Introduction
The kinesin superfamily proteins (Kifs) are
microtubule-dependent molecular motors and contain a conserved
motor catalytic domain that binds to and hydrolyzes adenosine
triphosphate (ATP) to produce chemical and mechanical energy.
Kinesins play important roles in intracellular transport, mitosis,
cellular morphogenesis and cellular functions. Kinesins not only
transport various cargos, such as membranous organelles, protein
complexes and mRNAs for the maintenance of basic cellular activity,
but also play significant roles in brain growth, memory and the
activity of neurons. It can be said that kinesins form the basis to
living systems (1).
Kif4, a Kif member classified in kinesin-4 (2), is strongly expressed in juvenile
tissues, including differentiated young neurons and is decreased
considerably in adult mice, apart from the spleen (3). Kif4 has been reported as an
essential factor involved in multiple cellular process, such as
cell proliferation, DNA damage response, viral protein
intracellular trafficking, immune cell activation and neuronal
survival in brain development (4–9).
It has also been suggested that Kif4 may be a primary initiating
trigger for tumorigenesis (10).
Angiogenesis is a pathological process which is
related to a wide range of diseases, from atherosclerosis to cancer
(11). Central to the
physiological regulation of angiogenesis is vascular endothelial
growth factor (VEGF)-A and its receptors, involving VEGF receptor 1
(VEGFR1) and 2 (VEGFR2). VEGF-A displays potent angiogenic and
vascular permeability activity. VEGFR2 is the crucial receptor for
the functions of vascular endothelial cells. VEGFR1 is a
high-affinity tyrosine kinase receptor for VEGF-A, a negative
regulator of VEGFR2 signaling capacity (12). VEGFR1 can be also generated as a
short soluble form (sVEGFR1), consisting of only the extracellular
ligand binding domain. Due to the lack of the transmembrane region,
sVEGFR1 has the potential to act as a decoy receptor for VEGF-A by
inhibiting the activation of VEGFR1 or VEGFR2 by binding to VEGF-A
(13). Therefore, it may
indirectly inhibit the pro-angiogenic activities VEGF-A.
It is known that sVEGFR1 is involved in the
pregnancy disorder of pre-eclampsia, which is characterized by
endothelial dysfunction (14) and
the therapeutic potential of sVEGFR1 as an anti-angiogenic agent is
gaining increasing attention in pre-clinical models of cancer.
Thus, sVEGFR1 is a diagnostic marker and therapeutic target in
angiogenesis-dependent diseases (15). In addition, VEGFR1-dependent
disease reactions are also regulated by a balance of expression
between the full-length and soluble forms of VEGFR1 (11).
Monocytes/macrophages represent immune effector
cells, equipped with chemokine receptors and pathogen recognition
receptors that mediate the migration from blood to tissues during
infection (16). VEGFR1, which is
well-expressed in monocytes/macrophages, is an important cell
surface marker, as well as a biologically functional molecule for
monocyte/macrophage lineages (17,18). Through the receptor VEGFR1,
monocytes/macrophages stimulate vascular biologyical responses in
various tissues and promote a variety of diseases, such as tumor
growth via pro-angiogenesis, tumor metastasis, lymphangiogenesis,
arthritis, atherosclerosis and wound healing (18–26). Therefore, it is important to
examine the expression of VEGFR1 in monocytes/macrophages and the
underlying mechanisms.
In this study, we investigated whether Kif4
regulates VEGFR1, sVEGFR1 and VEGF-A expression in RAW264.7
monocytes/macrophages, and explored the underlying signaling
mechanisms. Furthermore, we aimed to elucidate the regulatory
mechanisms of VEGF-A and VEGFR1.
Materials and methods
Cell line and cell culture
The murine monocyte/macrophage cell line, RAW264.7,
was obtained from the American Type Culture Collection (ATCC,
Manassas, VA, USA) and routinely cultured in Dulbecco's modified
Eagle's medium (DMEM) supplemented with 10% (v/v) heat-inactivated
fetal bovine serum (FBS) (both from Gibco-BRL, Carlsbad, CA, USA)
at 37°C in a humidified air atmosphere and 5% CO2. The
RAW264.7 cells were plated at 5×105 cells/well into
6-well plates and then treated with 1.5 ml of medium.
Silencing of mouse Kif4 gene using small
interfering RNA (siRNA)
A siRNA expression vector was constructed by
introducing synthetic double-stranded oligonucleotides as follows:
Kif4 sense, 5′-GCUGAAGUUUAGGCAAUTT-3′ and antisense,
5′-AUUGCCUAAACUUCUCAGCTT-3′; negative control (NC) sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′ synthesized by GeneChem, Co., Ltd.
(Shanghai, China) and green fluorescent FAM-labeled negative
control siRNA (Shanghai GenePharma Co., Ltd., Shanghai, China) was
used to detect the transfection efficiency. The RAW264.7 cells at
70% confluency were transfected with the Kif4 siRNA (si-Kif4) or NC
siRNA (si-NC) or FAM labeled NC siRNA (6 µl) premixed with
Lipofectamine™ 2000 (Invitrogen Life Technologies, Carlsbad, CA,
USA) (4.5 µl) in Opti-MEM in 6-well plates. The
mock-transfected cells were transfected only with Lipo fectamine™
2000 and were included as a control. After 6 h, the cells were
placed in fresh complete medium. In order to confirm whether siRNA
was able to transfect into cells, the cells transfected with FAM
labeled NC siRNA were observed under an inverted fluorescence
microscope (serial no. 321463; Leica, Wetzlar, Germany). Cells were
observed and photographed at ×100 magnification under light field
and fluorescence (520 nm), respectively. The transfection
efficiency was determined by the positive rate of FAM green
fluorescence from 5 different fields. To examine the effects of
si-Kif4 on macrophage morphology, RAW 264.7 cells were examined
using an inverted microscope (serial no. 321463; Leica). After
being transfected with the si-NC or si-Kif4 for 48 h, RAW264.7
cells were observed under light field at ×200 magnification. Mouse
insulin-like growth factor-1 (IGF-1) (100 ng/ml) was used to treat
the cells for 36 h in si-NC and si-Kif4 groups, after 48-h
transfection with si-NC or si-Kif4. IGF-1 was purchased from
ProSpec-Tany TechnoGene, Ltd. (Ness Ziona, Israel).
