Introduction
The liver represents the central organ for drug
metabolism and also a main target organ for drug-associated
toxicity. Therefore, the parenchymal cells of the liver, the
hepatocytes, are of special interest for pharmacological and
toxicological investigations.
Primary human hepatocytes (PHH) are considered to be
the gold standard for such investigations, however their
availability is limited and their metabolic activity varies,
primarily due to the mostly unknown genetic backgrounds of the
donors (1). Primary hepatocytes
are also isolated from animals, such as rats, but they have a
limited predictability due to cross-species differences in drug
metabolism and sensitivity (2).
Therefore, the development of in vitro culture models using
easy accessible cells of human origin is gaining increasing
scientific interest.
Pluripotent stem cells (PSC) constitute a promising
cell source for the generation of hepatocytes, due to their
capacity to differentiate into all cell types of the organism and
their ability to replicate while maintaining pluripotency. The
innovation of induced pluripotent stem cell (iPSC) technology
opened up the possibility of deriving pluripotent cells from
different donors (3,4) thereby circumventing the ethical
concerns associated with the use of human embryonic stem cells.
Thus, pluripotent cell lines with distinct genotypes can be
generated, which are of interest in relation to specific disease
mechanisms, and to the development of drugs (5,6).
These properties of PSC in combination with the increasing
knowledge of the in vivo embryonic development of
hepatocytes (7) have led to the
establishment of several protocols for the in vitro
differentiation of PSC into hepatocyte-like cells (HLCs) (8–10).
Current protocols mimic the different stages of the in vivo
development of hepatocytes by the sequential addition of specific
growth factors, like activin A, Wnt3a, hepatocyte growth factor
(HGF) and oncostatin M (OSM) (8,11).
Small chemical molecules, such as dimethyl sulfoxide (DMSO),
bromo-indirubin-3′-oxim and SB431542 (12) can be applied as well. The
generated HLCs demonstrate some characteristics of hepatocytes,
such as susceptibility to hepatitis C virus infection (13), secretion of hepatic proteins
(14,15) and activity of metabolic enzymes
(16,17). However, the drug metabolizing
capabilities of HLCs obtained with current protocols are still
below those of PHH (18). Recent
findings suggested that HLCs resemble immature or fetal hepatocytes
rather than adult hepatocytes (19,20).
In order to increase the functionality and the
maintenance of HLCs, the use of extracellular matrices (21,22), transcription factor overexpression
(23,24) or modified cultivation media
(25) were suggested. Further
approaches focus on complex culture systems to provide an
organotypic environment that better approximates the in vivo
situation. Cultivation of cells in a 3D environment facilitates the
formation of physiological cell-cell-contacts, which have been
demonstrated to be crucial for the preservation of a mature hepatic
phenotype (26). Different 3D
culture systems were investigated for hepatic differentiation of
PSC, including scaffold-based technologies (27–29) or scaffold-free culture systems,
which rely on the self-assembly of the cells (17,30). However, due to the lack of
standardized methods to characterize the HLCs after hepatic
differentiation, it is difficult to compare the results from
different approaches and culture models.
In the present study, the authors investigated the
hepatic differentiation of human iPSCs (hiPSCs) in two different 3D
culture systems, a scaffold-free microspheroid culture system and a
3D hollow-fiber perfusion bioreactor (31). The differentiation outcome in
these 3D systems was compared with that in conventional 2D
cultures. All culture systems were treated with the same
differentiation protocol, allowing a comparative analysis of the
generated HLCs at mRNA, protein and metabolic level. In addition,
data from hiPSC-derived differentiated cells were compared to those
from PHH. Based on the results, promising approaches for the
development of physiologically relevant in vitro liver
models were identified.
Materials and methods
Culture of hiPSCs
The generation and characterization of the hiPSC
line SB Adult3 clone 4 (AD3C4) is described by van de Bunt et
al (32). The hiPSC lines
AD2C3, AD3C1 and AD4C1 were generated and characterized in the same
