Introduction
Colorectal cancer (CRC) is the most common
gastrointestinal malignancy, and is one of the leading causes of
cancer-related deaths worldwide. Approximately 50,000 individuals
die of CRC each year. The estimated number of deaths are 26,020 and
23,170 for men and women, respectively, in 2016 (1).
Surgery, chemotherapy and irradiation remain the
most common treatments of this devastating disease. Yet, the
therapeutic effect of these traditional treatments is not as
effective as expected (2).
Therefore, improvement in the therapeutic approach is imperative
for more efficacious treatment of CRC. Targeted therapy is a new
treatment for human cancers and is a developing future trend.
Several targeted drugs have been used clinically for CRC therapy,
such as cetuximab (3) and
panitumumab (4), monoclonal
antibodies which target the epidermal growth factor receptor
(EGFR). EGFR is overexpressed in 60–70% of CRC cases, and these
patients could benefit from EGFR-targeted therapy (5,6).
Unfortunately, cetuximab is ineffective for patients without EGFR
overexpression or with EGFR extracellular domain mutations. Thus,
it is necessary to identify new targets of therapy in EGFR
signaling pathways.
The Ras gene, which locates at the downstream
of EGFR in the RAS/RAF/MAPK pathway plays a major role in the
development of CRC (7).
K-ras mutation is an early event in colorectal tumorgenesis
(8–10), and ocurs in 30–60% of all CRC
cases (11–18). However, N-ras mutations are
rare in CRC (19). Glarakis et
al found that the incidence of N-ras mutations is 1.3%
in CRC (1/76) (20). H-ras
mutations are far less common. From COSMIC database, among 765
colon adenocarcinomas only 1% were found to harbor H-ras
mutation (21). Mutant p21Ras
resulting from mutations of the ras gene abolish GAP-induced
GTP hydrolysis of Ras proteins, leading to constitutive activation
of ras (GTP-bound active form). Such activating mutations
result in constitutive signaling, and thereby cause an increase in
proliferation and in malignant transformation (22).
As known, most oncogenes play a carcinogenic role by
gene amplification and the overexpression of wild-type proteins
(23,24). Ras proteins, p21Ras, are
overexpressed in 29–76% of CRC (25), but the subtype of p21Ras proteins
that are overexpressed in CRC and mutation status remain unknown,
limiting the development of therapeutic antibodies targeting the
ras gene. Thus, the present study was performed to
investigate the subtypes of p21Ras proteins and mutation status in
CRC by immunohistochemistry and direct sequence analysis to explore
whether or not wild-type p21Ras could be a target for CRC
therapy.
Materials and methods
Samples
All samples were collected from archives at the
Kunming General Hospital between April 2009 and May 2015. This
study was approved by the Ethics Committee of Kunming General
Hospital. Written informed consent was provided by all patients. A
total of 378 samples were used, including 45 cases of normal
colorectal tissues (with a 5-cm distance from the tumor margin), 73
cases of colorectal inflammatory polyps, 48 cases of colorectal
low-grade intraepithelial neoplasia, 83 cases of colorectal
high-grade intraepithelial neoplasia and 129 cases of colorectal
cancers. All of the samples were formalin-fixed and paraffin
embedded (FFPE).
Among the 129 CRC patients, 85 were males and 44
were females with an average age of 58.06 years (range 30–93
years). Microscopically, 48 were poorly differentiated, 58 were
moderately differentiated, and 13 were well differentiated.
Seventy-eight patients presented with regional lymph node
metastasis. All of the patients receive no radiation therapy or
neoadjuvant chemotherapy prior to surgery.
Antibodies
Monoclonal antibody KGH-R1 which is able to react
with all three subtypes of p21Ras, H-p21Ras, N-p21Ras and K-p21Ras,
was prepared in our laboratory (26). Monoclonal antibody 60309-1-Ig to
K-p21Ras was purchased from ProteinTech Group (Chicago, IL, USA),
and monoclonal antibody sc-31 to N-p21Ras and monoclonal antibody
sc-29 to H-p21Ras were purchased from Santa Cruz Biotechnology,
Inc. (Santa Cruz, CA, USA).
Tissue microarray (TMA) construction
FFPE colorectal tissue samples were selected for TMA
construction (Boyikang, Beijing, China). Briefly, the areas
containing cancer tissues were annotated on hematoxylin and eosin
(H&E) slides and identified by two pathologists.
Four-millimeter cores were removed from the selected area (region
of interest) with a needle punch. These 4-mm donor cores were
subsequently embedded in previously arranged recipient paraffin
blocks through a precisely spaced 36-hole array pattern. Core
positions in the recipient paraffin block were noted on a TMA map.
After paraffin was intenerated and cooling, the recipient blocks
were cut using a microtome and used for immunohistochemistry.
Immunohistochemistry
Sections (4-μm) from the TMA containing CRC
and normal tissues were collected on glass slides coated with
adhesive polylysine. The sections were incubated in an oven at 60°C
for 3 h, then deparaffinized in xylene, hydrated in descending
concentrations of ethanol (2 × 100% for 3 min, 95% for 3 min and
85% for 3 min) and washed in double-distilled water three times.
Subsequently, the slides were subjected to specific epitope
unmasking by an autoclave (1600 W, 2 min) in citrate acid buffer
(0.01 M pH 6.0), and then exposed to 3% H2O2
to block endogenous peroxidase reactivity for 10 min, followed by
washing in distilled water. To avoid unspecific staining, 10% BSA
was used to block the sections for 40 min at 37°C. Then the
sections were incubated in the p21Ras monoclonal antibody at 4°C
overnight. Controls were obtained by incubating serial sections
with the blocking solution but incubated in phosphate-buffered
solutions (PBS, pH 7.2) instead of the primary antibodies, and then
washed in 0.01 mol/l PBS. The sections were then sequentially
exposed to horseradish peioxidase secondary antibody (ZSGB-BIO,
Beijing, China) for 30 min, and washed with PBS three times. To
visualize the sections, diaminobenzidine as a chromogen was applied
for 5–7 min and hematoxylin counterstain for 1 min. Finally, all
slides were dehydrated and mounted.