Flow cytometric analysis
The RAW264.7 cells were collected and washed with
cold phosphate-buffered saline (PBS). The cells were then incubated
with monoclonal antibodies against CD11c (M10118-02B) and CD80
(M10801-09B) (both from Tianjin Sungene Biotech Co., Ltd., Tianjin,
China) for 20 min in the dark. The cells were washed with cold PBS
again and analyzed with a flow cytometer (FACSCalibur;
Becton-Dickinson, Mountain View, CA, USA). A minimum of 10,000
events/sample were collected for analysis.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) assays
Total RNA was extracted from the cells using an
RNeasy Mini kit (Qiagen, Dusseldorf, Germany) in accordance with
the RNeasy Mini Handbook. RNA was reverse transcribed using the
ReverTra Ace qPCR RT kit (Toyobo Co., Ltd., Osaka, Japan). PCR
analysis was performed with cDNA and the following primers: mouse
VEGFR1 forward, 5′-CTTTCTCAAGTGCAGAGGGG-3′ and reverse,
5′-TCATGTGCACAAGTTTGGGT-3′; mouse VEGF-A forward,
5′-GGGACCCCTTCGTCCTCTC-3′ and reverse,
5′-GTCTCCTGGGGACAGAATTAGTG-3′; and mouse glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) forward, 5′-CCCACTAACATCAAATGGGG-3′ and
reverse, 5′-CCTTCCACAATGCCAAAGTT-3′.
The cDNA sample amplification was carried out using
SYBR PremixEx Taq™ II (Tli RNaseH Plus; Takara, Dalian, China). The
RT-PCR conditions were as follows: 3 sec at 95°C, followed by 40
cycles consisting of denaturing at 95°C for 5 sec, annealing at
60°C for 10 sec, and extending at 72°C for 15 sec. Relative
quantification values were calculated using the ΔΔCqmethod.
Western blot analysis
The cells were collected and lysed in ice-cold
radioimmunoprecipitation assay (RIPA) lysis buffer containing 1 mM
phenylmethanesulfonyl fluoride (Solarbio, Beijing, China). The
supernatant were collected following centrifugation at 10,000 rpm
for 15 min at 4°C. The protein concentration was determined using a
Micro BCA™ Protein Assay kit (Pierce, Rockford, IL, USA). Equal
amounts of protein were loaded, separated by SDS-PAGE and
transferred onto PVDF membranes (Invitrogen Life Technologies). For
2 h, the membranes were blocked with 5% (w/v) fat-free milk
dissolved in Tris-buffered saline containing 0.05% Tween-20 (TBST).
The membranes were then hybridized to a mouse anti-AKT antibody
(1:1,000 #9272), a phosphorylated Akt (p-Akt) antibody (1:1,000;
#9271) or GAPDH antibody (1:2,000; #97166) (all from Cell Signaling
Technology, Inc., Boston, MA, USA) at room temperature for 2 h. The
membranes were washed in TBST and then incubated with
HRP-conjugated secondary antibody (1:2,000; ZDR-5307; ZSGB-BIO,
Beijing, China). Blots were visualized using an ECL
chemiluminescent kit (Millipore, Billerica, MA, USA). The bands
were processed and analyzed using the FluorChem system (Alpha
Innotech, Cell Biosciences Inc., Santa Clara, CA, USA).
Enzyme-linked immunosorbent assay
(ELISA)
The concentrations of mouse sVEGFR1 and VEGF-A in
the cell culture medium were detected using ELISA kits (R&D
Systems, Inc., Minneapolis, MN, USA), according to the
manufacturer's instructions. Briefly, 50 µl assay diluent
was added to each well and the same volume of serially diluted
standards and samples were loaded on the plate followed by
incubation for 2 h at room temperature on the shaker at 200 rpm.
Excess antibodies were then removed 5 times with wash buffer, and
incubated for 2 h with conjugate. The reaction was developed by
substrate and the optical density was measured using a
spectrophotometer (GeneQuant 100; Biochrom, Holliston, MA, America)
at 450 nm (correcting at 540 nm).
Statistical analysis
The values are expressed as the means ± standard
deviation (SD). Data were analyzed using the Student's t-test or by
analysis of variance (ANOVA). A P-value <0.05 was considered to
indicate a statistically significant difference.