way from fibroblasts 24245, 23447 and 23801 (Lonza CC-2511, tissue
acquisition numbers are given; Lonza Group, Ltd., Basel,
Switzerland), respectively. Cells were seeded on culture plates
coated with growth-factor-reduced Matrigel (Corning Inc., Corning,
NY, USA). For expansion, hiPSCs were maintained at 37°C, 5%
CO2 using mTeSR™1 medium (Stemcell Technologies, Inc.,
Vancouver, BC, Canada) supplemented with 100,000 U/l penicillin and
100 mg/l streptomycin (Thermo Fisher Scientific, Inc., Waltham, MA,
USA). The cells were passaged with 0.5 mM EDTA (Thermo Fisher
Scientific, Inc.) every 3–5 days, after reaching a confluence of
~70%.
Hepatocyte-like cell differentiation in
2D cultures
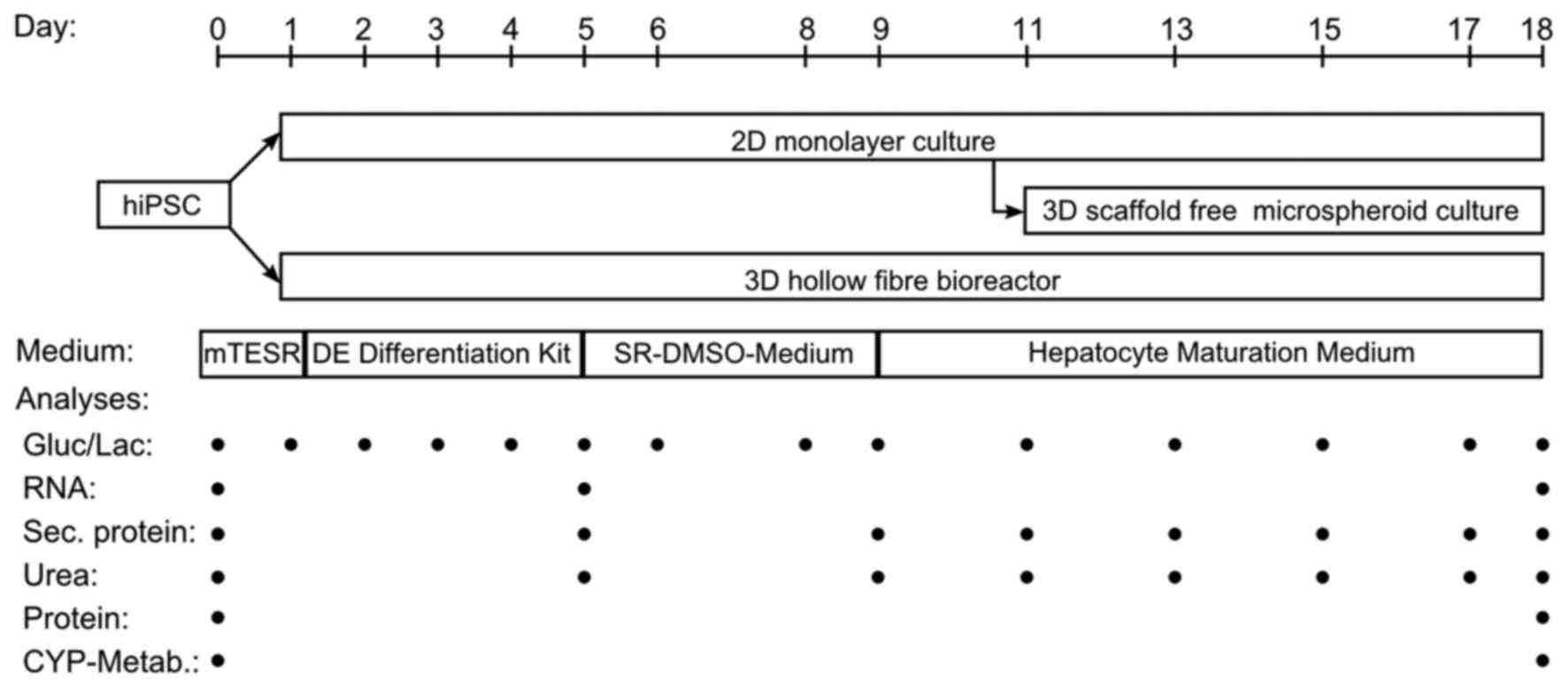
Differentiation of hiPSCs in 2D monolayer cultures
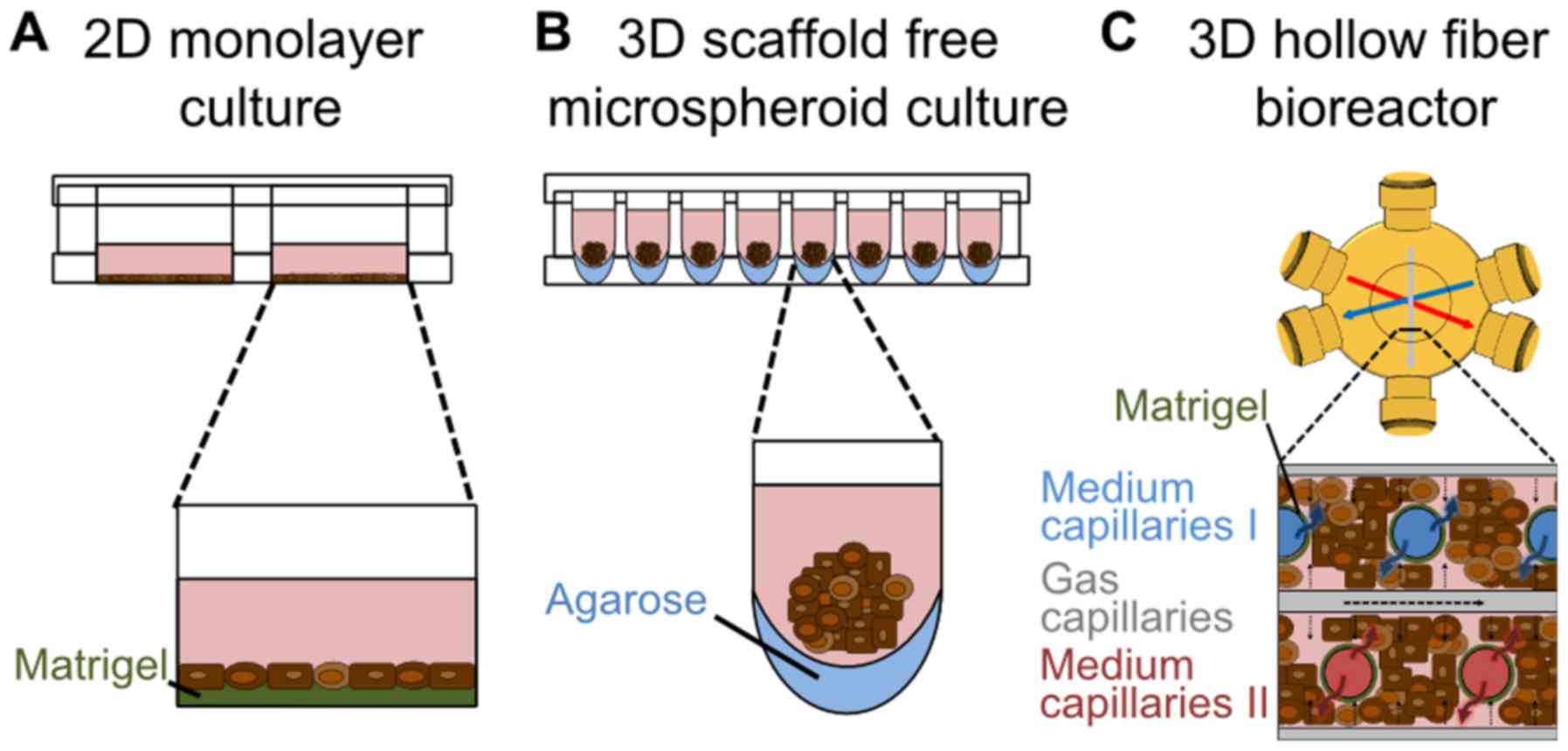
(Fig. 1A) was performed according
to Szkolnicka et al (11)
with minor changes. In detail, hiPSCs were plated onto
Matrigel-coated [1:20 diluted in Dulbecco's modified Eagle's medium
(DMEM)/F12 medium] 24-well plates and differentiated into
definitive endodermal (DE) cells using the STEMdiff™ Definitive
Endoderm kit (Stemcell Technologies, Inc.) according to the
manufacturer's instructions until day 5. From day 5 to 8 cultures
were maintained in SR-DMSO-Medium [knockout DMEM supplemented with
20% knockout serum replacement medium, 0.5% GlutaMAX, 1%
non-essential amino acids, 0.1 mM β-mercaptoethanol, DMSO
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and 1%
penicillin/streptomycin]. From day 9 on, hepatocyte maturation
medium [HepatoZYME-SFM, 1% GlutaMAX, 1% penicillin/streptomycin, 10
µM hydrocortisone 21-hemisuccinate sodium salt
(Sigma-Aldrich; Merck KGaA), 10 ng/ml human HGF and 20 ng/ml human
OSM (both from PeproTech EC Ltd., London, UK)] was used and renewed
every other day. All reagents were purchased from Thermo Fisher
Scientific, Inc., if not stated otherwise.
Adaptations for differentiation using 3D
microspheroids
For the 3D microspheroid differentiation (Fig. 1B), the first steps of
differentiation were performed in conventional 2D cultures using
6-well plates as described above. On day 11 of the differentiation
process, the cells were detached enzymatically with TrypLE™ (Thermo
Fisher Scientific, Inc.) and plated onto low attachment 96-well
plates. These were prepared by adding 50 µl of 60°C warm
1.5% agarose (Serva Electrophoresis GmbH, Heidelberg,
Germany)-DMEM/F12 (Thermo Fisher Scientific, Inc.) solution into
each well of the plate (33).
Cells were seeded in these 96-well plates at a density of 10,000
cells/well in 150 µl of hepatocyte maturation medium. Every
other day 100 µl of the medium were renewed. The method of
spheroid formation was proven with additional hiPSC lines, showing
the robust generation of one spheroid of constant size per well
(Fig. 2).
Adaptations for differentiation using
perfused 3D bioreactors
For hepatic differentiation under dynamic
conditions, a hollow-fiber bioreactor technology was used (Fig. 1C). The bioreactor (StemCell
Systems GmbH, Berlin, Germany) consists of independent yet
interwoven hollow-fiber capillary systems, which serve for
counter-current medium perfusion via two medium capillary systems
and decentralized oxygenation via one gas capillary system. The
cells are cultured in the extra-capillary space (cell compartment)
(31). The cell compartment
volume of the used bioreactors was 2 ml and they were integrated
into a perfusion circuit (StemCell Systems GmbH) with a total
volume of 20 ml. Electronic control of system functions was
provided by a perfusion device (StemCell Systems GmbH) that
contained two modular pump units, a heating unit and a gas-mixing
unit.
Prior to cell inoculation the bioreactors were
flushed with 3 mg Matrigel in 5 ml DMEM/F-12 medium and incubated
at RT for 1 h. Afterwards, 1×108 hiPSCs were seeded into
each bioreactor. Cultures were maintained at 37°C, the medium
recirculation rate was 10 ml/min and the feed rate was 1 ml/h. A
mixture of 95% air and 5% CO2 was supplied at a flow
rate of 20 ml/min. CO2 perfusion rates were adjusted, if
required, to maintain a stable pH between 7.2 and 7.4. After an
adaptation phase of two days with mTeSR™1, differentiation of the
cells was performed with the same media compositions as used for 2D
cultures. After each differentiation step, the culture medium was
rinsed out by flushing the perfusion circuit with 60 ml of the
culture medium used in the next differentiation step.
