembrane protease serine 4 (TMPRSS4) is a novel
TTSP expressed at the cell surface. It is overexpressed in
pancreatic, thyroid, colon, lung and gastric cancer tissues and is
associated with poor patient prognosis (12), suggesting a possible role for
TMPRSS4 in tumor development and progression. Recent studies
suggest that TMPRSS4 can induce epithelial-mesenchymal transition
(EMT), accompanied by increased invasive activity and malignant
transformation in human epithelial cancer cells (13–15). Given these findings, TMPRSS4
appears to play important roles in carcinogenesis and may represent
a new therapeutic target for cancers. It has also been reported
that TMPRSS4 is overexpressed in BC tissue, but few studies have
documented the function of TMPRSS4 and its underlying mechanism in
BC cell lines. In the present study, we examined TMPRSS4 expression
in BC tissues and its correlation with clinicopathological
parameters and prognosis. Moreover, we detected the function of
TMPRSS4 in breast cancer cell lines and key biomarkers in EMT to
further investigate the relationship between TMPRSS4 and EMT in
BC.
Materials and methods
Clinical specimens
One hundred and seven formalin-fixed,
paraffin-embedded (FFPE) breast cancer (BC) specimens and 52 normal
breast tissue samples were obtained from the Department of
Pathology, Taian Central Hospital between January 2008 and December
2009. All patients included in the present study received no
radiotherapy or chemotherapy prior to surgical resection. Major
patient demographic and clinicopathological characteristics were
available, including age, menopausal status, tumor size, lymph node
(LN) status, histological subtype, tumor grade, clinical stage, and
ER, PR and HER2 status. Males were excluded and all patients were
females. TNM stage and histological grade were classified according
to the 7th edition of American Joint Committee on Cancer (AJCC) TNM
system (16) and the 4th edition
of WHO histological grade (17).
The study protocol was approved by the Institutional Ethics
Committee of Taian Central Hospital and informed consent was
obtained from all patients. The study was undertaken according to
the ethical standards of the World Medical Association Declaration
of Helsinki.
Follow-up information was obtained from the medical
records or by phone call. All patients had follow-up records for
>5 years. The follow-up deadline was June 2015. Disease-free
survival (DFS) time and overall survival (OS) time were calculated
from the date of surgery to the date of first recurrence or death,
which were the two assessments used for prognostic analyses.
Immunohistochemistry and evaluation
Immunohistochemical analysis of breast tissues was
performed on formalin-fixed, paraffin-embedded, 4-μm-thick
tissue sections using the (ABC) avidin-biotin-peroxidase complex
method. Briefly, the sections were deparaffinized and dehydrated
using a graded series of ethanol solutions. Antigen retrieval was
carried out by treatment in a microwave in a 0.01 M citrate buffer
(pH 6.0). The sections were incubated with the primary rabbit
anti-TMPRSS4 antibody (1:200; ab150595, Abcam, Cambridge, UK)
overnight at 4°C followed by the secondary antibody. The primary
antibody was replaced with Tris-buffered saline to act as the
negative control. The results were visualized with
diaminobenzidine, and all sections were counterstained with
hematoxylin and differentiated by hydrochloric acid alcohol.
The immunostaining results were determined
independently by two expert pathologists (X.-M.L., X.C.) in a
double-blind manner. The staining intensity was classified as
follows: 0, no staining; 1, weak staining; 2, moderate staining; 3,
strong staining. The percentage of positive-stained tumor cells was
scored as follows: 0, none; 1, <15% positive tumor cells; 2,
15–50% positive tumor cells; and 3, >50% positive tumor cells.
The staining index (SI) was calculated as staining intensity score
x proportion of positive tumor cells. SI scores of 0–3 were
considered low expression and >3 were considered high
expression.
Cell culture
All five human breast cancer cell lines, MCF-7,
T47D, ADM, MDA-MB-468 and MDA-MB-231 were purchased from Shanghai
Cancer Institute and preserved at the Department of Pathology of
Shandong University. MCF-7 cells were maintained in Dulbecco's
Modified Eagle's Medium (DMEM) with 10% fetal bovine serum (FBS).
T47D and ADM cells were cultured in RPMI-1640 medium containing 10%
FBS. MDA-MB-468 and MDA-MB-231 cells were cultured in L-15 medium
containing 10% FBS. All media and FBS were purchased from Gibco
(Los Angeles, CA, USA). Cells were incubated at 37°C with 5%
CO2.
Transient transfection
Commercial siRNA targeting TMPRSS4 was purchased
from RiboBio Co., Ltd.(Guangzhou, China). Cells were seeded in
6-well culture plates (Nest Biotechnology Co., Ltd., Wuxi, China)
at a density of 5×105 cells/well and transfected with
siRNA and negative controls (NCs) by X-tremeGENE transfection
reagent 12 h later (Roche Applied Science, Indianapolis, IN, USA).
Forty-eight hours after transfection, quantitative real-time PCR
(RT-qPCR) and western blot analysis were used to examine
transfection efficiency. Cell function assays, RNA isolation and
total cell protein extraction were performed 48 h after
transfection.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated using the TRIzol agent
(Invitrogen, Carlsbad, CA, USA) and RNA samples (1 μg) were
reverse transcripted to cDNA using PrimeScript RT Master Mix
(538100; Toyobo, Osaka, Japan). The qPCR was performed using
SYBR-Green PCR Master Mix (15153900; Roche, Indianapolis, IN, USA)
on the Applied Biosystems 7900HT Real-time PCR system.
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was used as
an internal control for each sample, and the expression of each
sample was normalized to GAPDH mRNA. Primers used were as follows:
TMPRSS4 forward, 5′-CCGATGTGTTC AACTGGAAG-3′ and reverse,
5′-GAGAAAGTGAGTGG GAACTG-3′; GAPDH forward, 5′-GCACCGTCAAGGCTG
AGAAC-3′ and reverse, 5′-TGGTGAAGACGCCAGTGGA-3′. After denaturation
for 10 min at 95°C, the reaction was continued for 40 cycles at
94°C for 10 sec, 60 or 62°C for 20 sec and 72°C for 20 sec. The
relative expression of each miRNA was calculated using the
2−ΔΔCt method.