Results
Silencing of Kif4 decreases VEGF-A mRNA
expression, but increases VEGF-A levels in supernatant of RAW264.7
cells
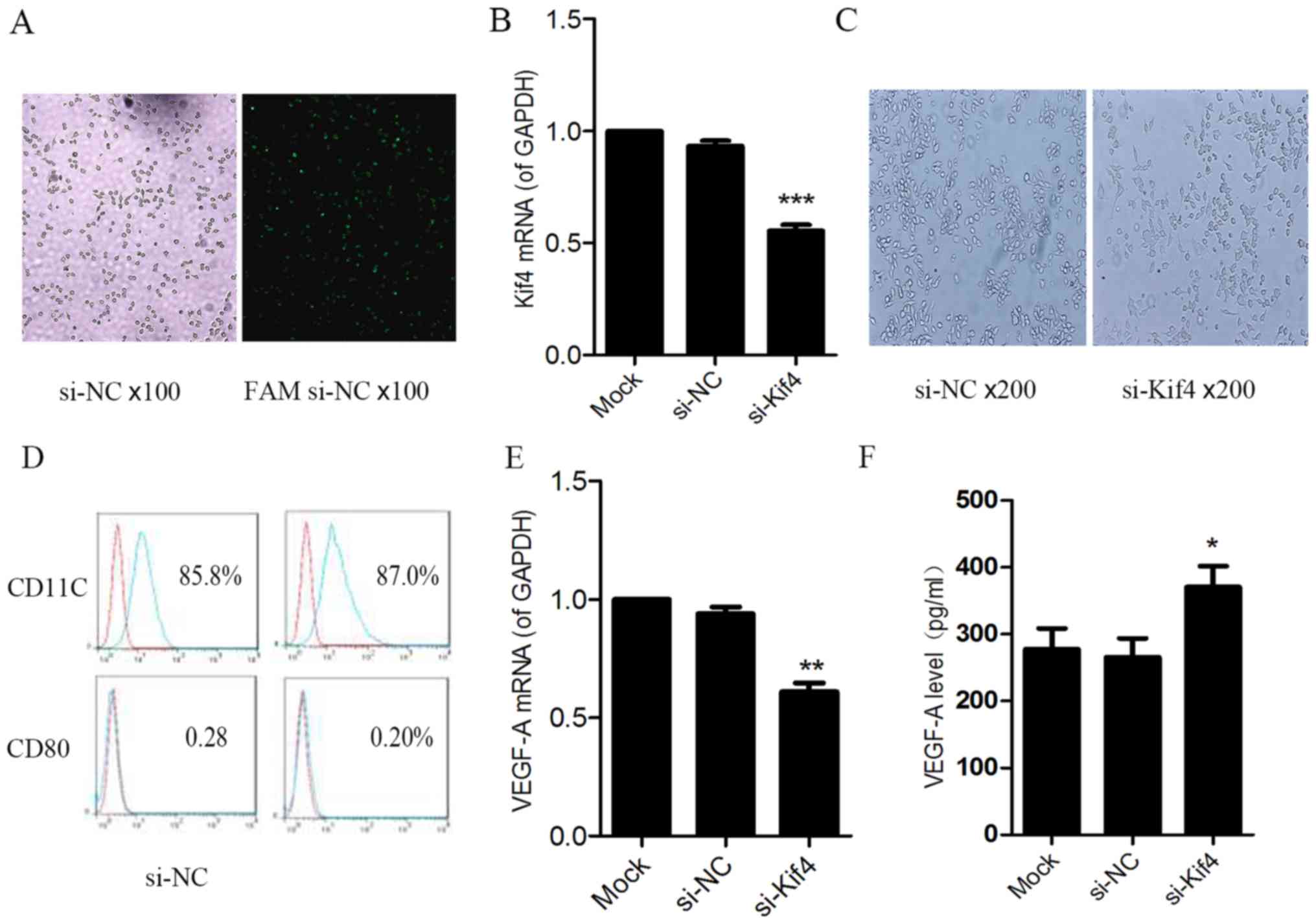
The RAW264.7 cells were transfected with si-NC or
FAM si-NC for 6 h, and were observed under a fluorescence
microscope. We observed green fluorescence in the RAW264.7 cells.
This suggests that the FAM si-NC with green fluorescent tags was
successfully transfected into the cells (Fig. 1A).
RNA interference targeting Kif4 was determined by
RT-PCR. The reduction of Kif4 mRNA expression by ~60% was observed
in the si-Kif4 transfected cells compared to the si-NC- and
Lipofectamine 2000 only-transfected cells (Fig. 1B). No morphological changes in the
cells were observed at 48 h following transfection (Fig. 1C). In both the si-NC group and
si-Kif4 group, RAW264.7 cell bodies were plump, oval and thin at
the cell ends. Most of the cells exhibited two or fewer extended
dendritic pseudopods. Flow cytometric analysis indicated that the
expression of the costimulatory molecules, CD11c and CD80, was not
obviously altered (Fig. 1D).
The results of RT-PCR revealed that at 24 h
following transfection of the RAW264.7 cells with si-Kif4, VEGF-A
mRNA expression was markedly decreased (Fig. 1E). However, at 48 h
post-transfection, the level of VEGF-A was increased in the cell
supernatant, as shown by ELISA (Fig.
1F), which was inconsistent with the decreased gene level. We
hypothesized that this was due to a decrease in the level of
sVEGFR1 in the cell supernatant, which is a decoy receptor of
VEGF-A, which kept the levels of free VEGF-A from decreasing. Thus,
we examined whether Kif4 regulates the levels of VEGFR1 and sVEGFR1
in the RAW264.7 cells.