Culture of primary human hepatocytes
PHH were isolated from macroscopically healthy
tissue from resected human livers of patients with informed consent
of the patients according to the ethical guidelines of the Charité
Universitätsmedizin Berlin (Berlin, Germany). Cell isolation was
performed according to Pfeiffer et al (34). Hepatocytes were seeded at a
density of 2.0×105 cells/cm2 in 24-well
plates (BD Biosciences, Franklin Lakes, NJ, USA) coated with
rat-tail collagen. Cells were cultivated using Heparmed Vito 143
supplemented with 0.8 mg/l insulin, 5 mg/l transferrin, 0.003 mg/l
glucagon, 100,000 U/l penicillin and 100 mg/l streptomycin (all
from Merck KGaA), and 10% FCS (GE Healthcare Life Sciences,
Chalfont, UK). PHH were either used directly after isolation (PHH 0
h) or after 24 h of cultivation in 2D culture plates (PHH 24
h).
Glucose and lactate measurements
The metabolic activity of the cells was assessed by
measuring glucose and lactate concentrations with a blood gas
analyzer (ABL 700; Radiometer, Copenhagen, Denmark).
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated using TRIzol™ (Thermo Fisher
Scientific, Inc.), following the manufacturer's instructions. The
High Capacity cDNA reverse transcription kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.) was used to convert 1 µg of
RNA to cDNA following the manufacturer's instructions. The
quantitative validation of the expression of selected genes was
performed by RT-qPCR, as previously described (35). In detail, the Applied Biosystems
StepOne real-time PCR system was applied using custom PrimerDesign
primers (Primerdesign Ltd., Chandler's Ford, UK) and the SYBR-Green
PCR master mix (cat. no. 4368577; Applied Biosystems; Thermo Fisher
Scientific, Inc.), following the manufacturer's instructions.
Primers are listed in Table I.
Reactions were run in triplicate on a StepOne Plus instrument
(Applied Biosystems; Thermo Fisher Scientific, Inc.). Running
conditions were: 95°C for 10 min, followed by 40 cycles of 95°C for
15 sec and 60°C for 60 sec.
 | Table IPrimer sequences for the custom
real-time PCR (Primerdesign Ltd.). |
Table I
Primer sequences for the custom
real-time PCR (Primerdesign Ltd.).
| Gene symbol | Gene name | Forward primer
5′→3′ sequence | Reverse primer
3′→5′ sequence | Amplicon size
(bp) |
|---|
| AFP | α-fetoprotein |
CAGTAATTCTAAGAGTTGCTAAAGGAT |
CCTGGATGTATTTCTGTAATTCTTCTT | 117 |
| AHR | Aryl hydrocarbon
receptor |
AATTTTGACCCTGGTTTTTGGATT |
TGGTTTGGAATAATTGTGAATAGCA | 129 |
| ALB | Albumin |
TGACAAATCACTTCATACCCTTTTT G |
CATTCATTTCTCTCAGGTTCTTG | 118 |
| CXCR4 | C-X-C motif
chemokine receptor 4 |
CCAAAGAAGGATATAATGAAGTCACT |
GGGCTAAGGGCACAAGAGA | 88 |
| CYP1A2 | Cytochrome P450
family 1 subfamily A member 2 |
GCCTTCATCCTGGAGACCTT |
TCAGCGTTGTGTCCCTTGT | 82 |
| CYP3A4 | Cytochrome P450
family 3 subfamily A member 4 |
ACCGTAAGTGGAGCCTGAAT |
AAGTAATTTGAGGTCTCTGGTGTT | 90 |
| CYP3A7 | Cytochrome P450
family 3 subfamily A member 7 |
AGAGAGATAAGGAAGGAAAGTAGTGA |
TGTGTACGGGTTCCATATAGATAGA | 114 |
| HNF4A | Hepatocyte nuclear
factor 4α |
GACCTCTACTGCCTTGGACAA |
GATGAAGTCGGGGGTTGGA | 87 |
| NANOG | Nanog homeobox |
GCTGTGTGTACTCAATGATAGATTT |
GAGGTTCAGGATGTTGGAGAG | 85 |
| SOX9 | SRY-box 9 |
GGACCAGTACCCGCACTTG |
AATCCGGGTGGTCCTTCTTG | 143 |
| SOX17 | SRY-box 17 |
GTAGAAGGGGATGTCCAAGTAAT |
TGTGAAGATTAAGGTAAACTGAATGT | 144 |
Data from RT-qPCR were normalized to multiple
internal control genes (18S, EIF 2A4, β-actin and SDHA) with the
geNorm algorithm as described by Vandesompele et al
(36). Results are presented as
fold-changes in gene expression relative to 2D cultures on day 18
calculated with the ΔΔCq method (37).
Enzyme-linked immunosorbent assay
(ELISA)
Cell culture supernatants were clarified by
centrifugation and stored at −20°C until assayed. The secretion of
α-fetoprotein (AFP), albumin (ALB) and α-1-antitrypsin (A1AT) was
quantified with an ELISA, using the antibodies provided in Table II and the protocol as described
by Liu et al (38). ELISA
plates were read at 490 nm with a reference wavelength of 630 nm
using a MRX II plate reader (Dynex Technologies, Chantilly, VA,
USA) and the concentration of the appropriate protein in each
sample was calculated from standard curves using MRX II Endpoint
software 2.02 (Dynex Technologies).
 | Table IIAntibodies used for analysis of
hepatic export proteins using ELISA. |
Table II
Antibodies used for analysis of
hepatic export proteins using ELISA.
| Species and antigen
name | Type | Target | Provider, catalog
no. | Dilution |
|---|
| Rabbit anti-albumin
(capture antibody) | Polyclonal | Anti-human | Agilent
Technologies, Inc., A0001 | 1:1,000 |
| Mouse anti-albumin
antibody (intermediate antibody) | Monoclonal | Anti-human | Sigma-Aldrich,
A6684 | 1:1,000 |
| Rabbit anti-IgGs,
HRP conjugated (detection antibody) | Polyclonal | Anti-mouse | Agilent
Technologies, Inc., P0260 | 1:1,000 |
| Rabbit anti-AFP
(capture antibody) | Polyclonal | Anti-human | Agilent
Technologies, Inc., A0008 | 1:2,000 |
| Rabbit anti-AFP,
HRP conjugated (detection antibody) | Polyclonal | Anti-human | Agilent
Technologies, Inc., P0128 | 1:2,500 |
| Sheep anti-A1AT,
HRP conjugated (detection antibody) | Polyclonal | Anti-human | Abcam, ab 8768 | 1:1,500 |
Urea analysis
Cell culture supernatants were clarified by
centrifugation and stored at −20°C until assayed. Urea was measured
in the cell culture supernatant without any additional treatment,
using the QuantiChrom™ Urea assay kit (DIUR-500; BioAssay Systems,
Hayward, CA, USA) according to the manufacturer's instructions.
Immunofluorescence analysis
The 2D cultures were fixed in 4% paraformaldehyde
(PFA; Electron Microscopy Science, Hatfield, PA, USA) in
phosphate-buffered saline (PBS) at room temperature for 10 min.