Western blot analysis
Total protein was extracted from the transfected
cells 48 h after transfection and protein concentration was
determined by the BCA protein assay kit (Blue Skies, Shanghai,
China). Primary antibodies including TMPRSS4 (1:1,000; ab82176,
Abcam), E-cadherin, claudin-1, vimentin, Slug and ZEB1 (1:1,000;
mAb 3195, mAb 13255, mAb 5741, mAb 9585, mAb 3396, Cell Signaling
Technology, Danvers, MA, USA) from the EMT antibody sampler kit
(1:1,000; Cell Signaling Technology) were incubated overnight at
4°C, washed, and then incubated with horseradish
peroxidase-labelled secondary anti-rabbit IgG (1:5,000; antibody
7074, Cell Signaling Technology) for 30 min. Immunoreactive bands
were detected with a chemiluminescence kit (Millipore, Billerica,
MA, USA) according to the manufacturer's procedure.
Cell proliferation assay
Cell Titer 96 non-radioactive cell proliferation
(MTS) (Promega BioSciences, Madison, WI, USA) and Cell-Light™ EdU
cell proliferation detection (EdU) assays (RiboBio Co., Ltd.) were
performed to test the proliferation ability of the cells following
the manufacturer's protocol. Briefly, for the MTS assay,
transiently transfected cells were planted in 96-well plates with
6,000 cells/well and incubated for 24, 48 and 96 h. The viability
of the cells was determined with MTS. The absorbance value at a
wavelength of 490 nm was used as an indicator of cell viability.
For the EdU assay, 24 h after transfection, the cells were cultured
in triplicate at 6,000 cells/well in 96-well plates the day before
EdU incubation. After EdU labeling, the cells were treated with 100
μl of Apollo reaction cocktail. Then nucleic acids in all
cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) and
visualized under a fluorescence microscope (Olympus, Tokyo, Japan).
The percentage of EdU-positive cells was defined as the
proliferation rate. Data were obtained from three independent
experiments.
Cell migration and invasion assays
Migration assays were performed with Transwell
inserts containing a polycarbonate membrane with 8.0 μm
pores (Corning, New York, NY, USA). To measure the invasion of
cancer cells, membranes were coated with Matrigel matrix (BD
Biosciences, Bedford, MA, USA). Forty-eight hours after
transfection with the siRNA or the negative control,
1×105 cells in 200 μl serum-free media were added
into the inside chamber and 600 μl medium with 10% FBS was
added to the outside chamber. After incubated for 24 h at 37°C in a
CO2 incubator, cells on the inner surface were removed
softly, while the migrated or invaded cells that attached to the
bottom of the membrane insert were fixed and stained with 1%
crystal violet. Then the cells were counted under a microscope in 5
different fields.
Statistical analysis
All statistical analyses were performed using SPSS
19.0 software (SPSS, Inc., Chicago, IL, USA). A Student's t-test
was used to analyze difference between two groups. The Chi-square
test and Fisher's exact test were used to analyze the relationship
between TMPRSS4 expression and the clinicopathological parameters.
OS and DFS were evaluated with the Kaplan-Meier method, and
differences were compared by log-rank test. Analyses of predictive
factors for OS and DFS were performed with univariate and
multivariate Cox proportional hazards regression method. Data
represent the means ± standard deviation (SD) from 3 independent
experiments. A two-tailed value of P<0.05 was considered
statistically significant.
Results
TMPRSS4 is overexpressed in breast cancer
tissues
The characteristics of the study population are
summarized in Table I. Patient
age ranged from 25 to 78 years, with a mean age of 51.6 years.
Median follow-up time was 65.2 months. TMPRSS4-positive staining
was located mainly in the cytoplasm or cell membrane of the tumor
cell nests. The positive TMPRSS4 expression rate was 65.4% (70/107)
in BC cases and 17.6% (9/52) in normal breast tissues (Fig. 1). A significant statistical
difference was found between the two groups (P<0.05).
Furthermore, the expression of TMPRSS4 was increased along with
grade, with the highest expression in grade III BC tissues and the
lowest expression in grade I BC tissues (P<0.05). TMPRSS4 was
also overexpressed in invasive mucinous carcinoma and
micropapillary carcinoma tissues (Fig. 1E and F) with no statistically
significant differences between them.
 | Table IAssociation of TMPRSS4 expression with
clinicopathological features of the patients with BC. |
Table I
Association of TMPRSS4 expression with
clinicopathological features of the patients with BC.
| Variables | No. (n=107) | TMPRSS4 expression
| P-value |
|---|
| Low | High |
|---|
| Age (years) | | | | 0.474 |
| ≤50 | 47 | 18 | 29 | |
| >50 | 60 | 19 | 41 | |
| Menopausal
status | | | | 0.158 |
| Premenopausal | 58 | 20 | 38 | |
|
Postmenopausal | 49 | 17 | 32 | |
| Tumor size
(cm) | | | | 0.044 |
| ≤2 | 41 | 19 | 22 | |
| >2 | 66 | 18 | 48 | |
| Histological
subtype | | | | 0.717 |
| Ductal | 70 | 22 | 48 | |
| Lobular | 13 | 6 | 7 | |
| Mucinous | 12 | 4 | 8 | |
|
Micropapillary | 12 | 5 | 7 | |
| Grade of ductal
cancer | | | | 0.006 |
| I | 16 | 9 | 7 | |
| II | 35 | 10 | 25 | |
| III | 19 | 3 | 16 | |
| LN metastasis | | | | 0.002 |
| Negative | 59 | 28 | 31 | |
| Positive | 48 | 9 | 39 | |
| Clinical stage | | | | 0.015 |
| I, II | 58 | 26 | 32 | |
| III, IV | 49 | 11 | 38 | |
| ER statusa | | | | 0.292 |
| Negative | 45 | 13 | 32 | |
| Positive | 62 | 24 | 38 | |
| PR statusa | | | | 0.428 |
| Negative | 49 | 15 | 34 | |
| Positive | 58 | 22 | 36 | |
| HER2 statusa | | | | 0.362 |
| Negative (IHC
0–2+) | 67 | 21 | 46 | |
| Positive (IHC
3+) | 40 | 16 | 24 | |
Correlation of TMPRSS4 with different
clinicopathological parameters
Next, we evaluated the associations between TMPRSS4
protein expression and a series of clinicopatho-logical
characteristics including age, menopausal status, tumor size,
histological type, histological grade, lymph node stage, clinical
stage, and status of ER, PR and HER2 in the BC patients. As shown
in Table I, high expression of
TMPRSS4 was positively correlated with tumor size (P=0.044),
histological grade (P=0.006), lymph node metastasis (P=0.002),
clinical stage (P=0.015), but was not correlated with other
clinicopathological parameters, including patient age (P=0.474),
menopausal status (P=0.158), histological subtype (P=0.717), and
status of ER (P=0.292), PR (P=0.428) and HER2 (P=0.362).