Silencing of Kif4 using siRNA (si-Kif4)
regulates the expression of sVEGFR1 and VEGFR1 in RAW264.7
cells
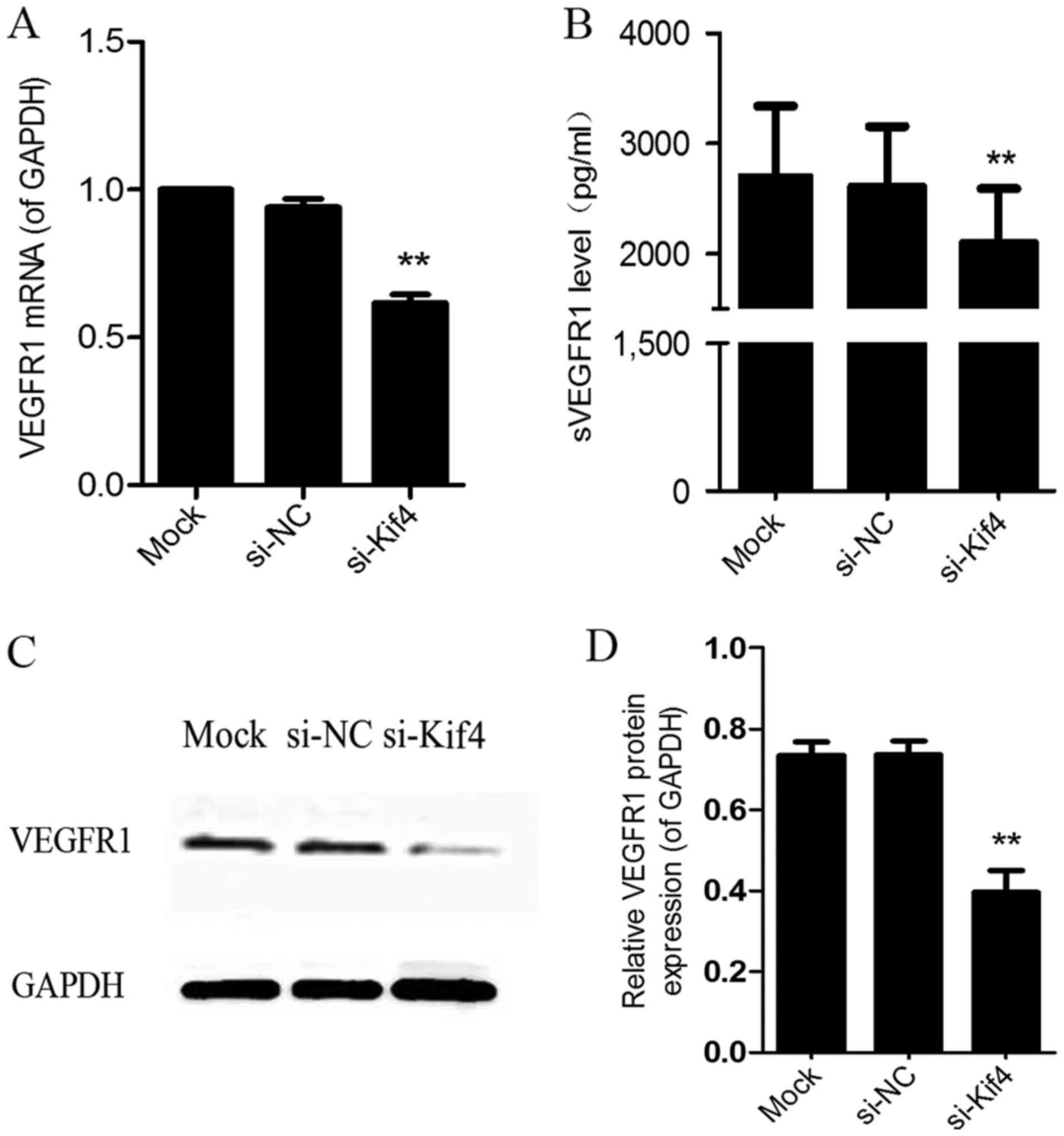
The results of RT-PCR revealed that at 24 h
post-transfection, VEGFR1 mRNA expression was decreased (Fig. 2A). The results of ELISA also
revealed that the level of sVEGFR1 was decreased in the cell
supernatant following transfection with si-Kif4 (Fig. 2B). In addition, the results of
western blot analysis indicated that the protein expression of
VEGFR1 was decreased following transfection with si-Kif4 (Fig. 2C).
Silencing of Kif4 using siRNA (si-Kif4)
regulates the levels of VEGFR1 and sVEGFR1 in RAW264.7 cells
through the inhibition of the PI3K/Akt signaling pathway
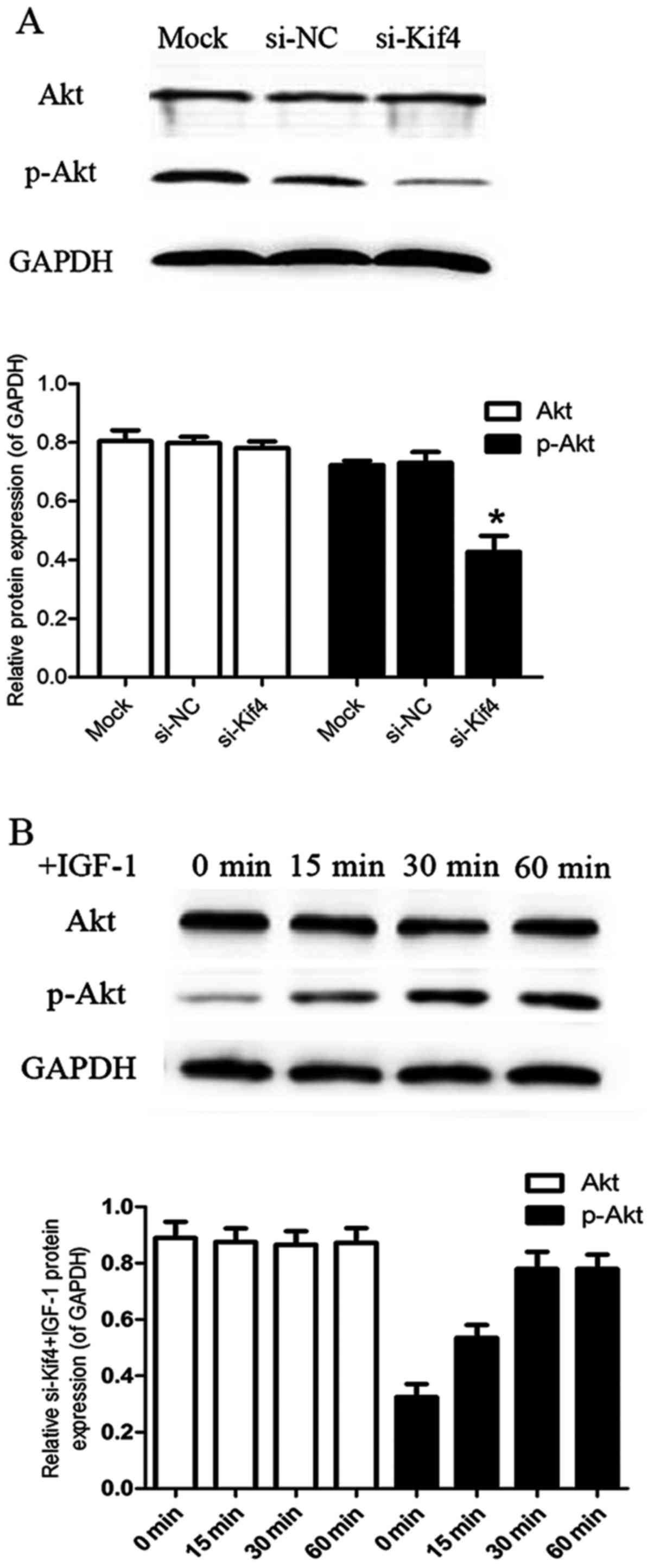
We then explored the signaling mechanisms involved
in the regulatory effects of si-Kif4 on the levels VEGFR1 and
sVEGFR1 in RAW264.7 cells. We found that the levels of p-Akt
significantly decreased at 48 h post-transfection in the
si-Kif4-transfected group compared with the mock- and
si-NC-transfected groups (Fig.