The expression of p21Ras was evaluated by the
percentage of positive cells and histological score (Hscore)
(27). Briefly, at least 300
cells were counted in every component on every slide. The staining
patterns were graded as membranous or cytoplasmic. Protein
immunoreactivity was scored according to the intensity of staining,
which was graded on an arbitrary scale ranging from 0 to 3: 0,
negative (no stained cells); 1, low (primrose yellow cells); 2,
medium (yellow cells); and 3, high expression (tawny cells). A mean
percentage of positive tumor cells was determined in at least 3
areas at ×40 magnifications and ranged from 0 to 100%.
DNA extraction
Tumor areas in the FFPE tissue blocks were circled
by two experienced pathologists. FFPE serial sections
(10-μm) were used for DNA extraction by means of the QIAamp
DNA FFPE tissue kit according to the QIAamp DNA FFPE tissue
handbook. The purity of the extracted DNA was tested using an
ultraviolet spectrophotometer.
PCR amplification reaction
Primer 5.0 and oligo softwares were used to design
primers for K-ras, N-ras and H-ras
amplication. Primer sequences were as follows K-ras exon 2
sense, 5′-TTATTATAAGGCCTGCTG-3′ and antisense, 5′-TGTATCAAAGA
ATGGTCC-3′; K-ras exon 3 sense, 5′-GTGTTTCTCCCTTCTCAG-3′ and
antisense, 5′-GGCATTAGCAAAGACTCA-3′; K-ras exon 4 sense,
5′-TGTTACTAATGACTGTGCTA-3′ and antisense, 5′-TAACAGTTATGATTTTGC-3′;
K-ras exon 5 sense, 5′-ACATGGCTTTCCCAGTAA-3′ and antisense,
5′-GTTGCCACCTTGTTACCT-3′; N-ras exon 2 sense,
5′-AATTAACCCTGATTACTGG-3′ and antisense, 5′-TAAAGATGATCCGAC AAG-3′;
N-ras exon 3 sense, 5′-TAACCTTGGCAATAGCAT-3′ and antisense,
5′-TAACCTCATTTCCCCATA-3′; N-ras exon 4 sense,
5′-CATGAGCCACTGTACCCA-3′ and antisense, 5′-TTGCACAAATGCTGAAAG-3′;
N-ras exon 5 sense, 5′-GAGATACAAATGCAAGAG-3′ and antisense,
5′-AAACACCAGCACTCCT-3′; H-ras exon 2 sense,
5′-AGACCCTGTAGGAGGACCC-3′ and antisense, 5′-CTGCTGGCACCTGGAC-3′;
H-ras exon 3 sense, 5′-CACGGAAGGTCCTGAGGGG-3′ and antisense,
5′-GCCTGGCCCCACCTGTG-3′; H-ras exon 4 sense,
5′-CTCTCGCTTTCCACCTCT-3′ and antisense, 5′-AGCTGTGGGGTGGAGA-3′;
H-ras exon 5 sense, 5′-GGCAGGCGGCCACAGG-3′ and antisense,
5′-ATCCGGTGGGCGTGGC-3′. Human K-ras, N-ras and
H-ras gene sequences were obtained from GeneBank AF493917,
AF493919 and AF493916, respectively. Exon 1 is an untranslated
region (UTR). The amplification was performed in a final volume of
25 μl containing 2.5 μl 10X PCR buffer, 2 μl
dNTP mixture, 1.0 μl Taq enzyme, 1.0 μl
forward primer (10 μmol/l), 1.0 μl reverse primer (10
μmol/l), and at least 800 ng DNA. PCR reaction conditions
were as follows: initial denaturation at 95°C for 4 min, 30 cycles
at 95°C for 30 sec, suited annealing temperature for 30 sec,
amplification at 72°C for 30 sec; and a final extension at 72°C for
10 min. The PCR amplification products were separated by 1% agarose
gel electrophoresis (AGE) for 30 min and imaged using the Syngene
imaging system (Synoptics Ltd., Cambridge UK).
DNA sequence analysis
Twenty microliters of PCR products were sent to the
Beijing Genomics Institute (BGI) for sequencing. The sequencing was
conducted in both directions (forward and reverse), and then
analyzed by Align X (Invitrogen, Carlsbad, CA, USA) and Chromas
(Technelysium Pty Ltd., Queensland, Australia) softwares.
Statistical analysis
The statistical analysis was performed using the
SPSS software package, standard version 22.0 (SPSS Inc., Chicago,
IL, USA). Data are expressed as mean ± SD. Statistical significance
was determined by the Student's t-test. A value of P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of p21Ras
p21Ras expression was detected in none of the normal
colorectal epithelium (0/45), in 9.59% of the inflammatory polyps
(7/73), in 64.58% of the low-grade intraepithelial neoplasia
samples (31/48), in 60.24% of the high-grade intraepithelial
neoplasia samples (50/83) and 64.89% of invasive colorectal
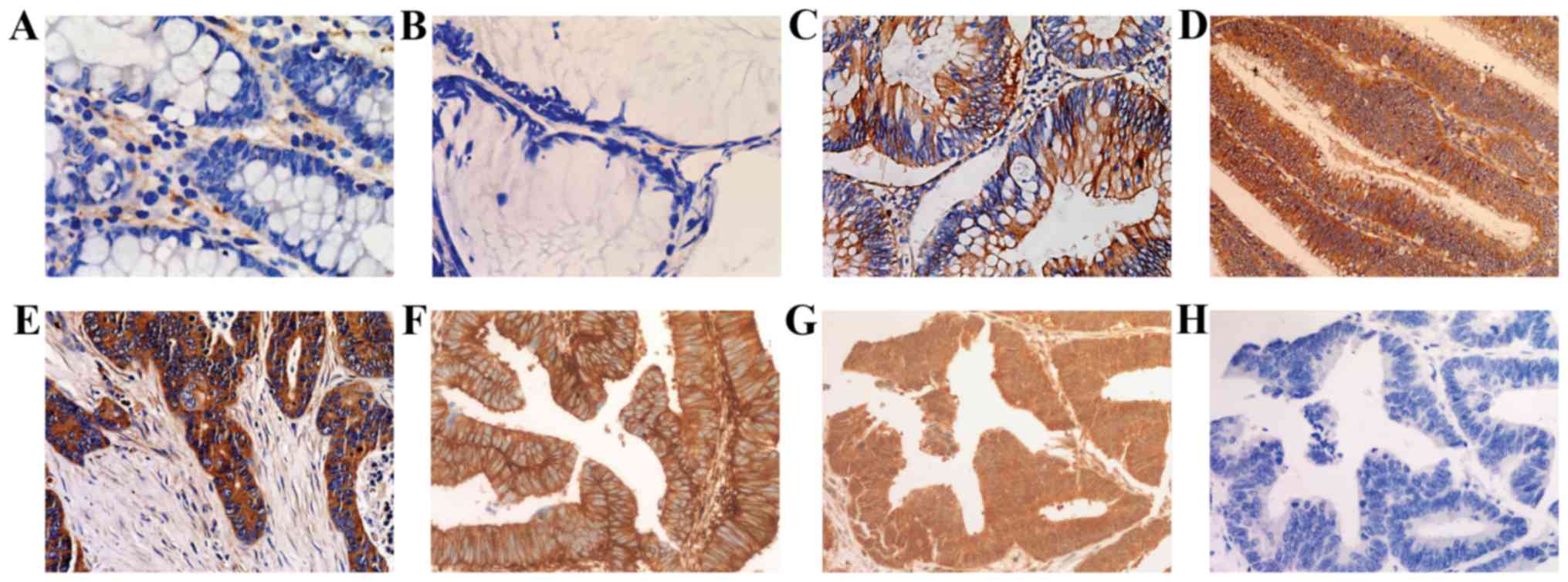
carcinoma samples (61/94) (Fig. 1
and Table I). The expression
products were localized in the cytoplasm and cell membrane.