Overexpression of TMPRSS4 is correlated
with poor outcomes
We then investigated whether the expression of
TMPRSS4 is associated with clinical outcomes in BC. The OS and DFS
indicated by the Kaplan-Meier survival curves of the BC patients
according to low and high TMPRSS4 expression are shown in Fig. 2. Patients with high expression of
TMPRSS4 had shorter OS (P=0.036) and DFS (P=0.010) when compared
with the patients with low TMPRSS4 expression. Univariate and
multivariate analyses were carried out using Cox proportional
hazard model to evaluate the influence of TMPRSS4 expression and
pathological factors on the prognosis of BC patients. In Table II, the univariate analysis
revealed that tumor size (OS, P=0.042; DFS, P=0.047), lymph node
metastasis (OS, P=0.000; DFS, P=0.001), histological grade (OS,
P=0.036; DFS, P=0.039), clinical stage (OS, P=0.009; DFS, P=0.012),
and TMPRSS4 expression (OS, P=0.015; DFS, P=0.018) were prognostic
factors for BC patients. The multivariate analysis indicated that
TMPRSS4 expression was one of the independent prognostic factors,
along with clinical stage and lymph node metastasis (Table II).
 | Table IIUnivariate and multivariate Cox
regression analyses of overall survival and disease-free survival
in patients with BC. |
Table II
Univariate and multivariate Cox
regression analyses of overall survival and disease-free survival
in patients with BC.
| Variables | Univariate analysis
| Multivariate
analysis
|
|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| OS |
| Age (≤50 vs.
>50 years) | 1.362 | 1.282–2.530 | 0.533 | | | |
| Menopausal status
(pre vs. post) | 1.342 | 0.601–2.993 | 0.473 | | | |
| Histological
subtype (ductal vs. lobular) | 1.087 | 0.932–1.397 | 0.710 | | | |
| Tumor size (≤2 vs.
>2 cm) | 1.549 | 1.247–1.222 | 0.042 | 2.168 | 0.724–2.854 | 0.068 |
| Grade of ductal