Primary and secondary antibodies were applied in PBS with 0.5%
Triton X-100 and 1% BSA (both from Sigma-Aldrich; Merck KGaA) and
incubated at 4°C overnight or at room temperature for 1.5 h,
respectively. All primary and secondary antibodies are provided in
Table III. Finally, a nuclear
counter stain was performed with 0.8 µg/ml Hoechst 33342
(Thermo Fisher Scientific, Inc.) in PBS at room temperature for 30
min. The quantification of immunoreactive cells was performed with
the Cellomics Array Scan VTi and Cellomics Scan and View
Software (version 6.3.1) (both from Thermo Fisher Scientific,
Inc.).
 | Table IIIPrimary and secondary antibodies used
for immunofluorescence analysis. |
Table III
Primary and secondary antibodies used
for immunofluorescence analysis.
| Species and antigen
name | Type | Target | Provider, catalog
no. | Dilution |
|---|
| Mouse
anti-α-fetoprotein | Monoclonal | Anti-human | Thermo Fisher
Scientific, Inc., 180003 | 1:1,000 |
| Mouse
anti-cytokeratin 18 | Monoclonal | Anti-human | Santa Cruz
Biotechnology, Inc., sc-6259 | 1:100 |
| Rabbit
anti-albumin | Polyclonal | Anti-human | Dako Cytomation,
A0001 | 1:2,000 |
| Rabbit
anti-hepatocyte nuclear factor 4α | Polyclonal | Anti-human | Santa Cruz,
sc-8987 | 1:100 |
| A488 goat
anti-mouse | Polyclonal | Anti-mouse | Thermo Fisher
Scientific, Inc., A11029 | 1:500 |
| A594 goat
anti-rabbit | Polyclonal | Anti-rabbit | Thermo Fisher
Scientific, Inc., A11037 | 1:500 |
The microspheroids were collected in a 1.5 ml
reaction tube and fixed in 4% PFA (Electron Microscopy Science) in
PBS at 4°C overnight. An ~60°C warm 2% agarose solution in PBS was
prepared, added to the microspheroids and centrifuged at 20,000 × g
for 1 sec. The resulting agarose plug was dehydrated in a tissue
processor (Tissue-Tek VIP; Sakura Finetek Europe B.V., Flemingweg,
The Netherlands), paraffinized and cut into slides of 4.0 µm
thickness.
For immunohistochemical staining of the bioreactor
cultures the hollow-fiber bed was excised en bloc, fixed with 4%
formaldehyde solution (Herbeta Arzneimittel Detlef Karlowski e.K.,
Berlin, Germany) at room temperature for 1 h, dehydrated,
paraffinized and cut into slides of 4.0 µm thickness.
Paraffin sections were rehydrated and boiled in 10
mM citrate buffer (pH 6.0) for 12 min. Pre-blocking was performed
with PBS containing 0.5% Triton X-100 and 1% BSA at room
temperature for 1 h. Antibody staining was performed as described
above. The primary and secondary antibodies used are detailed in
Table III. Finally, the cells
were embedded in Roti-Mount FluorCare DAPI (Carl Roth GmbH + Co.
KG, Karlsruhe, Germany). The quantification of marker positive
cells was performed with the Opera Phenix and the Harmony software
(version 4.1) (both from PerkinElmer, Inc., Waltham, MA, USA).
Cytochrome P450 (CYP) analysis
CYP iso-enzyme activities were analyzed based on
assays established in previous studies (39,40). A substrate mix containing
midazolam, phenacetin and bupropion was prepared in the respective
culture medium without pretreatment for induction. Details of the
used substrates, their final concentrations and information on the
LC-MS analysis are provided in Table
IV. A medium blank was used for normalization. In 2D cultures,
the assay was performed on four wells of a 24-well plate and the
supernatants were pooled per time-point. In microspheroids, the
assay was performed in 8 wells of a 96-well plate by collecting ~10
microspheroids/well. Supernatants of the wells were pooled per
time-point. For the bioreactors, 1 ml of a 20-fold concentrated
substrate mix was applied to the bioreactor system. For all culture
systems, samples were taken after 0, 1, 2 and 6 h.
 | Table IVApplied substrates and their
corresponding CYP isoenzymes with resulting products and applied
concentrations. |
Table IV
Applied substrates and their
corresponding CYP isoenzymes with resulting products and applied
concentrations.
| Substrate | Corresponding CYP
isoenzyme | Provider | Final
concentration | Solutions for
elution | Recorded
transitions |
|---|
| Midazolam | CYP3A4/5 | Roche Diagnostics
GmbH | 25 µM | Double distilled
water containing 0.1% formic acid and acetonitrile containing 0.1%
formic acid | 1′-Hydroxymidazolam
342.1–324.0 m/z |
| Phenacetin | CYP1A2 | Sigma-Aldrich | 200 µM | Double distilled
water containing 0.1% formic acid and methanol containing 0.1%
formic acid | Paracetamol
152.1–110.1 m/z |
| Bupropion | CYP2B6 | Sigma-Aldrich | 75 µM | Double distilled
water containing 0.1% formic acid and methanol containing 0.1%
formic acid | Hydroxybupropion
256.1–238.1 m/z |
The supernatants were diluted with quench solution
(20% methanol or acetonitrile with 0.1% formic acid) containing the
stable isotope-labeled metabolite as an internal standard. LC-MS
analysis was performed using the following equipment: HTS-xt PAL
autosampler (CTC Analytics AG, Zwingen, Switzerland), 1290 infinity
G4220A binary pump and degasser (Agilent Technologies, Inc., Santa
Clara, CA, USA), MistraSwitch column oven (Maylab Analytical
Instruments GmbH, Vienna, Austria) and a YMC C18 Triart, 30×2 mm,
1.9 µm (YMC Europe GmbH, Dinslaken, Germany) column. All ion
chromatograms were recorded on a 6500 Triple Quad (QqQ)
LC-MS/MS-system hybrid mass spectrometer (AB Sciex Pte, Concord,
ON, Canada) equipped with an Iondrive™ Turbo V ion source operated
in the positive electrospray ionization mode. Integration of
chromatograms as well as determination of peak areas was performed
by Analyst software version 1.6.2 (Applied Biosystems/MDS
Sciex).
DNA isolation
RLT buffer (Qiagen GmbH, Hilden, Germany) was added
to a defined number of wells, number of microspheroids or volume of
the cell compartment of bioreactors. The DNA was isolated from the
cell extracts by use of the QIAamp DNA micro kit (Qiagen GmbH)
according to the manufacturer's protocol.
Statistical analysis
Data evaluation and graphical illustration were
performed with GraphPad Prism 5.0 and 7.0 (GraphPad Software, Inc.,
San Diego, CA, USA). Experiments were performed in triplicate,
unless stated otherwise, and results are presented as median ±
interquartile range. Data for energy metabolism and secretion of
proteins were normalized to the initial cell number and area under
curve (AUC) was calculated. Data for CYP activities were normalized
against the DNA content at day 18. Differences between the
different culture systems or to PHH were detected applying the
Mann-Whitney test or the unpaired, two-tailed t-test.
Results
Differentiation of hiPSCs to HLCs
The hiPSC line AD3C4 was selected from different
previously tested hiPSC lines generated within the StemBANCC
consortium. The comparative study of a 2D cell culture
differentiation system (Fig. 1A)
with a 3D scaffold-free microspheroid system (Fig. 1B) and a perfused 3D hollow-fiber
bioreactor (Fig. 1C) was
performed using three independent batches of AD3C4 hiPSCs at the
same passage number. To compare the different culture systems, mRNA
levels and secretion of stage specific markers as well as
immunohistochemical staining and CYP activity were analyzed
(Fig. 3).