 | Table Ip21Ras expression in benign lesions
and malignant tumors of colorectal cancer. |
Table I
p21Ras expression in benign lesions
and malignant tumors of colorectal cancer.
| Tissue | n | Positive | Percentage of
positive tissues (%) | Intensity | Percentage of
positive cells (%) | Hscore |
|---|
| Normal
epithelium | 45 | 0 | 0 | 0.0123 | 1.23±2.98 | 1.23±2.98 |
| Inflammatory
polyps | 73 | 7 | 9.59 | 0.0741 | 7.06±15.64 | 7.41±16.74 |
| Low-grade
intraepithelial neoplasia | 48 | 31 | 64.58 | 0.6654 | 53.33±42.36 | 66.54±59.91 |
| High-grade
intraepithelial neoplasia | 83 | 50 | 60.24 | 0.942 | 57.28±48.73 | 94.2±87.46 |
| Colorectal
carcinoma | 94 | 61 | 64.89 | 1.161 | 63.11±47.11 | 116.14±103.30 |
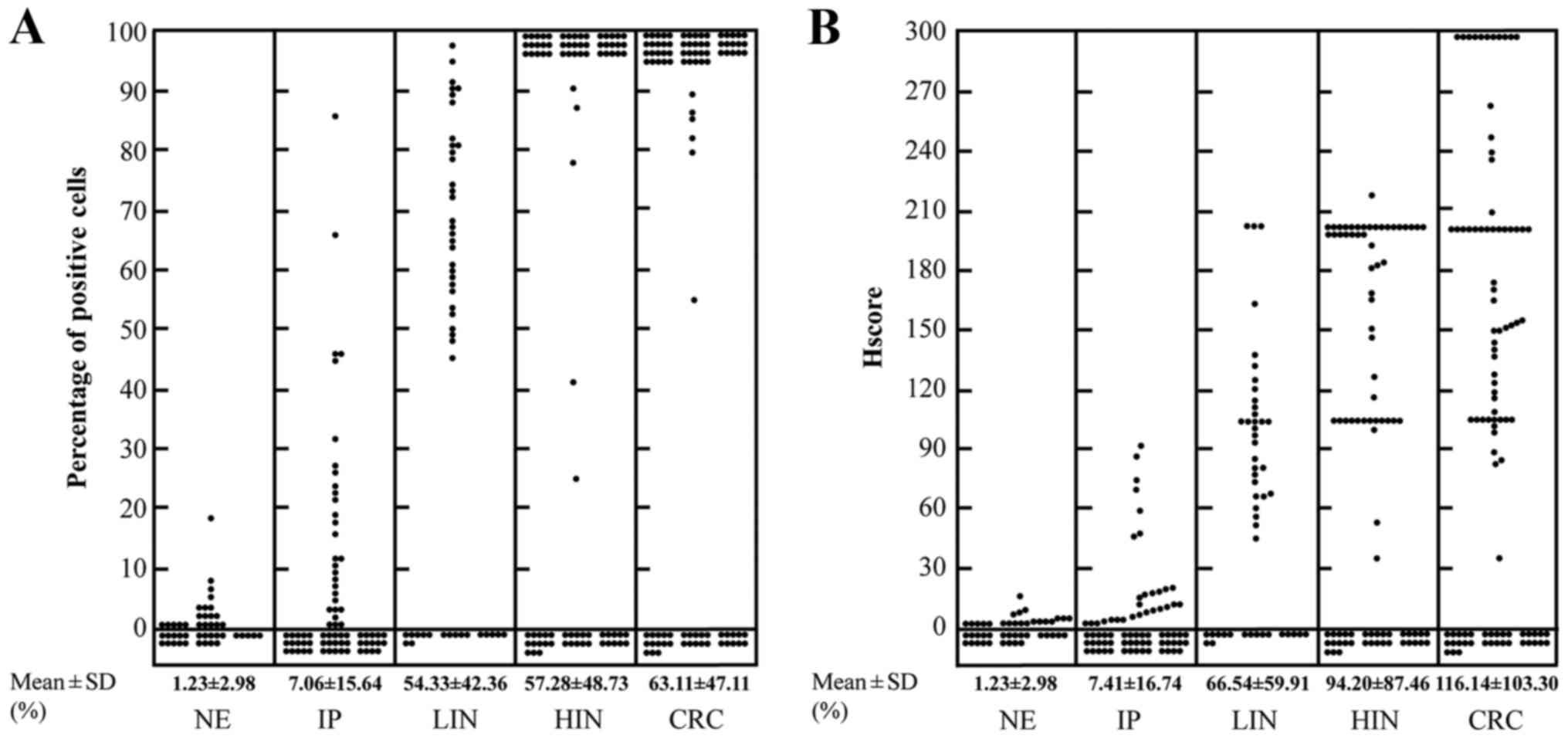
We used the percentage of positive cells and Hscore
to evaluate the expression level of p21Ras. In normal colorectal
epithelium, inflammatory polyps, low-grade intraepithelial
neoplasia, high-grade intraepithelial neoplasia and invasive
colorectal carcinoma samples, the mean percentages of positive
cells were 1.23±2.98, 7.06±15.64, 53.33±42.36, 57.28±48.73 and
63.11±47.11%, and the mean Hscores were 1.23±2.98, 7.41±16.74,
66.54±59.91, 94.20±87.46 and 116.14±103.30, respectively (Fig. 2 and Table I), which indicated a gradual
increase from normal colorectal epithelium to CRC.