cancer (I, II vs. III) | 2.372 | 1.532–3.278 | 0.036 | 2.874 | 1.153–3.238 | 0.057 |
| LN metastasis
(negative vs. positive) | 1.114 | 1.034–2.382 | 0.000 | 1.136 | 1.040–1.460 | 0.001 |
| Clinical stage (I,
II vs. III, IV) | 2.166 | 1.420–3.218 | 0.009 | 2.039 | 1.027–3.995 | 0.015 |
| ER status
(negative vs. positive) | 1.458 | 1.032–2.688 | 0.297 | | | |
| PR status
(negative vs. positive) | 1.362 | 1.067–2.876 | 0.374 | | | |
| HER2 status
(negative vs. positive) | 1.299 | 0.998–1.745 | 0.107 | | | |
| TMPRSS4 expression
(low vs. high) | 1.265 | 1.091–1.777 | 0.015 | 1.289 | 1.098–1.850 | 0.016 |
| DFS |
| Age (≤50 vs.
>50 years) | 1.562 | 1.187–2.623 | 0.624 | | | |
| Menopausal status
(pre vs. post) | 1.447 | 0.648–3.232 | 0.367 | | | |
| Histological
subtype (ductal vs. lobular) | 1.396 | 0.993–2.056 | 0.802 | | | |
| Tumor size (≤2 vs.
>2 cm) | 1.567 | 1.278–1.793 | 0.047 | 2.272 | 0.927–3.568 | 0.073 |
| Grade of ductal
cancer (I, II vs. III) | 2.569 | 1.587–2.763 | 0.039 | 2.942 | 1.278–3.524 | 0.066 |
| LN metastasis
(negative vs. positive) | 2.054 | 1.029–3.398 | 0.001 | 2.143 | 1.042–3.489 | 0.002 |
| Clinical stage (I,
II vs. III, IV) | 3.971 | 1.355–5.637 | 0.012 | 2.873 | 1.964–5.761 | 0.018 |
| ER status
(negative vs. positive) | 1.545 | 1.058–2.734 | 0.301 | | | |
| PR status
(negative vs. positive) | 1.378 | 1.075–2.888 | 0.381 | | | |
| HER2 status
(negative vs. positive) | 1.282 | 0.989–1.763 | 0.147 | | | |
| TMPRSS4 expression
(low vs. high) | 1.278 | 1.137–1.821 | 0.018 | 1.355 | 1.185–1.971 | 0.019 |
TMPRSS4 knockdown suppresses the
proliferation of BC cells in vitro
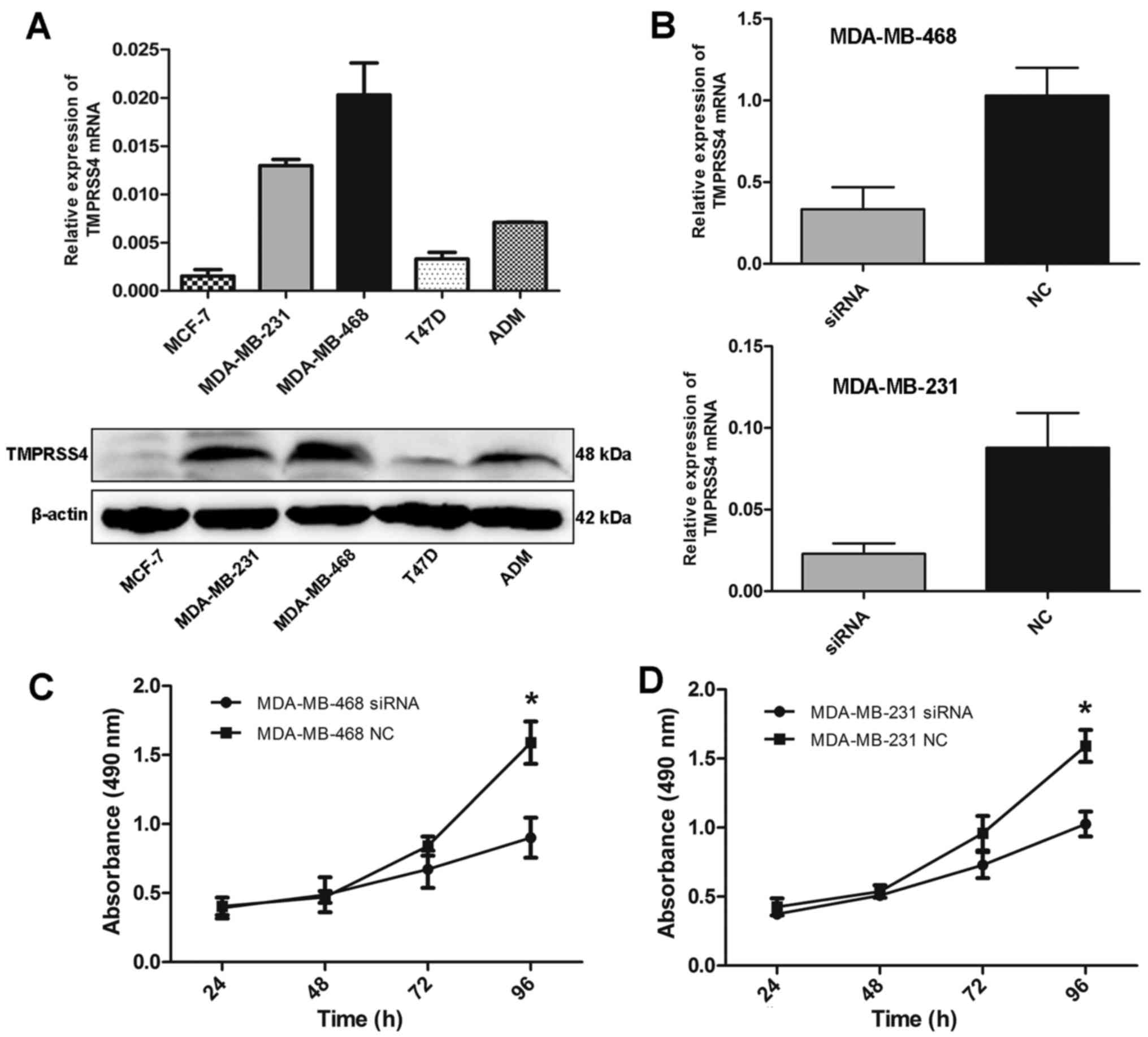
As TMPRSS4 was highly expressed in BC tissues, we
further detected the expression level of TMPRSS4 in different BC
cell lines. RT-qPCR and western blot analysis showed that TMPRSS4
was differentially expressed in five BC cell lines, with the