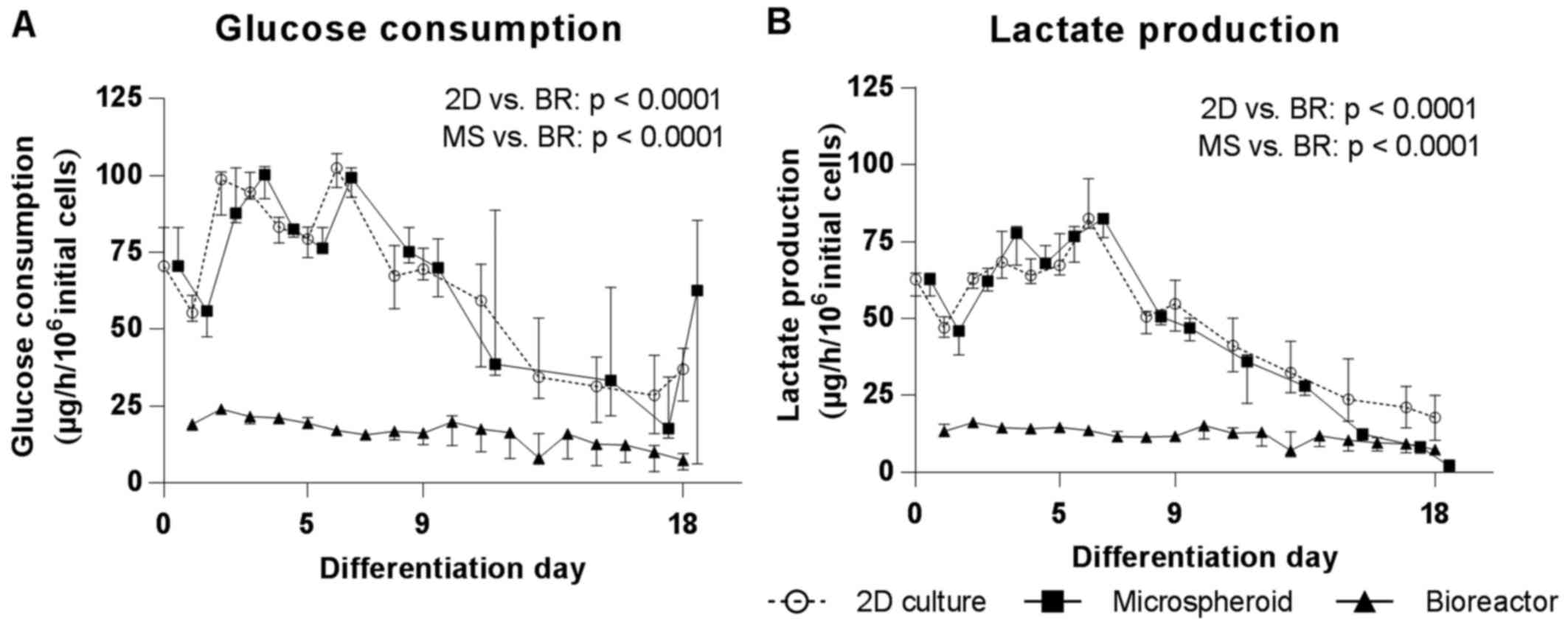
Energy metabolism
In order to evaluate the energy metabolism during
hepatic differentiation of hiPSCs, the glucose consumption rates
and lactate production rates were determined at the time of medium
exchange (2D culture, microspheroids) or daily (bioreactor).
Glucose consumption and lactate production rates increased in 2D
cultures and microspheroids during the first 6 days of
differentiation. Following this, rates continuously declined in
both culture systems during the whole differentiation period
(Fig. 4A). The bioreactor
cultures showed a slight increase of glucose consumption and
lactate production rates during the first 2 days of differentiation
and afterwards also a continuous but slight decrease until the end
of hepatic differentiation. At the end of differentiation, these
values were comparable to 2D cultures and microspheroids (Fig. 4A). The AUC for glucose consumption
was significantly higher in 2D cultures and microspheroids compared
with bioreactors (p<0.0001). The time course of lactate
production mirrored that of glucose consumption and the AUC for
lactate production was also significantly higher in 2D cultures and
microspheroids compared with bioreactors (p<0.0001) (Fig. 4B)
Gene expression profiling of
hiPSC-derived HLCs
To evaluate the maturation state of the HLCs
obtained in the different culture systems compared to freshly
isolated and cultured PHH, the mRNA expression of stage-specific
genes was analyzed and calculated in relation to 2D differentiated
HLCs at day 18. The expression of the pluripotency-associated
homeobox gene Nanog (NANOG) was downregulated in HLC relative to
undifferentiated hiPSCs, irrespective of the culture system
(Fig. 5A). Within the HLC and PHH
groups, the 2D differentiated HLCs expressed the highest amount of
NANOG. The DE markers SRY-box 17 (SOX17) and C-X-C
motif chemokine receptor 4 (CXCR4) peaked at day 5 in 2D
cultures and showed subsequently a downregulation (Fig. 5B and C). The significantly higher
expression of AFP in the HLCs compared to cultured PHH
(p<0.0001) demonstrated the immature properties of the in
vitro generated cells. Within the in vitro generated
cells, the 2D HLCs displayed a significantly higher expression of
AFP compared to HLCs derived in the bioreactor (p=0.0477),
and a higher, although not significant, expression than the
microspheroids. There was also a significantly lower AFP expression
in cultured PHH compared to freshly isolated PHH (p=0.032)
(Fig. 5D). The cholangiocyte
marker SRY-box 9 (SOX9) showed the highest expression in
HLCs from 2D cultures and bioreactors (Fig. 5E). Microspheroids and cultured PHH
expressed significantly lower amounts of SOX9. Expression of
the hepatocyte marker ALB was highest in freshly isolated
PHH and showed a significant drop after 24 h of cultivation
(p=0.0028) (Fig. 5F). HLCs
derived in the bioreactor expressed ALB at a higher amount
compared to 2D HLCs and microspheroids indicating a higher degree
of maturation in the bioreactor system. The expression of
hepatocyte nuclear factor 4α (HNF4A) was significantly
higher in HLCs as compared to cultured PHH (p≤0.0003) (Fig. 5G). Among the HLCs, cells derived
in the bioreactor expressed significantly more HNF4A than
cells generated in 2D or in microspheroids (p≤0.0059). Furthermore,
there was a significant drop of HNF4A expression in cultured
PHH compared to freshly isolated PHH (p≤0.0001). Regarding the
expression of metabolic enzymes, the CYP3A4 expression
presented a significant decrease from freshly isolated PHH to
cultured PHH and a further decrease from cultured PHH to all in
vitro generated HLCs (p≤0.0142) (Fig. 5H). The highest CYP3A7 expression
among the HLCs could be detected in the bioreactor system. There
was again a significant decrease of CYP3A7 expression from
freshly isolated PHH to PHH cultured over 24 h (p<0.0001)
(Fig. 5I). CYP1A2
indicated only a marginal expression in HLCs in all culture
systems. In contrast, freshly isolated PHH demonstrated a high
expression, which however significantly decreased within 24 h of
cultivation (Fig. 5J). The
nuclear receptor aryl hydrocarbon receptor (AHR) was
expressed in the in vitro systems at levels positioned
between freshly isolated PHH (high expression) and cultivated PHH
(low expression) (Fig. 5K). In
conclusion, among the in vitro systems the bioreactor system
showed the highest maturation stage on mRNA expression level, but
was still lower as compared to freshly isolated PHH.