Correlation between p21Ras expression and
clinicopathologic variables
A total of 94 CRC cases were adopted here. There was
a statistical correlation between histologic type and Hscores, and
also lymph node metastasis and Hscores (P<0.05). However, no
correlation was observed between Hscore and the other patient
clinicopathologic parameters (P>0.05), which suggests that the
expression of p21Ras and indices such as gender, age, invasive
depth are independent events. Furthermore, the same results were
found between the percentage of positive cells and the
clinicopathologic variables (Table
II).
 | Table IICorrelation between p21Ras expression
and the clinicopathologic features of the invasive colorectal
adenocarcinoma patients. |
Table II
Correlation between p21Ras expression
and the clinicopathologic features of the invasive colorectal
adenocarcinoma patients.
| Clinicopathologic
features | Cases (N=94) | Hscore | P-value | Percentage of
positive cells (%) | P-value |
|---|
| Gender | | | >0.05 | | >0.05 |
| Male | 64 | 114.29±103.76 | | 63.33±46.78 | |
| Female | 30 | 120.10±103.97 | | 62.63±48.62 | |
| Age (years) | | | >0.05 | | >0.05 |
| ≤50 | 28 | 105.07±104.43 | | 55.95±49.55 | |
| <50 | 66 | 120.84±103.26 | | 66.14±46.09 | |
| Histologic
type | | | <0.05 | | <0.05 |
| Non-mucinous
adenocarcinoma | 84 | 121.43±101.70 | | 66.40±46.20 | |
| Mucinous
adenocarcinoma | 10 | 35.4±66.99 | | 25.40±42.77 | |
|
Differentiation | | | <0.01 | | <0.01 |
| Well | 13 | 40.92±73.16 | | 28.77±45.08 | |
| Moderate | 34 | 102.78±90.99 | | 63.75±47.95 | |
| Poor | 47 | 182.08±91.68 | | 87.47±31.42 | |
| Invasive depth | | | >0.05 | | >0.05 |
| Superficial
muscle | 7 | 83.71±83.61 | | 57.14±53.45 | |
| Deep muscle | 17 | 103.65±108.56 | | 57.59±49.91 | |
| Full
thickness | 70 | 122.42±104.20 | | 65.04±46.37 | |
| Tumor size
(cm) | | | >0.05 | | >0.05 |
| <2 | 12 | 72.17±93.12 | | 45.96±48.44 | |
| 2–5 | 59 | 123.94±101.51 | | 67.81±45.85 | |
| >5 | 22 | 119.09±111.27 | | 60.00±49.36 | |
| Lymph node
metastasis | | | <0.05 | | <0.05 |
| − | 63 | 131.21±104.57 | | 68.97±45.88 | |
| + | 31 | 85.52±95.06 | | 51.19±48.12 | |
Expression of p21Ras subtypes in CRC
Three Mabs, each of which is able to recognize one
of the p21Ras subtypes were used to detect the expression of the
three p21Ras subtypes by immunohistochemistry (Fig. 1). It was demonstrated that
K-p21Ras was expressed in all 35 CRC, N-p21Ras was expressed in
30/35 of CRC samples, and H-p21Ras was not expressed in all of the
CRCs tested (Table III).
Notably, overexpression of both K-p21Ras and N-p21Ras were detected
in 30 cases (Table III).
Analysis of the immunohistochemical staining of K-p21Ras, N-p21Ras
and H-p21Ras was also evaluated according to the percentage of
positive cells and Hscore, which were 92.08±10.98, 77.00±33.21, 0%
and 180.08±50.81, 154.04±92.26, 0, respectively.
 | Table IIIp21Ras expression and ras
mutations in CRC. |
Table III
p21Ras expression and ras
mutations in CRC.
| Patients | K-ras
| H-ras
| N-ras
|
|---|
| Expression | Exon | Mutation | Expression | Mutation | Expression | Exon | Mutation |
|---|
| 201503220 | + | Exon 4 |
c.436G>A→p.A146T | - | - | - | Exon 2 | c.81T>C |