3A). IGF-1 is the specific agonist of PI3K/Akt. Thus, we also
used IGF-1 to examine its effects on the transfected cells. The
results revealed a high level of p-Akt at 30 min after the addition
of IGF-1 (100 ng/ml) in the si-Kif4-transfected group; the level
remained high at 60 min (Fig.
3B).
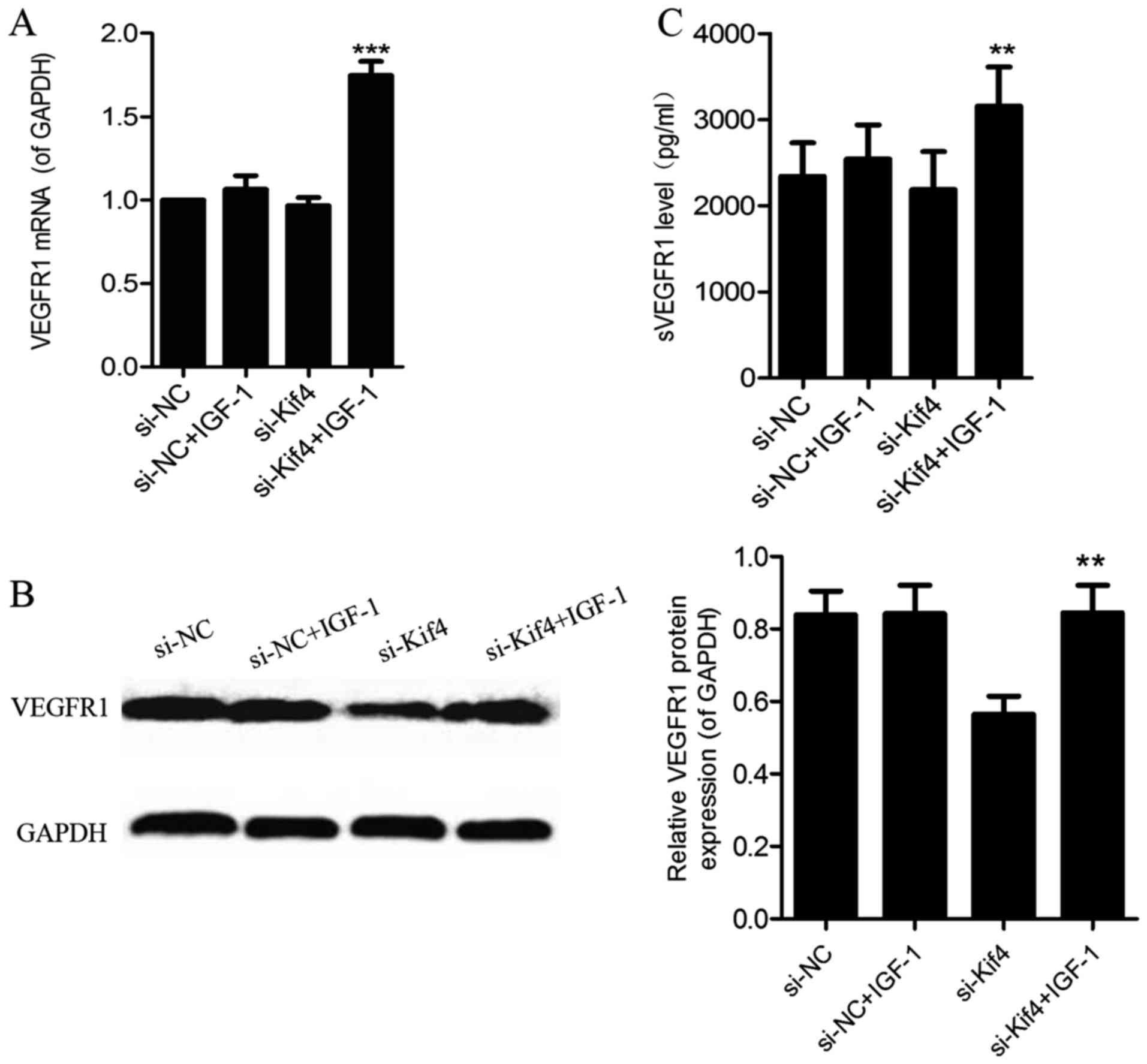
The suppressive effects of the silencing
of Kif4 on VEGFR1 mRNA expression and the level of sVEGFR1 are
abrogated by IGF-1
Following the addition of IGF-1 (100 ng/ml) for 36 h
in the si-NC- and si-Kif4-transfected groups, we detected a
significant increase in the VEGFR1 mRNA (Fig. 4A), and VEGFR1 protein levels
(Fig. 4B); the levels of sVEGFR1
also increased in the cell supernatant in the si-Kif4-transfected
group (Fig. 4C).