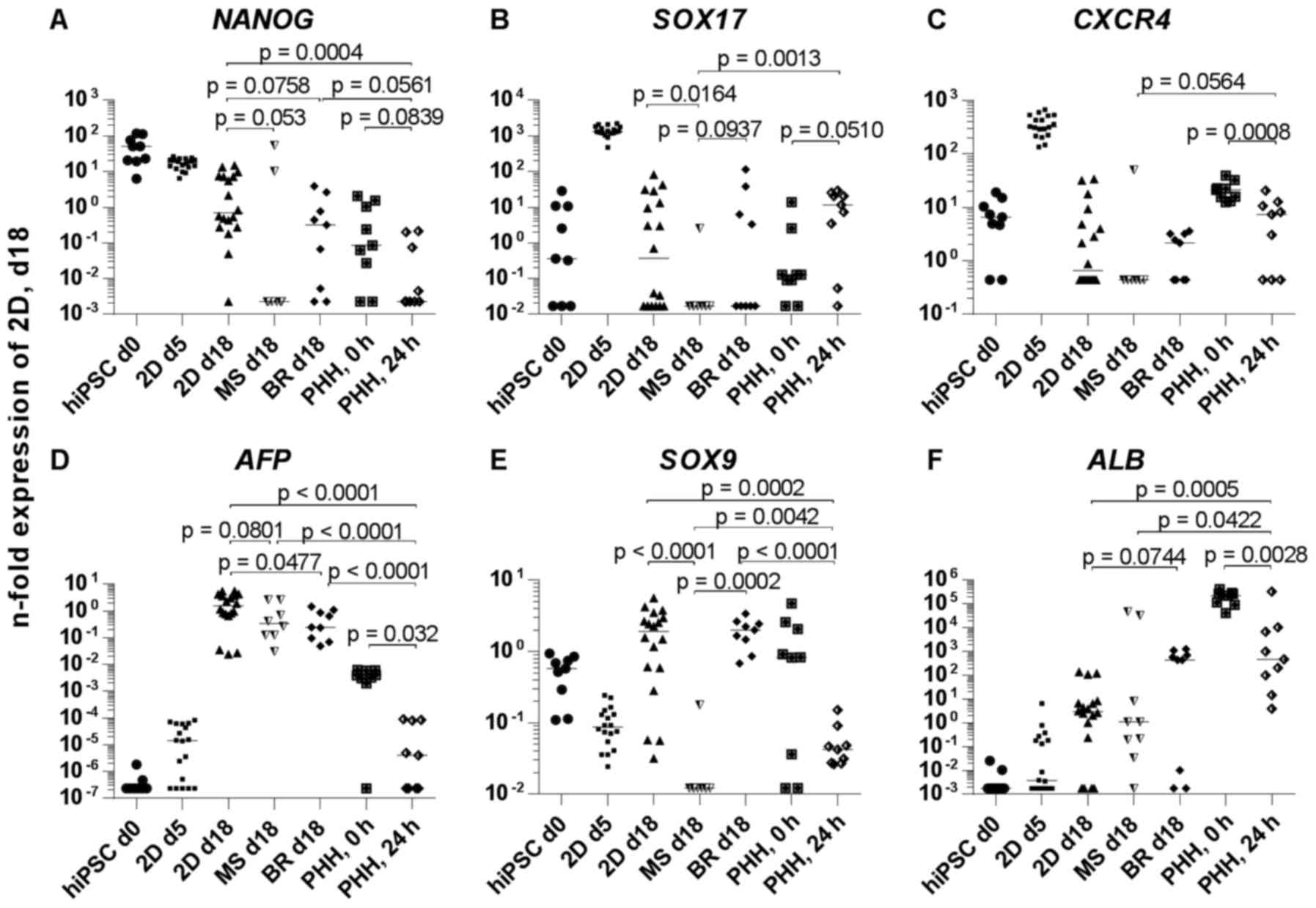 | Figure 5Gene expression of hiPSC derived
hepatocyte-like cells in 2D cultures, microspheroids, bioreactors
or in PHH. The mRNA expression of (A) NANOG, (B)
SOX17, (C) CXCR4, (D) AFP, (E) SOX9,
(F) ALB, (G) HNF4A, (H) CYP3A4, (I)
CYP3A7, (J) CYP1A2 and (K) AHR is shown.
Samples for expression analysis were taken before (hiPSC day 0) and
after hepatic differentiation of hiPSC in 2D cultures (2D day 18),
microspheroids (MS day 18) or bioreactors (BR day 18). In addition,
samples were taken after definitive endodermal differentiation in
2D cultures (2D day 5). Further, mRNA samples from freshly isolated
(0 h) or 2D cultured PHH (24 h) were used for expression analyses.
Differences in gene expression between groups were calculated using
the Mann-Whitney test. Data from day 0 and 5 were not included in
comparative statistics because the aim was to compare the different
culture systems among each other and to the current
'gold-standard', the cultured PHH. p<0.1 are given in the graphs
[median and data points of 3 resp. 6 (for 2D cultures) independent
experiments plus technical replicates (three per experiment) are
shown]. hiPSC, human induced pluripotent stem cells; PHH, primary
human hepatocytes; MS, microspheroids; BR, bioreactors; NANOG,
Nanog homeobox; SOX17, SRY-box 17; CXCR4, C-X-C motif chemokine
receptor 4; AFP, α-fetoprotein; SOX9, SRY-box 9; ALB, albumin;
HNF4A, hepatocyte nuclear factor 4α; CYP3A4, cytochrome P450 family
3 subfamily A member 4; CYP3A7, cytochrome P450 family 3 subfamily
A member 7; CYP1A2, cytochrome P450 family 1 subfamily A member 2;
AHR, aryl hydrocarbon receptor; d, day. |
Secretion of hepatic proteins and
metabolites by HLCs
The detection of secreted hepatic proteins allows an
estimation of the differentiation status of the cells over time.
The secretion of AFP increased in all culture systems from day 9
until day 13 and subsequently decreased until day 18 reaching basal
levels (Fig. 6A). Secretion of
ALB and A1AT could be detected only in the bioreactors from
differentiation day 9 and 11 onwards, respectively (Fig. 6B and C). Urea production was
significantly higher in the microspheroids than in 2D cultures
(p=0.0027) or bioreactors (p=0.0022) and peaked at day 13 of
differentiation (Fig. 6D).
However, the strong production of urea decreased subsequently to a
lower level as compared to the 2D cultures and bioreactors, which
produced a constant level of urea from day 11 onwards (Fig. 6D). Compared to the 2D cultures,
cells in the bioreactor produced significantly less urea
(p=0.0021).