| 201503213 | + | - | - | - | - | - | Exon 2 | c.81T>C |
| 201503102 | + | - | - | - | - | - | - | - |
| 201502928 | + | Exon 4 |
c.436G>A→p.A146T | + | - | - | - | - |
| 201502903 | + | - | - | + | - | - | Exon 2 | c.81T>C |
| 201502056 | + | Exon 2 |
c.35G>A→p.G12D | + | - | - | - | - |
| 201501338 | + | - | - | + | - | - | Exon 2 | c.81T>C |
| 201501304 | + | - | - | + | - | - | - | - |
| 201500934 | + | - | - | + | - | - | Exon 2 | c.81T>C |
| 201500667 | + | - | - | + | - | - | - | - |
| 201412412 | + | Exon 2 |
c.38G>A→p.G13D | + | - | - | - | - |
| 201410010 | + | - | - | + | - | - | - | - |
| 201409737 | + | Exon 2 |
c.38G>A→p.G13D | + | - | - | - | - |
| 201408315 | + | Exon 2 |
c.35G>C→p.G12A | + | - | - | Exon 2 | c.81T>C |
| 201407762 | + | - | - | + | - | - | - | - |
| 201406231 | + | Exon 4 |
c.436G>A→p.A146T | | | | | |
| | Exon 5 |
c.526>T→p.E176Stop | - | - | - | - | - |
| 201405694 | + | - | - | + | - | - | Exon 2 | c.81T>C |
| 201405647 | + | Exon 2 |
c.35G>A→p.G12D | + | - | - | Exon 2 | c.81T>C |
| 201503581 | + | - | - | + | - | - | - | - |
| 201503452 | + | - | - | - | - | - | - | - |
| 201502977 | + | Exon 5 |
c.467T>C→p.F156S | + | - | - | - | - |
| 201502204 | + | - | - | + | - | - | - | - |
| 201501593 | + | Exon 2 |
c.35G>T→p.G12V | + | - | - | Exon 2 | c.81T>C |
| 201501337 | + | - | - | + | - | - | - | - |
| 201501079 | + | - | - | + | - | - | - | - |
| 201407828 | + | Exon 2 |
c.38G>A→p.G13D | + | - | - | - | - |
| 201407425 | + | - | - | + | - | - | Exon 2 | c.81T>C |
| 201407236 | + | Exon 2 |
c.35G>A→p.G12D | + | - | - | - | - |
| 201405983 | + | Exon 2 |
c.38G>A→p.G13D | + | - | - | Exon 2 | c.81T>C |
| 201405149 | + | - | - | + | - | - | - | - |
| 201404719 | + | - | - | + | - | - | - | - |
| 201404718 | + | Exon 2 |
c.38G>A→p.G13D | + | - | - | Exon 2 | c.81T>C |
| 201404238 | + | - | - | + | - | - | - | - |
| 201401791 | + | - | - | + | - | - | - | - |
| 201401610 | + | - | - | + | - | - | Exon 2 | c.81T>C |
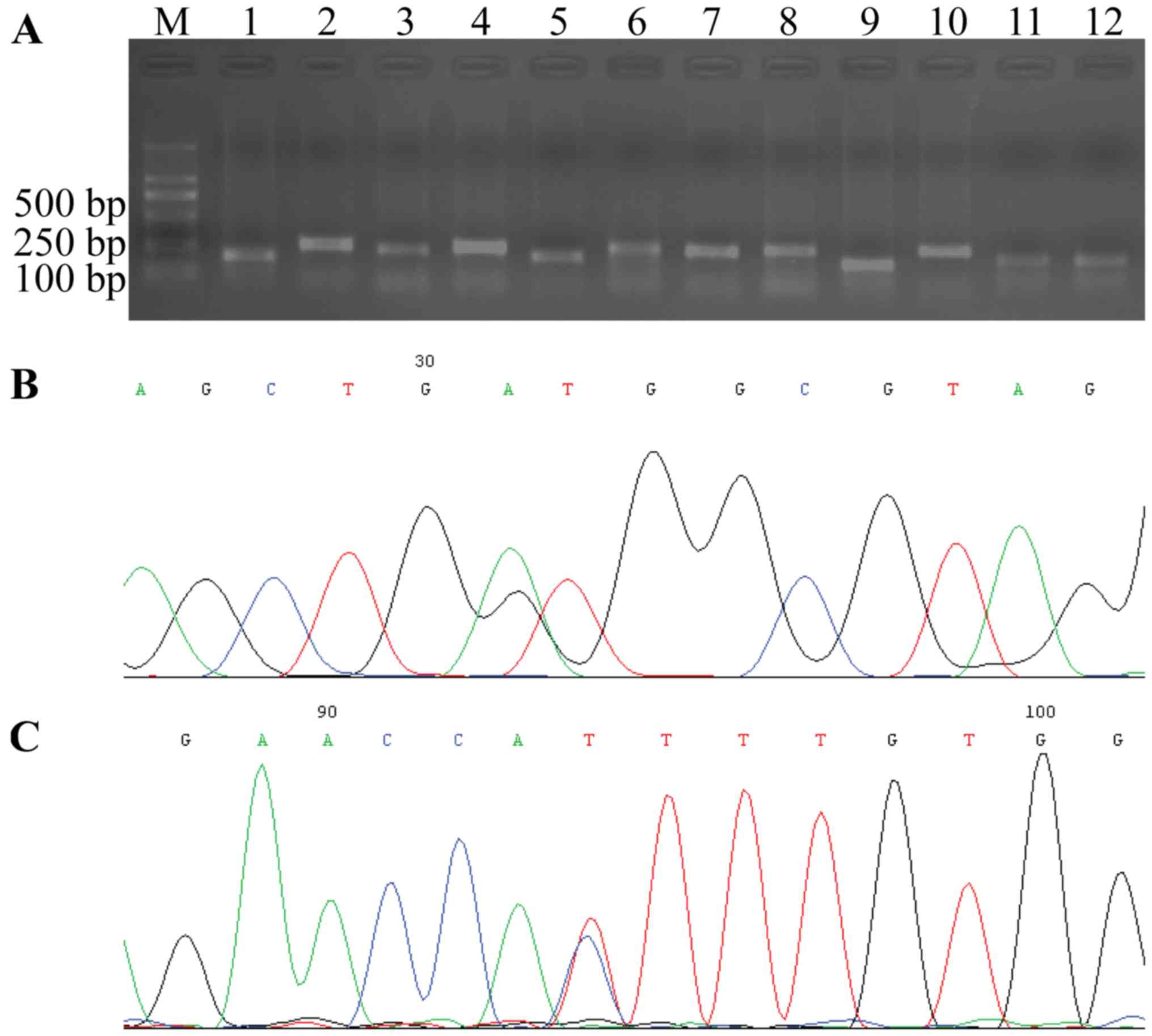
Ras mutation status in CRC
All 12 exons of K-ras, N-ras and
H-ras in 35 of the CRC cases were amplified successfully by
designed primers. Each PCR product was confirmed by agarose gel
electrophoresis with expected sizes of 185, 274, 255, 263, 217,
339, 255, 286, 187, 276, 207 and 215 bp, respectively (Fig. 3).
Among the 35 cases of CRC with K-p21Ras
overexpression, K-ras mutations were detected in 40% of the
cases (14/35), however, K-ras mutations were not detected in
the other 60% of CRC cases which indicated that the overexpression
of p21Ras in these 60% CRC were wild-type. K-ras mutation
was present in codon 12 (5 cases), 13 (5 cases), 146 (3 cases), 156
(1 case) and 176 (1 case). The most frequent K-ras mutation
was transition of base G/A (5/14, 35.7%) in codon 13, which
resulted in the substitution of glycine with aspartate.
N-ras mutations were not found in all 30 of the
N-p21Ras-overexpressing CRC cases (Table III). In CRC without N-p21Ras
expression no N-ras mutation was detected, and in CRC
without H-p21Ras expression only a H-ras nonsense mutation
at codon 27 was found (Fig.
3).
Discussion
Amplification of oncogenes and protein
overexpression have been identified in various solid tumors.