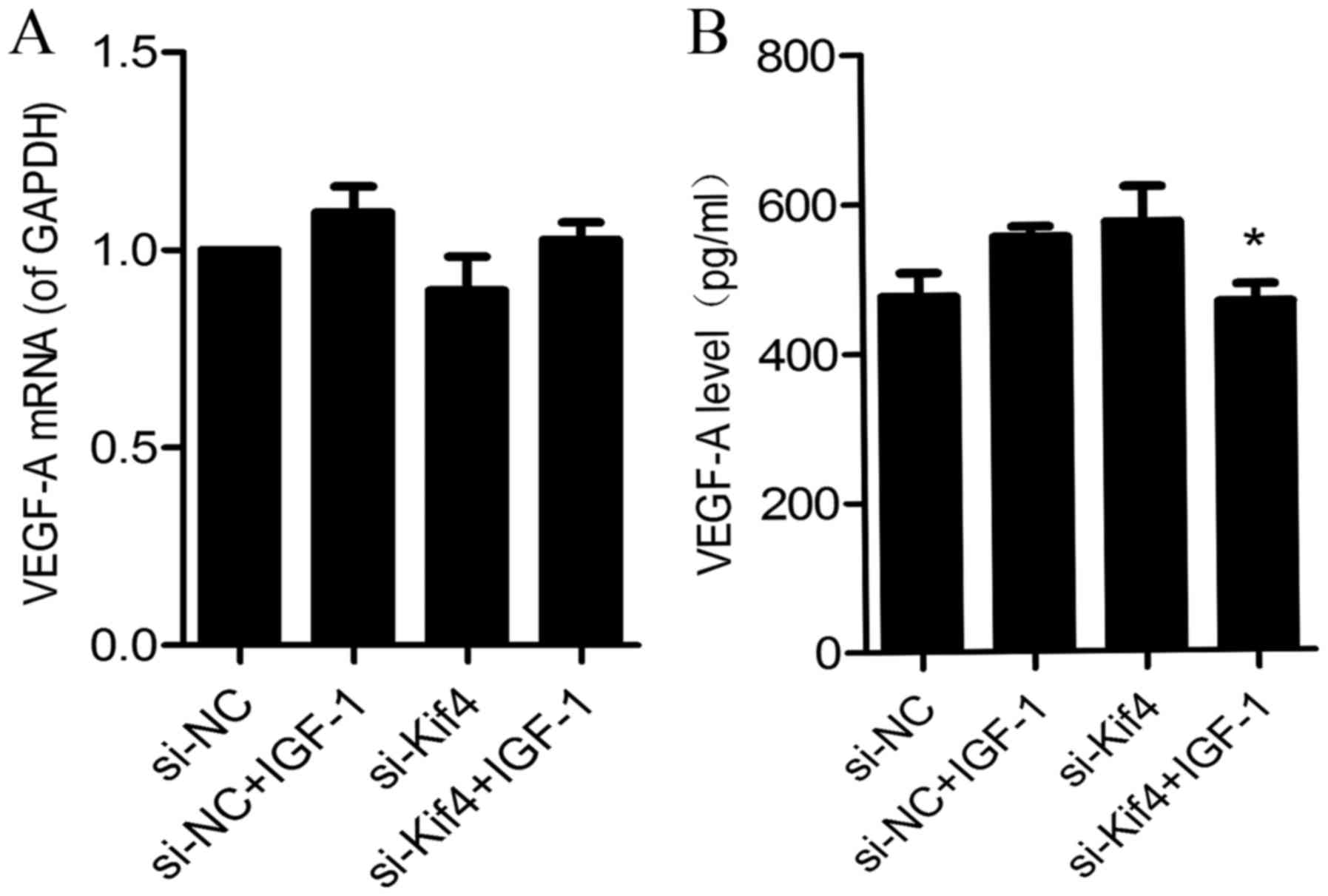
The addition of IGF-1 alters the mRNA
expression of VEGF in si-Kif4-transfected cells, as well as the
level of VEGF-A in the cell supernatant
Following the addition of IGF-1 (100 ng/ml) for 36
h, we found that VEGF-A mRNA expression was did significantly
altered between the si-NC- and si-Kif4-transfected groups (Fig. 5A); however, the expression of
VEGF-A in the cell supernatant was decreased in the
si-Kif4-transfected group (Fig.
5B).
Discussion
Kinesin superfamily motor proteins can not only
directionally transport various cargos, but also play significant
roles in various processes fundamental for life, such as
angiogenesis (1). KIF13B has been
shown to be an essential molecular motor to the trafficking of
VEGFR2 from the Golgi apparatus to the endothelial cell surface to
mediate angiogenesis (27).
Kif11/Eg5 plays a role in endothelial cell adhesion, migration and
angiogenesis (28).
In this study, our results demonstrated that the
silencing of Kif4 regulated the expression of VEGFR1, sVEGFR1 and
VEGF, and inhibited the activation of the PI3K/Akt signaling
pathway in RAW264.7 cells, which may be one of the mechanisms
responsible for Kif4 participating in the regulation of
angiogenesis.