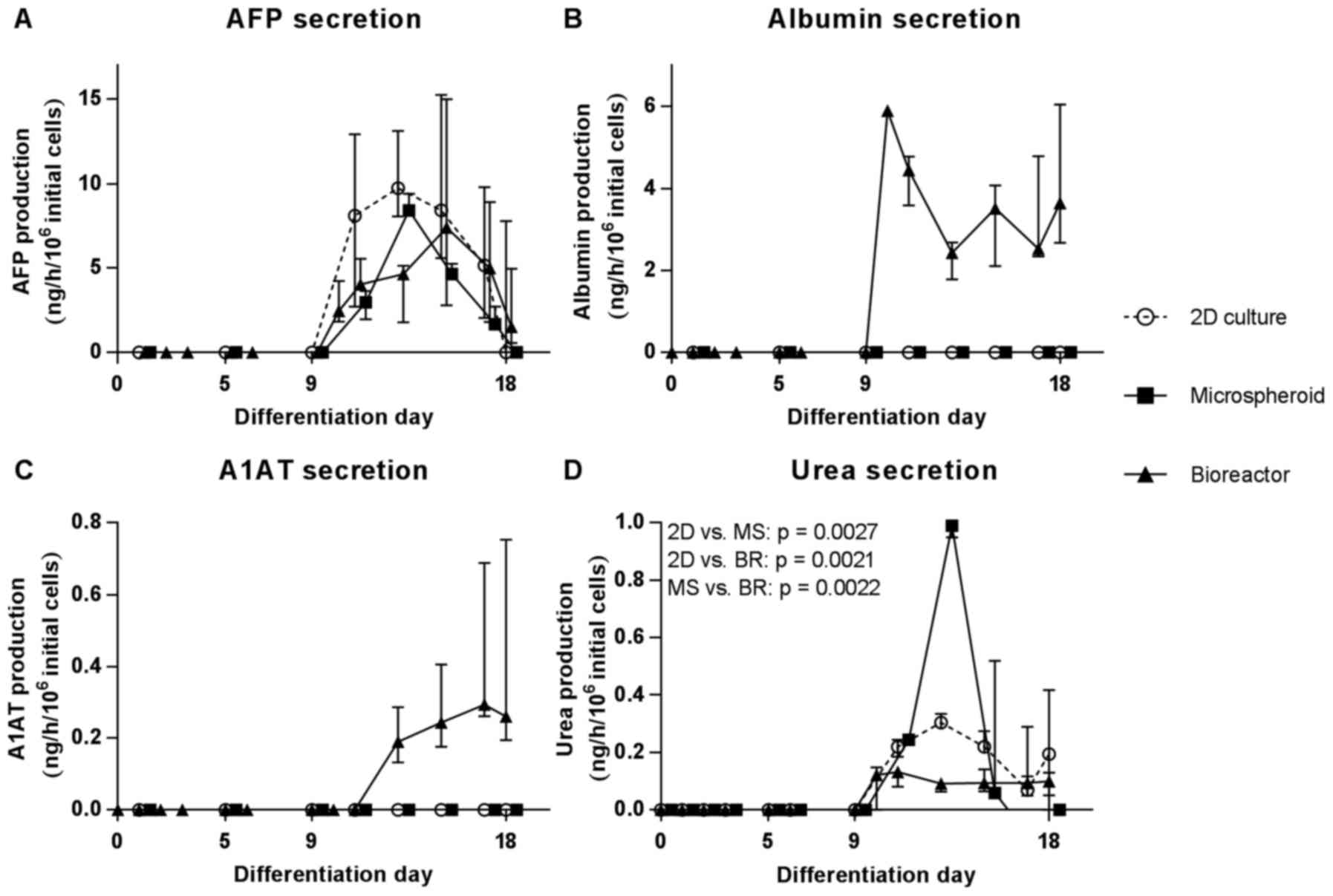 | Figure 6Secretion of specific proteins and
metabolites by hiPSCs during hepatic differentiation in 2D cultures
(2D, circles), MS (squares) or bioreactors (BR, triangles). (A)
Secretion of AFP, (B) albumin (ALB), (C) A1AT and (D) urea is
shown. Values are normalized to 106 initial cells on day
0 (2D cultures and bioreactors) or day 11 (microspheroids). Areas
under curve were calculated for each dataset and differences
between groups were determined with the unpaired, two-tailed t-test
(2D cultures: n=6; microspheroids and bioreactors: n=3, median of
biological replicates ± interquartile range). MS, microspheroids;
BR, bioreactors; AFT, α-fetoprotein; A1AT, α-1-antitrypsin; hiPSCs,
human induced pluripotent stem cells. |
Immunohistochemical characterization of
HLCs
Immunohistochemical analysis of liver-specific
markers was performed to characterize the cell composition in the
different culture systems at day 18 (Fig. 7A–C). The hepatocyte marker HNF4A
was detectable in half of the cells in the 2D cultures, whereas
this proportion was lower in the microspheroids and bioreactors
(Fig. 7D–F and J). A shift from
the nuclear staining to a diffuse cytoplasmic and weak nuclear
staining was obvious, implying a possible downregulation of HNF4A.
In contrast, cytokeratin 18 (CK18) was detectable at similar levels
in all culture systems (Fig. 7D–F and
K). The fetal hepatic marker AFP was present in three quarters
of the cells in the 2D culture system, which was significantly
higher compared to the microspheroids (p<0.0001) and the
bioreactors (p=0.0004 (Fig. 7G–I and
L). The number of ALB-positive cells was significantly
higher in 2D cultures (p=0.0017) and bioreactors (p=0.0476)
compared with microspheroids (Fig.
7G–I and M). Cells double-positive for AFP and ALB, which
indicate a transition from fetal-like to mature hepatocytes, were
present in all in vitro culture systems, with a significant
lower amount in microspheroids compared to 2D cultures (p=0.0254)
(Fig. 7N). In addition, the
immunohistochemical analysis revealed an irregular distribution of
cells positive for HNF4A or ALB in 3D culture systems. Furthermore,
the microspheroids showed a decrease in size with increasing
culture time and holes could be detected within the spheroids and
in the aggregates from the bioreactor at day 18.
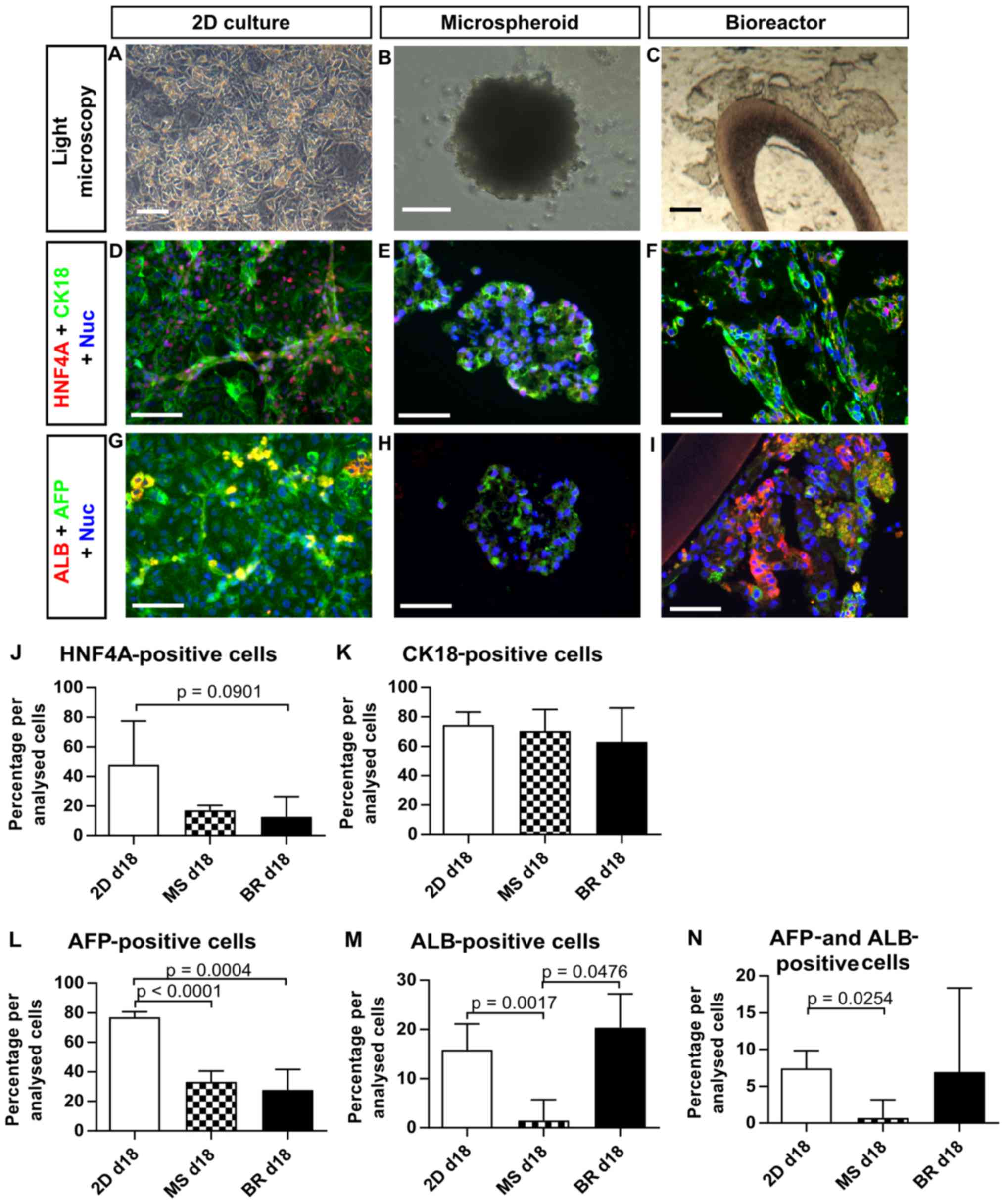 | Figure 7Expression of liver-specific
immunohistochemical markers in human induced pluripotent stem
cell-derived hepatocyte-like cells in 2D cultures (2D day 18), MS
(day 18) or BR (day 18). (A–C) Bright-field images of the three
culture systems at the day of analysis. (D–N) Expression of (D–F
and J) HNF4A, (D–F and K) cytokeratin 18, (G–I, L and N) AFP and
(G–I, M and N) ALB was determined by quantitative analysis of
immunofluorescence pictures. Differences between groups were
determined with the unpaired, two-tailed t-test (2D cultures: n=6;
MS and BR: n=3, median of biological replicates ± inter-quartile
range). p<0.1 are given in the graphs. Scale bars correspond to
100 µm. d, day; MS, microspheroids; BR, bioreactors; HNF4A,
hepatocyte nuclear factor 4α; CK18, cytokeratin 18; AFP,
α-fetoprotein; ALB, albumin. |
CYP3A4 activity of HLCs in different
culture systems
The basal capacity of CYP-dependent drug metabolism
was examined in the HLC cultures by the application of substrates
for CYP1A2 (phenacetin), CYP2B6 (bupropion) and CYP3A4/5
(midazolam). Activity of CYP2B6 could not be detected in HLCs
irrespective of the cultivation system (data not shown). Data,
normalized against the DNA content, measured at the day of the CYP
analysis, showed a turnover of phenacetin in the linear range of
the quantification method only for HLCs from the bioreactor and in
PHH 2D-cultured for 24 h (Fig.