highest expression in MDA-MB-468 cells (Fig. 3A).
To inhibit endogenous high TMPRSS4 levels, two cell
lines (MDA-MB-468 and MDA-MB-231) were transfected with a small
interfering RNA (siRNA) duplex and transfection efficiency was
evaluated by RT-qPCR. Substantial knockdown of TMPRSS4 mRNA was
observed in the MDA-MB-468 and MDA-MB-231 cells (all P<0.001)
(Fig. 3B). Following siRNA
transfection, the cell proliferation rate was evaluated by MTS and
EdU assays. MTS assay showed that the cell proliferation rate was
significantly decreased in both the MDA-MB-468 and MDA-MB-231 cell
lines 96 h after trans-fection (all P<0.05) (Fig. 3C and D). Similar changes were
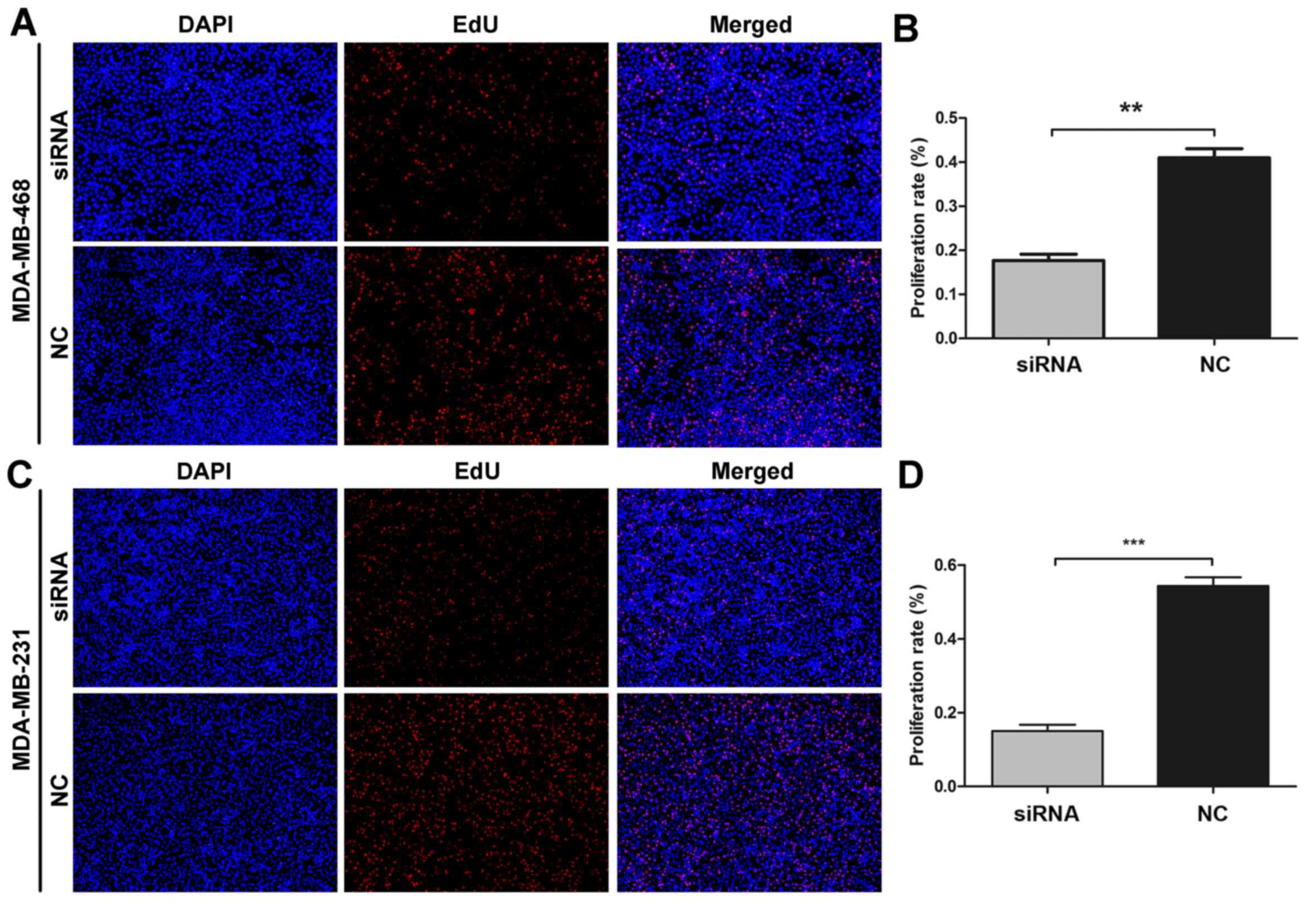
further observed in the EdU assay (Fig. 4A and C). The cell proliferation
rate was significantly decreased by 42.97±3.04 and 27.53±4.60% in
the MDA-MB-468 and MDA-MB-231 cells (all P<0.01) (Fig. 4B and D).
TMPRSS4 knockdown inhibits the migration
and invasion abilities of BC cells in vitro
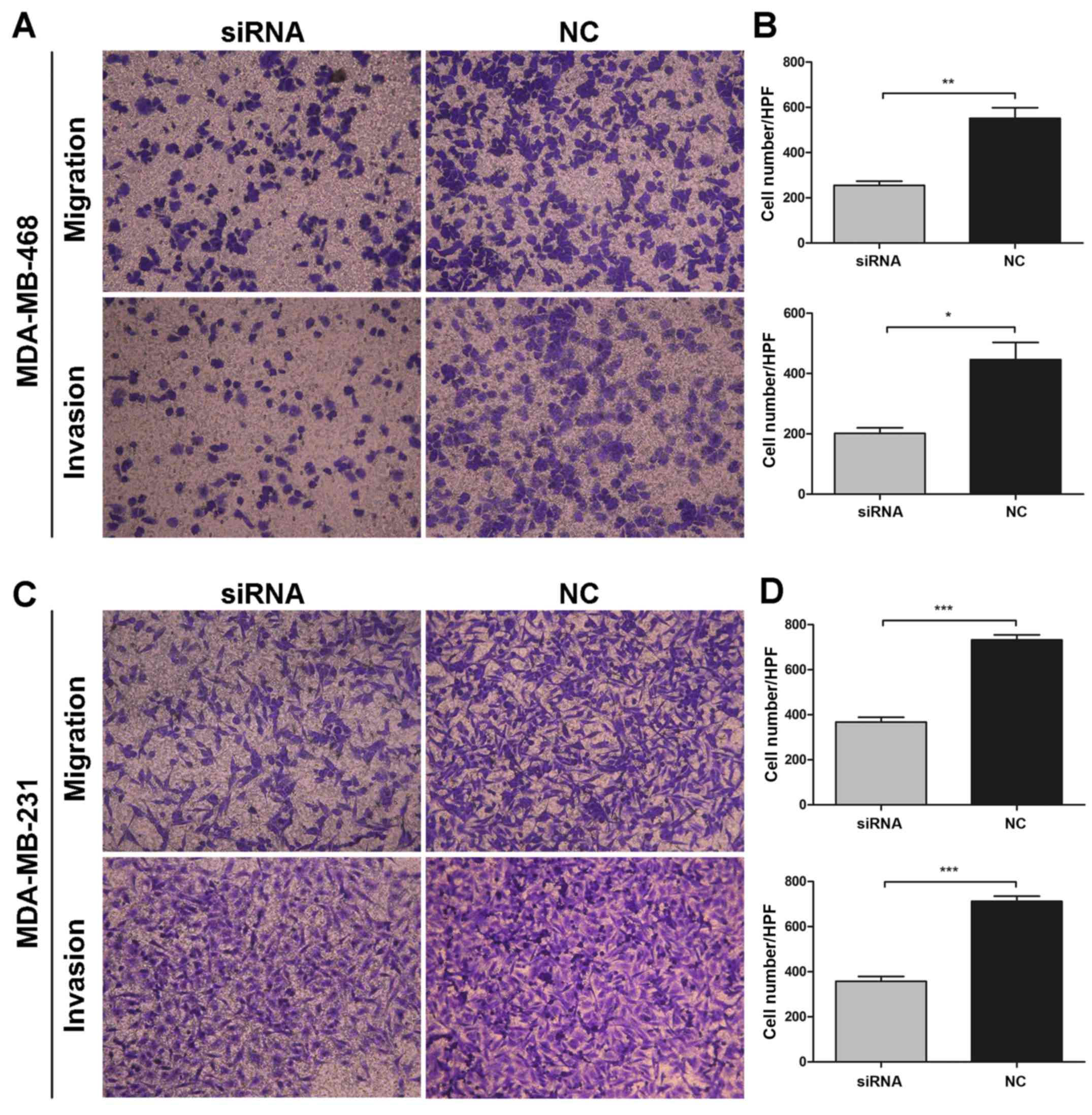
Given that TMPRSS4 was highly expressed in
metastatic breast cancer tissues and cell lines, the roles of
TMPRSS4 in cell migration and invasion in breast cancer were also
investigated. As shown in Fig. 5A and
C, MDA-MB-468 and MDA-MB-231 cells transfected with siRNA
showed a significantly decreased migration and invasion capability,
compared with the untreated cells. Transfected cells showed a
considerable decrease in migration activity by 46.40±1.68 and
50.12±2.41% and a decrease in invasion capacity by 45.59±2.75 and
50.13±2.49% in the MDA-MB-468 (all P<0.001) (Fig. 5B) and MDA-MB-231 (P<0.05 and
P<0.001) (Fig. 5D) cells,
respectively, when compared with these abilities noted in the
negative control group. Collectively, these results suggest that
knockdown of TMPRSS4 significantly inhibited the migration and
invasion activity of BC cells in vitro.