Overexpression of the human epidermal growth factor receptor 2
(HER2) gene occurs in 15–25% of human breast cancers (28). EGFR is overexpressed in 40–60% of
non-small cell lung cancer cases (23). Overexpression is considered to be
the main activation mechanisms of oncogenes, and oncogene proteins
could be potential targets for cancer therapy. Trastuzumab
(29), pertuzumab (30) and lapatinib (31) targeting HER2 protein have been
approved as standard care for inhibiting HER2 activity in the
treatment of HER2-positive breast cancer. Cetuximab (3,32),
panitumumab (4,33) and nimotuzomab (34) targeting EGFR protein have been
used to treat human cancers with EGFR overexpression, such as CRC
and non-small cell lung cancer.
Ras gene protein p21Ras was found to be
overexpressed in most human tumors, including CRC (35), bladder cancer (36), breast cancer (37,38), stomach adenocarcinomas (39), thyroid cancer (40) and laryngeal cancer (41). However, no targeted drugs that
target against p21Ras directly have been exploited. Recently, we
prepared a novel anti-p21Ras Mab, KGH-R1, which can recognize and
react with three type of p21Ras, including H-p21Ras, N-p21Ras and
K-p21Ras, and the single chain antibody derived from this Mab could
regress p21Ras-overexpressing tumors in vitro and in
vivo (26,42). In this study, the Mab was employed
to examine p21Ras expression in normal colorectal epithelium,
inflammatory polyps, low-grade intraepithelial neoplasia,
high-grade intraepithelial neoplasia and invasive CRC. The results
showed that there was almost no p21Ras expression in normal
colorectal mucosa, but high level expression of p21Ras in CRC and
colorectal intraepithelial neoplasia. Together with the reported
data (15,43–46), we confirmed that p21Ras
overexpression is an important event in colorectal carcinogenesis
and plays a major role in the development of CRC.
Subsequently, we evaluated expression of p21Ras
subtypes by immunohistochemistry using anti-K-ras, anti-N-ras or
anti-H-ras Mab, and found that K-p21Ras was expressed in all of the
tested CRC tissues, which was significantly higher than the results
of Elsabah and Adel (42.3%) (47). The different frequency of Ras
expression probably resulted from region variations, and the
difference in sample sources (48,49). Additionally, we found that
N-p21Ras was expressed in 85.7% of the CRC cases, but H-p21Ras was
not expressed in any tested CRC case. Our data indicated that
K-p21Ras and N-p21Ras are deeply involved in CRC development.
Furthermore, DNA sequencing was used to reveal the
mutation status of the overexpressed p21Ras, and found that 60% of
K-p21Ras-overexpressing CRC samples did not harbor K-ras
mutation. N-ras mutation was not found in any of the
N-p21Ras-overexpressing CRCs. Thus, overexpression of the wild-type
p21Ras may be another important mechanism in CRC development, and
the therapeutic antibodies targeting wild-type p21Ras may have
better prospect for the therapy of CRC. To date, few studies have
reported the overexpression of wild-type p21Ras in cancers. To the
best of our knowledge, this is the first time to reveal wild-type
p21Ras expression in CRC. The mechanism involved in the induction
of tumorigenesis by the overexpression of wild-type p21Ras remains
unclear. Zheng et al reported that overexpression of the
wild-type N-p21Ras induces IL-8 by binding and activating the
cytoplasmic pool of JAK2. IL-8 then acts on tumor cells and
promotes the progression of cancer (50). In addition, we speculated that
overexpression of the wild-type p21Ras leads to the excessive
GTP-bound active form that cannot be completely hydrolyzed, and
finally stimulates persistent cell proliferation and tumorigenesis.
However, on the other hand, Spandidos and Wilkie reported that
after rat 208F cells (a derivative of Rat-1 cells) were transfected
with T24 mutant H-ras (51) or the mutant N-ras (52), the expression level of normal
H-ras1 gene was elevated, leading to suppression of the
transformed and tumorigenic phenotypes induced by mutant ras
genes. Thus, wild-type H-p21Ras plays a complex role in the
development of cancers, and further studies are needed to clarify
the mechanisms of wild-type p21Ras overexpression in cancer
development.
In conclusion, we detected the expression level of
p21Ras in benign and malignant CRC, as well as the p21Ras subtypes
and mutation status of the ras gene in CRC. We conclude that
the overexpression of wild-type p21Ras, especially wild-type
K-p21Ras and N-p21Ras play a prominent role in the development of
CRC. This also implies that wild-type p21Ras is a promising target
for CRC therapy and it is feasible to develop the antibody drugs
against wild-type p21Ras.
Acknowledgments
The present study was supported by grants from the
National Natural Science Foundation of China (no. 81460464) and the
Applied Foundation Key Project of Yunnan Province (no.
2013FA059).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cunningham D, Atkin W, Lenz HJ, Lynch HT,
Minsky B, Nordlinger B and Starling N: Colorectal cancer. Lancet.
375:1030–1047. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jonker DJ, O'Callaghan CJ, Karapetis CS,
Zalcberg JR, Tu D, Au HJ, Berry SR, Krahn M, Price T, Simes RJ, et
al: Cetuximab for the treatment of colorectal cancer. N Engl J Med.
357:2040–2048. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sartore-Bianchi A, Moroni M, Veronese S,
Carnaghi C, Bajetta E, Luppi G, Sobrero A, Barone C, Cascinu S,
Colucci G, et al: Epidermal growth factor receptor gene copy number
and clinical outcome of metastatic colorectal cancer treated with
panitumumab. J Clin Oncol. 25:3238–3245. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Del Vecchio Blanco G, Paoluzi OA, Sileri
P, Rossi P, Sica G and Pallone F: Familial colorectal cancer
screening: When and what to do? World J Gastroenterol.
21:7944–7953. 2015.PubMed/NCBI
|
|
6
|
Jonker DJ, Karapetis CS, Harbison C,
O'Callaghan CJ, Tu D, Simes RJ, Malone DP, Langer C, Tebbutt N,
Price TJ, et al: Epiregulin gene expression as a biomarker of
benefit from cetuximab in the treatment of advanced colorectal
cancer. Br J Cancer. 110:648–655. 2014. View Article : Google Scholar :
|
|
7
|
Adjei AA: Blocking oncogenic Ras signaling
for cancer therapy. J Natl Cancer Inst. 93:1062–1074. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bos JL, Fearon ER, Hamilton SR, Verlaan-de
Vries M, van Boom JH, van der Eb AJ and Vogelstein B: Prevalence of
ras gene mutations in human colorectal cancers. Nature.