Initially, we found that the mRNA expression of
VEGF-A was decreased in the RAW264.7 cells (Fig. 1E), but the protein level of VEGF-A
was significantly higher in the cell supernatant in the cells
transfected with si-Kif4. To explain these seemingly contradictory
results, we considered whether this occurred due to the fact that
the level of sVRGFR1, a decoy receptor of VEGF-A, was decreased in
the cell supernatant. The silencing of Kif4 decreased the VEGFR1
mRNA, sVEGFR1 and VEGFR1 protein expression (Fig. 2A–C). By contrast, the
transmembrane form of VEGFR1 was significantly increased (data not
shown). This indicated that the silencing of Kif4 not only
modulated the protein expression of VEGFR1 in RAW264.7 cells, but
also regulated the proportion of the trans-membrane form of VEGFR1
and secreted forms. However, the mechanisms involved require
further investigation.
The PI3K/Akt pathway is a cell survival pathway,
which regulates gene expression and cell metabolism, as well as
cytoskeletal rearrangements. The PI3K/Akt pathway has been
implicated in various human diseases, including diabetes and cancer
(29,30).
In the human placental hypoxia model, the PI3K/Akt
pathway has been shown to be significantly involved in the
regulation and expression sVEGFR1 (31). Granulocyte-macrophage
colony-stimulating factor (GM-CSF) can stimulate human monocytes to
produce sVEGFR1, inhibiting the biological effects of VEGF-A, by
activating the PI3K/Akt pathway (32). Human monocyte Fcγ receptor
activation can also promote the secretion of sVEGFR1 via the
PI3K/Akt pathway, thereby inhibiting human umbilical vein
endothelial cell (HUVEC) tube formation effect (33).
In order to understand the molecular mechanisms
responsible for the regulation of VEGFR1 and sVEGFR1 in RAW264.7
cells by the silencing of Kif4, we investigated PI3K/Akt signaling.
Transfection with si-Kif4 inhibited the PI3K/Akt signaling pathway
at 24 h (data not shown), and the inhibitory effect was more
obvious at 48 h (Fig. 3A). IGF-1
is the specific agonist of PI3K/Akt (34) (Fig.
3B). The addition of IGF-1 increased the expression of VEGFR1
mRNA, VEGFR1 protein andthe level of sVEGFR1 in the
si-Kif4-transfected cells and in the cell supernatant (Fig. 4).
We also found that following the activation of the
PI3K/Akt signaling pathway, the level of VEGF-A mRNA was not
significantly altered (Fig. 5A);
however, the level of VEGF-A was significantly decreased in the
cell supernatant (Fig. 5B). We
considered that this was due to the increased level of sVEGFR1
which combined with VEGF-A, so that free VEGF-A was reduced.
In conclusion, in this study, to the best of our
knowledge, we demonstrated for the first time that the silencing of
Kif4 regulated VRGFR1 mRNA transcription and sVEGFR1 production in
monocytes via the PI3K/Akt signaling pathway, which may be one of
the mechanisms responsible for these processes. Our results not
only provide a deeper understanding of the function of Kif4, but
also enhance our understanding of VEGFR1 and sVEGFR1 generation
mechanisms in monocytes. These data may help to explain the role of
monocytes/macrophages in angiogenesis and provide valuable
information on treatment strategies for diseases, such as cancer
and inflammation.
Acknowledgments
The present study was supported by the Natural
Science Foundation of China (grant nos. 81271105, 31470885 and
31270971).
References
|
1
|
Hirokawa N, Noda Y, Tanaka Y and Niwa S:
Kinesin super-family motor proteins and intracellular transport.
Nat Rev Mol Cell Biol. 10:682–696. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lawrence CJ, Dawe RK, Christie KR,
Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV,
Hirokawa N, Howard J, et al: A standardized kinesin nomenclature. J
Cell Biol. 167:19–22. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sekine Y, Okada Y, Noda Y, Kondo S, Aizawa
H, Takemura R and Hirokawa N: A novel microtubule-based motor
protein (KIF4) for organelle transports, whose expression is
regulated developmentally. J Cell Biol. 127:187–201. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kurasawa Y, Earnshaw WC, Mochizuki Y,
Dohmae N and Todokoro K: Essential roles of KIF4 and its binding
partner PRC1 in organized central spindle midzone formation. EMBO
J. 23:3237–3248. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhu C and Jiang W: Cell cycle-dependent
translocation of PRC1 on the spindle by Kif4 is essential for
midzone formation and cytokinesis. Proc Natl Acad Sci USA.
102:343–348. 2005. View Article : Google Scholar :
|
|
6
|
Wu G, Zhou L, Khidr L, Guo XE, Kim W, Lee
YM, Krasieva T and Chen PL: A novel role of the chromokinesin Kif4A
in DNA damage response. Cell Cycle. 7:2013–2020. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Martinez NW, Xue X, Berro RG, Kreitzer G
and Resh MD: Kinesin KIF4 regulates intracellular trafficking and
stability of the human immunodeficiency virus type 1 Gag
polyprotein. J Virol. 82:9937–9950. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bernasconi P, Cappelletti C, Navone F,
Nessi V, Baggi F, Vernos I, Romaggi S, Confalonieri P, Mora M,
Morandi L, et al: The kinesin superfamily motor protein KIF4 is
associated with immune cell activation in idiopathic inflammatory
myopathies. J Neuropathol Exp Neurol. 67:624–632. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Midorikawa R, Takei Y and Hirokawa N: KIF4
motor regulates activity-dependent neuronal survival by suppressing
PARP-1 enzymatic activity. Cell. 125:371–383. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mazumdar M, Lee JH, Sengupta K, Ried T,
Rane S and Misteli T: Tumor formation via loss of a molecular motor
protein. Curr Biol. 16:1559–1564. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Carmeliet P: Angiogenesis in life, disease
and medicine. Nature. 438:932–936. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Olsson AK, Dimberg A, Kreuger J and
Claesson-Welsh L: VEGF receptor signalling - in control of vascular
function. Nat Rev Mol Cell Biol. 7:359–371. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cao Y: Positive and negative modulation of
angiogenesis by VEGFR1 ligands. Sci Signal. 2:re12009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ahmad S and Ahmed A: Elevated placental
soluble vascular endothelial growth factor receptor-1 inhibits
angiogenesis in preeclampsia. Circ Res. 95:884–891. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu FT, Stefanini MO, Mac Gabhann F, Kontos
CD, Annex BH and Popel AS: A systems biology perspective on
sVEGFR1: its biological function, pathogenic role and therapeutic
use. J Cell Mol Med. 14:528–552. 2010.