TMPRSS4 knockdown influences the
expression of EMT- related genes
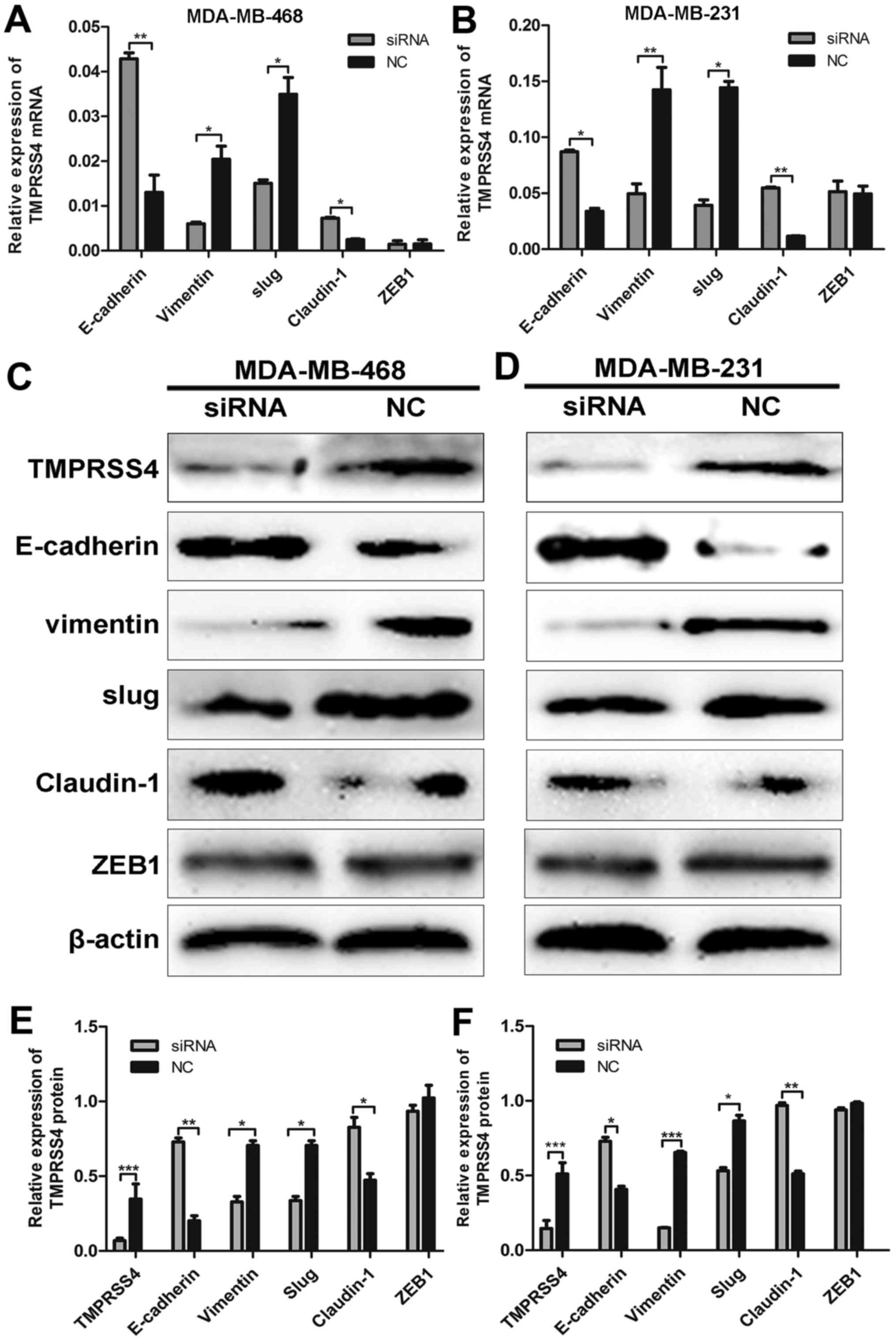
Following siRNA transfection, TMPRSS4 protein
expression was determined in the MDA-MB-468 and MDA-MB-231 cells by
western blot analysis (all P<0.05) (Fig. 6C and D). To verify the
relationship between TMPRSS4 expression and EMT, we observed the
expression of key biomarkers of EMT after TMPRSS4 was silenced.
RT-qPCR and western blot analysis revealed that TMPRSS4 knockdown
significantly enhanced the expression of E-cadherin and claudin-1
and inhibited the expression of vimentin and Slug in the MDA-MB-468
cells (all P<0.05) (Fig. 6A, C and
E), indicating suppression of EMT. Similar results were
observed in the MDA-MB-231 cells (all P<0.05) (Fig. 6B, D and F). The expression of ZEB1
did not show a significantly different change (P>0.05), and the
expression of Snail was undetectable in the two cell lines (data
not shown).
Discussion
The carcinogenesis and development of breast cancer
(BC) is linked to different molecular events, and the lack of
effective markers for the prediction of prognosis makes it
difficult to apply individualized treatment protocols to BC
patients. Proteases have been extensively studied as important
participants in the carcinogenesis of many types of tumors
(6–8). Recently, much attention has focused
on the role of TTSPs during tumor development and progression
(11,19). As a member of the family of cell
surface-associated proteases, TTSPs modulate a variety of normal
cellular activities as well as tumor invasion and metastasis
(19,20).