327:293–297. 1987. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ohnishi T, Tomita N, Monden T, Ohue M,
Yana I, Takami K, Yamamoto H, Yagyu T, Kikkawa N, Shimano T, et al:
A detailed analysis of the role of K-ras gene mutation in the
progression of colorectal adenoma. Br J Cancer. 75:341–347. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Puerta-García E, Cañadas-Garre M and
Calleja-Hernández MA: Molecular biomarkers in colorectal carcinoma.
Pharmacogenomics. 16:1189–1222. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu X, Jakubowski M and Hunt JL: KRAS gene
mutation in colorectal cancer is correlated with increased
proliferation and spontaneous apoptosis. Am J Clin Pathol.
135:245–252. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Andreyev HJ, Norman AR, Cunningham D,
Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N,
et al: Kirsten ras mutations in patients with colorectal cancer:
The 'RASCAL II' study. Br J Cancer. 85:692–696. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chang YS, Chang SJ, Yeh KT, Lin TH and
Chang JG: RAS, BRAF, and TP53 gene mutations in Taiwanese
colorectal cancer patients. Onkologie. 36:719–724. 2013.PubMed/NCBI
|
|
14
|
Palmirotta R, Savonarola A, Ludovici G, De
Marchis ML, Covello R, Ettorre GM, Ialongo C and Guadagni F:
Concurrent mutation in exons 1 and 2 of the K-ras oncogene in
colorectal cancer. Folia Histochem Cytobiol. 49:729–733. 2011.
View Article : Google Scholar
|
|
15
|
Salhab N, Jones DJ, Bos JL, Kinsella A and
Schofield PF: Detection of ras gene alterations and ras proteins in
colorectal cancer. Dis Colon Rectum. 32:659–664. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Thomas RJ, Liu YS, St Clair F, Norris PM,
Valentine R and Phillips WA: Frequency and clinico-pathological
associations of ras mutations in colorectal cancer in the Victorian
population. Aust NZ J Surg. 67:233–238. 1997. View Article : Google Scholar
|
|
17
|
Yaeger R, Cowell E, Chou JF, Gewirtz AN,
Borsu L, Vakiani E, Solit DB, Rosen N, Capanu M, Ladanyi M, et al:
RAS mutations affect pattern of metastatic spread and increase
propensity for brain metastasis in colorectal cancer. Cancer.
121:1195–1203. 2015. View Article : Google Scholar :
|
|
18
|
Kiaris H and Spandidos D: Mutations of ras
genes in human tumors (Review). Int J Oncol. 7:413–421.
1995.PubMed/NCBI
|
|
19
|
Irahara N, Baba Y, Nosho K, Shima K, Yan
L, Dias-Santagata D, Iafrate AJ, Fuchs CS, Haigis KM and Ogino S:
NRAS mutations are rare in colorectal cancer. Diagn Mol Pathol.
19:157–163. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Glarakis IS, Savva S and Spandidos DA:
Activation of the ras genes in malignant and premalignant
colorectal tumors. Oncol Rep. 5:1451–1454. 1998.PubMed/NCBI
|
|
21
|
Boidot R, Chevrier S, Julie V, Ladoire S
and Ghiringhelli F: HRAS G13D, a new mutation implicated in the
resistance to anti-EGFR therapies in colorectal cancer, a case
report. Int J Colorectal Dis. 31:1245–1246. 2016. View Article : Google Scholar
|
|
22
|
Spandidos DA, Sourvinos G, Tsatsanis C and
Zafiropoulos A: Normal ras genes: Their onco-suppressor and
pro-apoptotic functions (Review). Int J Oncol. 21:237–241.
2002.PubMed/NCBI
|
|
23
|
Xu N, Fang W, Mu L, Tang Y, Gao L, Ren S,
Cao D, Zhou L, Zhang A, Liu D, et al: Overexpression of wild-type
EGFR is tumorigenic and denotes a therapeutic target in non-small
cell lung cancer. Oncotarget. 7:3884–3896. 2016.
|
|
24
|
Lim SO, Park YM, Kim HS, Quan X, Yoo JE,
Park YN, Choi GH and Jung G: Notch1 differentially regulates
oncogenesis by wild-type p53 overexpression and P53 mutation in
grade III hepatocellular carcinoma. Hepatology. 53:1352–1362. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
McDermott U, Longley DB and Johnston PG:
Molecular and biochemical markers in colorectal cancer. Ann Oncol.
13(Suppl 4): 235–245. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang JL, Liu DX, Zhen SJ, Zhou YG, Zhang
DJ, Yang LY, Chen HB and Feng Q: A novel anti-p21R as scFv antibody
reacting specifically with human tumour cell lines and primary
tumour tissues. BMC Cancer. 16:1312016. View Article : Google Scholar
|
|
27
|
Budwit-Novotny DA, McCarty KS, Cox EB,
Soper JT, Mutch DG, Creasman WT, Flowers JL and McCarty KS Jr:
Immunohistochemical analyses of estrogen receptor in endometrial
adenocarcinoma using a monoclonal antibody. Cancer Res.
46:5419–5425. 1986.PubMed/NCBI
|
|
28
|
Ferretti G, Felici A, Papaldo P, Fabi A
and Cognetti F: HER2/neu role in breast cancer: From a prognostic
foe to a predictive friend. Curr Opin Obstet Gynecol. 19:56–62.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baselga J, Manikhas A, Cortés J, Llombart
A, Roman L, Semiglazov VF, Byakhov M, Lokanatha D, Forenza S,
Goldfarb RH, et al: Phase III trial of nonpegylated liposomal
doxorubicin in combination with trastuzumab and paclitaxel in
HER2-positive metastatic breast cancer. Ann Oncol. 25:592–598.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Swain SM, Kim SB, Cortés J, Ro J,
Semiglazov V, Campone M, Ciruelos E, Ferrero JM, Schneeweiss A,
Knott A, et al: Pertuzumab, trastuzumab, and docetaxel for
HER2-positive metastatic breast cancer (CLEOPATRA study): Overall
survival results from a randomised, double-blind,
placebo-controlled, phase 3 study. Lancet Oncol. 14:461–471. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guan Z, Xu B, DeSilvio ML, Shen Z,
Arpornwirat W, Tong Z, Lorvidhaya V, Jiang Z, Yang J, Makhson A, et
al: Randomized trial of lapatinib versus placebo added to
paclitaxel in the treatment of human epidermal growth factor
receptor 2-overexpressing metastatic breast cancer. J Clin Oncol.