|
|
16
|
Geissmann F, Manz MG, Jung S, Sieweke MH,
Merad M and Ley K: Development of monocytes, macrophages, and
dendritic cells. Science. 327:656–661. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Clauss M, Weich H, Breier G, Knies U,
Röckl W, Waltenberger J and Risau W: The vascular endothelial
growth factor receptor Flt-1 mediates biological activities.
Implications for a functional role of placenta growth factor in
monocyte activation and chemotaxis. J Biol Chem. 271:17629–17634.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sawano A, Iwai S, Sakurai Y, Ito M,
Shitara K, Nakahata T and Shibuya M: Flt-1, vascular endothelial
growth factor receptor 1, is a novel cell surface marker for the
lineage of monocyte-macrophages in humans. Blood. 97:785–791. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Crowther M, Brown NJ, Bishop ET and Lewis
CE: Micro-environmental influence on macrophage regulation of
angiogenesis in wounds and malignant tumors. J Leukoc Biol.
70:478–490. 2001.PubMed/NCBI
|
|
20
|
Hiratsuka S, Minowa O, Kuno J, Noda T and
Shibuya M: Flt-1 lacking the tyrosine kinase domain is sufficient
for normal development and angiogenesis in mice. Proc Natl Acad Sci
USA. 95:9349–9354. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lewis CE and Pollard JW: Distinct role of
macrophages in different tumor microenvironments. Cancer Res.
66:605–612. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kerber M, Reiss Y, Wickersheim A, Jugold
M, Kiessling F, Heil M, Tchaikovski V, Waltenberger J, Shibuya M,
Plate KH, et al: Flt-1 signaling in macrophages promotes glioma
growth in vivo. Cancer Res. 68:7342–7351. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Peppicelli S, Bianchini F and Calorini L:
Inflammatory cytokines induce vascular endothelial growth factor-C
expression in melanoma-associated macrophages and stimulate
melanoma lymph node metastasis. Oncol Lett. 8:1133–1138.
2014.PubMed/NCBI
|
|
24
|
Murakami M, Iwai S, Hiratsuka S, Yamauchi
M, Nakamura K, Iwakura Y and Shibuya M: Signaling of vascular
endothelial growth factor receptor-1 tyrosine kinase promotes
rheumatoid arthritis through activation of monocytes/macrophages.
Blood. 108:1849–1856. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Murakami M, Zheng Y, Hirashima M, Suda T,
Morita Y, Ooehara J, Ema H, Fong GH and Shibuya M: VEGFR1 tyrosine
kinase signaling promotes lymphangiogenesis as well as angiogenesis
indirectly via macrophage recruitment. Arterioscler Thromb Vasc
Biol. 28:658–664. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Matsui M, Takeda Y, Uemura S, Matsumoto T,
Seno A, Onoue K, Tsushima H, Morimoto K, Soeda T, Okayama S, et al:
Suppressed soluble Fms-like tyrosine kinase-1 production aggravates
atherosclerosis in chronic kidney disease. Kidney Int. 85:393–403.
2014. View Article : Google Scholar
|
|
27
|
Yamada KH, Nakajima Y, Geyer M, Wary KK,
Ushio-Fukai M, Komarova Y and Malik AB: KIF13B regulates
angiogenesis through Golgi to plasma membrane trafficking of
VEGFR2. J Cell Sci. 127:4518–4530. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Exertier P, Javerzat S, Wang B, Franco M,
Herbert J, Platonova N, Winandy M, Pujol N, Nivelles O, Ormenese S,
et al: Impaired angiogenesis and tumor development by inhibition of
the mitotic kinesin Eg5. Oncotarget. 4:2302–2316. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cantley LC: The phosphoinositide 3-kinase
pathway. Science. 296:1655–1657. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hemmings BA and Restuccia DF: The
PI3K-PKB/Akt pathway. Cold Spring Harb Perspect Biol. 7:1–3. 2015.
View Article : Google Scholar
|
|
31
|
Park JK, Jeong JW, Kang MY, Baek JC, Shin
JK, Lee SA, Choi WS, Lee JH and Paik WY: Inhibition of the PI3K-Akt
pathway suppresses sFlt1 expression in human placental hypoxia
models in vitro. Placenta. 31:621–629. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Eubank TD, Roberts R, Galloway M, Wang Y,
Cohn DE and Marsh CB: GM-CSF induces expression of soluble VEGF
receptor-1 from human monocytes and inhibits angiogenesis in mice.
Immunity. 21:831–842. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Justiniano SE, Elavazhagan S, Fatehchand
K, Shah P, Mehta P, Roda JM, Mo X, Cheney C, Hertlein E, Eubank TD,
et al: Fcγ receptor-induced soluble vascular endothelial growth
factor receptor-1 (VEGFR-1) production inhibits angiogenesis and
enhances efficacy of anti-tumor antibodies. J Biol Chem.
288:26800–26809. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Michell BJ, Griffiths JE, Mitchelhill KI,
Rodriguez-Crespo I, Tiganis T, Bozinovski S, de Montellano PR, Kemp
BE and Pearson RB: The Akt kinase signals directly to endothelial
nitric oxide synthase. Curr Biol. 9:845–848. 1999. View Article : Google Scholar : PubMed/NCBI
|