TMPRSS4, initially referred to as
TMPRSS3, is located on chromosome 11.q23.3 and encodes a
member of the type II TTSP family (21). Previous studies have shown that
TMPRSS4 is highly expressed in different types of cancer, such as
pancreatic cancer (21), thyroid
cancer (22), lung cancer
(23) and hepatocellular
carcinoma (15). Overexpression
of TMPRSS4 in non-small cell lung cancer is associated with poor
prognosis in patients with squamous histology (24). Huang et al (25) reported that TMPRSS4 is highly
expressed in colorectal cancer tissues both at the mRNA and protein
level and is correlated with pathological stage. In BC and
triple-negative BC (TNBC), high expression of TMPRSS4 was found to
be indicative of poor prognosis and related to tumor size, LN
metastasis and histological grade (26,27). Hence, TMPRSS4 appears to be a
factor regulating tumor development and progression. The present
study confirmed that TMPRSS4 was highly expressed in BC tissues
compared with normal breast tissues, and we further explored the
relationship between TMPRSS4 expression and histological subtypes.
Notably, TMPRSS4 was not only overexpressed in invasive ductal
carcinoma but also in invasive mucinous carcinoma and
micropapillary carcinoma. In addition, expression of TMPRSS4 was
increased along with grade, with the highest expression in grade
III BC tissues and the lowest in grade I BC tissues, suggesting a
positive correlation between TMPRSS4 and BC progression.
Our results also showed that TMPRSS4 overexpression
is significantly correlated with tumor size, histological grade,
lymph node metastasis, and clinical stage, suggesting that TMPRSS4
may be involved in the progression of BC. Survival analysis
demonstrated that high TMPRSS4 expression is associated with
shorter DFS and OS in BC patients. Univariate and multivariate Cox
regression analyses revealed that TMPRSS4 could serve as an
independent prognostic factor for BC. All these results confirmed
that TMPRSS4 is an important predictive indicator of BC
prognosis.
We identified the overexpression of TMPRSS4 in BC
tissues and different histological subtypes. Furthermore, RT-qPCR
and western blot analysis demonstrated that TMPRSS4 was
differentially expressed in five BC cell lines, with the highest
expression in MDA-MB-468 cells. Then silencing of TMPRSS4 in
MDA-MB-468 and MDA-MB-231 cells significantly inhibited cell
proliferation, migration and invasion in vitro. Here, we
show initial evidence that TMPRSS4 is highly expressed in BC cell
lines. However, the biological functions of TMPRSS4 and its
potential mechanisms in BC cells are not well understood. A recent
study reported that TFPI-2 negatively regulated cell growth by
inhibiting transcription of TMPRSS4 (28). Kim et al (14) also reported that TMPRSS4 induces
invasion, migration, and metastasis of cancer cells by facilitating
EMT events; TMPRSS4 induces invasion and EMT through upregulation
of integrin α5 and its signaling pathways.
EMT is a process implicated in the conversion of
early stage tumors to invasive malignancies. Induction of EMT
allows tumor cells to metastasize and establish secondary tumors at
a distant site due to weak intercellular adhesion and enhanced cell
motility (13,29,30). Loss of E-cadherin transcript, a
hallmark of EMT, plays a central role in the EMT process (13). In an attempt to determine the
mechanism by which E-cadherin is downregulated, we examined several
well-known E-cadherin transcriptional repressors/EMT-inducing
transcriptional repressors, including Slug, Snail, SIP1/ZEB1 and
E12/E47 (31). In this study,
western blot analysis showed that E-cadherin and claudin-1 were
upregulated after TMPRSS4 knockdown. On the contrary, Slug and
vimentin were downregulated, indicating that TMPRSS4 contributes to
tumor cell invasion and metastasis by promoting EMT. Cheng et
al (15) suggested that
TMPRSS4-induced EMT was mediated through Snail and Slug as a result
of Raf/MEK/ERK1/2 activation. However, the specific regulatory
mechanisms were not elucidated in our results and further in-depth
studies are required to validate these signaling pathways in BC
cells.
Taken together, our results demonstrated that the
upregulation of TMPRSS4 expression is a key event in BC progression
and it could promote cell proliferation, migration and invasion
abilities through possible induction of EMT. TMPRSS4 could be
regarded as a potential prognostic biomarker and a therapeutic
target for BC.
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (no. 81372856) and Taishan Scholars
Program of Shandong Province (no. ts201511096).
References
|
1
|
Miao H, Hartman M, Bhoo-Pathy N, Lee SC,
Taib NA, Tan EY, Chan P, Moons KG, Wong HS, Goh J, et al:
Predicting survival of de novo metastatic breast cancer in Asian
women: Systematic review and validation study. PLoS One.
9:e937552014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cummings MC, Chambers R, Simpson PT and
Lakhani SR: Molecular classification of breast cancer: Is it time
to pack up our microscopes? Pathology. 43:1–8. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ginsburg GS and Willard HF: Genomic and
personalized medicine: Foundations and applications. Transl Res.
154:277–287. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dechaphunkul A, Phukaoloun M,
Kanjanapradit K, Graham K, Ghosh S, Santos C and Mackey JR:
Prognostic significance of tissue inhibitor of metalloproteinase-1
in breast cancer. Int J Breast Cancer. 2012:2908542012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Geyer FC, Rodrigues DN, Weigelt B and
Reis-Filho JS: Molecular classification of estrogen
receptor-positive/luminal breast cancers. Adv Anat Pathol.
19:39–53. 2012. View Article : Google Scholar
|
|
6
|
Duffy MJ: Proteases as prognostic markers
in cancer. Clin Cancer Res. 2:613–618. 1996.PubMed/NCBI
|
|
7
|
Roy R, Yang J and Moses MA: Matrix
metalloproteinases as novel biomarkers and potential therapeutic
targets in human cancer. J Clin Oncol. 27:5287–5297. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Flores-Reséndiz D, Castellanos-Juárez E
and Benítez-Bribiesca L: Proteases in cancer progression. Gac Med
Mex. 145:131–142. 2009.In Spanish.
|
|
9
|
Netzel-Arnett S, Hooper JD, Szabo R,
Madison EL, Quigley JP, Bugge TH and Antalis TM: Membrane anchored
serine proteases: A rapidly expanding group of cell surface
proteolytic enzymes with potential roles in cancer. Cancer
Metastasis Rev. 22:237–258. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hooper JD, Clements JA, Quigley JP and
Antalis TM: Type II transmembrane serine proteases. Insights into
an emerging class of cell surface proteolytic enzymes. J Biol Chem.