31:1947–1953. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang F, Yu Y, Xing L and Chen M:
Cetuximab combined with chemotherapy is beneficial for patients
with advanced non-small cell lung cancer after EGFR-tyrosine kinase
inhibitors failure. Int J Clin Exp Med. 8:16140–16148.
2015.PubMed/NCBI
|
|
33
|
Socinski MA: Antibodies to the epidermal
growth factor receptor in non small cell lung cancer: current
status of matuzumab and panitumumab. Clin Cancer Res.
13:s4597–4601. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Boland W and Bebb G: The emerging role of
nimotuzumab in the treatment of non-small cell lung cancer.
Biologics. 4:289–298. 2010.PubMed/NCBI
|
|
35
|
Hand PH, Thor A, Wunderlich D, Muraro R,
Caruso A and Schlom J: Monoclonal antibodies of predefined
specificity detect activated ras gene expression in human mammary
and colon carcinomas. Proc Natl Acad Sci USA. 81:5227–5231. 1984.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Viola MV, Fromowitz F, Oravez S, Deb S and
Schlom J: ras oncogene P21 expression is increased in premalignant
lesions and high grade bladder carcinoma. J Exp Med. 161:1213–1218.
1985. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ohuchi N, Thor A, Page DL, Hand PH, Halter
SA and Schlom J: Expression of the 21,000 molecular weight ras
protein in a spectrum of benign and malignant human mammary
tissues. Cancer Res. 46:2511–2519. 1986.PubMed/NCBI
|
|
38
|
Ghosh AK, Moore M and Harris M:
Immunohistochemical detection of ras oncogene P21 product in benign
and malignant mammary tissue in man. J Clin Pathol. 39:428–434.
1986. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ohuchi N, Hand PH, Merlo G, Fujita J,
Mariani-Costantini R, Thor A, Nose M, Callahan R and Schlom J:
Enhanced expression of c-Ha-ras p21 in human stomach
adenocarcinomas defined by immunoassays using monoclonal antibodies
and in situ hybridization. Cancer Res. 47:1413–1420.
1987.PubMed/NCBI
|
|
40
|
Johnson TL, Lloyd RV and Thor A:
Expression of ras oncogene P21 antigen in normal and proliferative
thyroid tissues. Am J Pathol. 127:60–65. 1987.PubMed/NCBI
|
|
41
|
Scambia G, Catozzi L, Benedetti Panici P,
Ferrandina G, Almadori G, Paludetti G, Cadoni G, Distefano M,
Piffanelli A and Mancuso S: Expression of ras oncogene P21 protein
in normal and neoplastic laryngeal tissues: Correlation with
histopathological features and epidermal growth factor receptors.
Br J Cancer. 69:995–999. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang JL, Pan XY, Zhao WX, Hu QC, Ding F,
Feng Q, Li GY and Luo Y: The antitumor efficacy of a novel
adenovirus-mediated anti-p21Ras single chain fragment variable
antibody on human cancers in vitro and in vivo. Int J Oncol.
48:1218–1228. 2016.PubMed/NCBI
|
|
43
|
Sammoud S, Khiari M, Semeh A, Amine L,
Ines C, Amira A, Lilia K, Taher K, Sabeh M and Saadia B:
Relationship between expression of ras P21 oncoprotein and mutation
status of the K-ras gene in sporadic colorectal cancer patients in
Tunisia. Appl Immunohistochem Mol Morphol. 20:146–152. 2012.
View Article : Google Scholar
|
|
44
|
Allen DC, Foster H, Orchin JC and Biggart
JD: Immunohistochemical staining of colorectal tissues with
monoclonal antibodies to ras oncogene p21 product and carbohydrate
determinant antigen 19-9. J Clin Pathol. 40:157–162. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sun XF, Wingren S, Carstensen JM, Stål O,
Hatschek T, Boeryd B, Nordenskjöld B and Zhang H: ras P21
expression in relation to DNA ploidy, S-phase fraction and
prognosis in colorectal adenocarcinoma. Eur J Cancer. 27:1646–1649.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Morris VK, Lucas FA, Overman MJ, Eng C,
Morelli MP, Jiang ZQ, Luthra R, Meric-Bernstam F, Maru D, Scheet P,
et al: Clinicopathologic characteristics and gene expression
analyses of non-KRAS 12/13, RAS-mutated metastatic colorectal
cancer. Ann Oncol. 25:2008–2014. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Elsabah MT and Adel I: Immunohistochemical
assay for detection of K-ras protein expression in metastatic
colorectal cancer. J Egypt Natl Canc Inst. 25:51–56. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hirvikoski P, Auvinen A, Servomaa K, Kiuru
A, Rytömaa T, Makkonen K and Kosma VM: K-ras and P53 mutations and
overexpressions as prognostic factors in female rectal carcinoma.
Anticancer Res. 19:685–691. 1999.PubMed/NCBI
|
|
49
|
Okulczyk B, Kovalchuk O, Piotrowski Z,
Myśliwiec P and Chyczewski L: Clinical usefulness of K-RAS mutation
detection in colorectal cancer and in surgical margins of the
colon. Rocz Akad Med Bialymst. 49(Suppl 1): 52–54. 2004.
|
|
50
|
Zheng ZY, Tian L, Bu W, Fan C, Gao X, Wang
H, Liao YH, Li Y, Lewis MT, Edwards D, et al: Wild-type N-Ras,
overexpressed in basal-like breast cancer, promotes tumor formation
by inducing IL-8 secretion via JAK2 activation. Cell Rep.
12:511–524. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Spandidos A and Wilkie NM: The normal
human H-ras1 gene can act as an onco-suppressor. Br J Cancer Suppl.
9:67–71. 1988.PubMed/NCBI
|
|
52
|
Spandidos DA, Frame M and Wilkie NM:
Expression of the normal H-ras1 gene can suppress the transformed
and tumorigenic phenotypes induced by mutant ras genes. Anticancer
Res. 10:1543–1554. 1990.PubMed/NCBI
|