276:857–860. 2001. View Article : Google Scholar
|
|
11
|
Choi SY, Bertram S, Glowacka I, Park YW
and Pöhlmann S: Type II transmembrane serine proteases in cancer
and viral infections. Trends Mol Med. 15:303–312. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
de Aberasturi AL and Calvo A: TMPRSS4: An
emerging potential therapeutic target in cancer. Br J Cancer.
112:4–8. 2015. View Article : Google Scholar :
|
|
13
|
Jung H, Lee KP, Park SJ, Park JH, Jang YS,
Choi SY, Jung JG, Jo K, Park DY, Yoon JH, et al: TMPRSS4 promotes
invasion, migration and metastasis of human tumor cells by
facilitating an epithelial-mesenchymal transition. Oncogene.
27:2635–2647. 2008. View Article : Google Scholar
|
|
14
|
Kim S, Kang HY, Nam EH, Choi MS, Zhao XF,
Hong CS, Lee JW, Lee JH and Park YK: TMPRSS4 induces invasion and
epithelial-mesenchymal transition through upregulation of integrin
alpha5 and its signaling pathways. Carcinogenesis. 31:597–606.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang CH, Guo ZY, Chen ZT, Zhi XT, Li DK,
Dong ZR, Chen ZQ, Hu SY and Li T: TMPRSS4 facilitates
epithelial-mesenchymal transition of hepatocellular carcinoma and
is a predictive marker for poor prognosis of patients after
curative resection. Sci Rep. 5:123662015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: : AJCC Cancer Staging Manual. 7th edition.
Springer; New York, NY: pp. 237–246. 2010
|
|
17
|
Lakhani SR, Ellis IO, Schnitt SJ, Tan PH
and van de Vijver MJ: WHO classification of tumours of the breast.
World Health Organization classification of tumours. 4th edition.
IARC Press; Lyon: 2012
|
|
18
|
Wolff AC, Hammond ME, Hicks DG, Dowsett M,
McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M,
Fitzgibbons P, et al American Society of Clinical Oncology; College
of American Pathologists: Recommendations for human epidermal
growth factor receptor 2 testing in breast cancer: American Society
of Clinical Oncology/College of American Pathologists clinical
practice guideline update. J Clin Oncol. 31:3997–4013. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Szabo R and Bugge TH: Type II
transmembrane serine proteases in development and disease. Int J
Biochem Cell Biol. 40:1297–1316. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Antalis TM, Bugge TH and Wu Q:
Membrane-anchored serine proteases in health and disease. Prog Mol
Biol Transl Sci. 99:1–50. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wallrapp C, Hähnel S, Müller-Pillasch F,
Burghardt B, Iwamura T, Ruthenbürger M, Lerch MM, Adler G and Gress
TM: A novel transmembrane serine protease (TMPRSS3) overexpressed
in pancreatic cancer. Cancer Res. 60:2602–2606. 2000.PubMed/NCBI
|
|
22
|
Kebebew E, Peng M, Reiff E, Duh QY, Clark
OH and McMillan A: ECM1 and TMPRSS4 are diagnostic markers of
malignant thyroid neoplasms and improve the accuracy of fine needle
aspiration biopsy. Ann Surg. 242:353–363. 2005.PubMed/NCBI
|
|
23
|
Nguyen TH, Weber W, Havari E, Connors T,
Bagley RG, McLaren R, Nambiar PR, Madden SL, Teicher BA, Roberts B,
et al: Expression of TMPRSS4 in non-small cell lung cancer and its
modulation by hypoxia. Int J Oncol. 41:829–838. 2012.PubMed/NCBI
|
|
24
|
Larzabal L, Nguewa PA, Pio R, Blanco D,
Sanchez B, Rodríguez MJ, Pajares MJ, Catena R, Montuenga LM and
Calvo A: Overexpression of TMPRSS4 in non-small cell lung cancer is
associated with poor prognosis in patients with squamous histology.
Br J Cancer. 105:1608–1614. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang A, Zhou H, Zhao H, Quan Y, Feng B
and Zheng M: TMPRSS4 correlates with colorectal cancer pathological
stage and regulates cell proliferation and self-renewal ability.
Cancer Biol Ther. 15:297–304. 2014. View Article : Google Scholar :
|
|
26
|
Liang B, Wu M, Bu Y, Zhao A and Xie F:
Prognostic value of TMPRSS4 expression in patients with breast
cancer. Med Oncol. 30:4972013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cheng D, Kong H and Li Y: TMPRSS4 as a
poor prognostic factor for triple-negative breast cancer. Int J Mol
Sci. 14:14659–14668. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hamamoto J, Soejima K, Naoki K, Yasuda H,
Hayashi Y, Yoda S, Nakayama S, Satomi R, Terai H, Ikemura S, et al:
Methylation-induced downregulation of TFPI-2 causes TMPRSS4
overexpression and contributes to oncogenesis in a subset of
non-small-cell lung carcinoma. Cancer Sci. 106:34–42. 2015.
View Article : Google Scholar :
|
|
29
|
Brockhausen J, Tay SS, Grzelak CA,
Bertolino P, Bowen DG, d'Avigdor WM, Teoh N, Pok S, Shackel N,
Gamble JR, et al: miR-181a mediates TGF-β-induced hepatocyte EMT
and is dysregulated in cirrhosis and hepatocellular cancer. Liver
Int. 35:240–253. 2015. View Article : Google Scholar
|
|
30
|
Xu J, Li X, Yang H, Chang R, Kong C and
Yang L: SIN1 promotes invasion and metastasis of hepatocellular
carcinoma by facilitating epithelial-mesenchymal transition.
Cancer. 119:2247–2257. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Choi SY, Shin HC, Kim SY and Park YW: Role
of TMPRSS4 during cancer progression. Drug News Perspect.
21:417–423. 2008.PubMed/NCBI
|