8A). In comparison to PHH the CYP1A2 activity of HLCs was ~5
times lower in the bioreactor and not detectable in 2D cultures and
3D microspheroids, which is in line with the finding of marginal
mRNA expression of CYP1A2 (Fig. 5J). Activity of CYP3A4/5 was
detectable in all HLC samples, but also in undifferentiated hiPSCs
and PHH (Fig. 8B) which
corresponds to the mRNA expression data (Fig. 5J). The CYP3A4/5 activity of PHH
was significantly higher compared to all HLC culture systems
(p≤0.0007). Cells obtained in the microspheroids had a slightly
higher activity compared to the bioreactors and the 2D cultures.
The lowest activity was detectable in the undifferentiated
hiPSCs.
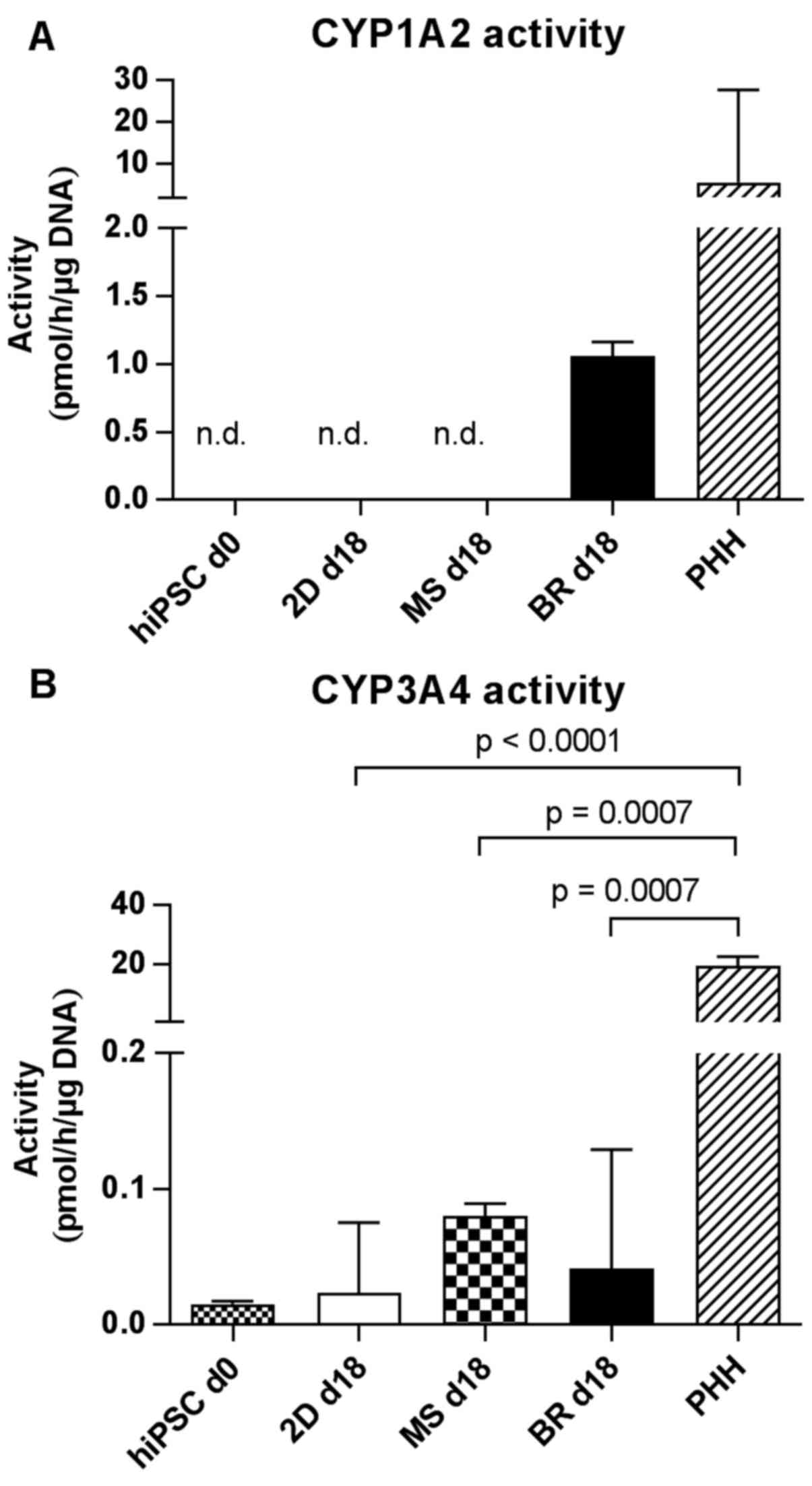 | Figure 8Activities of different cytochrome
P450 (CYP) isoenzymes in hiPSC-derived hepatocyte-like cells in 2D
cultures on day 18 (2D day 18), MS on day 18 (MS day 18), BR on day
18 (BR day 18), in undifferentiated hiPSCs on day 0 (hiPSC day 0)
or in PHH 24 h after seeding. CYP activities were determined by
measuring the formation of (A) acetaminophen from phenacetin via
CYP1A2 and (B) the formation of 1-OH-midazolam from midazolam via
CYP3A4/5. Differences in metabolic activity between
undifferentiated hiPSCs, 2D cultures, microspheroids, bioreactors
and PHH were calculated using the unpaired, two-tailed t-test
(hiPSC day 0 cultures and 2D day 18 cultures: n=6; microspheroids
day 18, bioreactors day 18 and adult PHH: n=3; median of biological
replicates ± interquartile range). p<0.1 are given in the
graphs. Values are normalized to the DNA content on day 18. n.d.,
not detected; d, day; hiPSCs, human induced pluripotent stem cells;
PHH, primary human hepatocytes; MS, microspheroids; BR,
bioreactors. |
Discussion
Human iPS cells are of interest as a source for
human hepatocytes including their possible use in pharmacological
analyses and toxicity testing, ideally to gain sophisticated data
in vitro. To date hiPSC-derived hepatocytes still show an
immature phenotype and lack the functional range of their in
vivo counterparts (19,20). In the present study, the
propensity of two different 3D culture systems to enhance hepatic
maturation of hiPSCs was investigated: i) Scaffold-free
microspheroids based on the self-aggregation of pre-differentiated
cells, and ii) a hollow-fiber bioreactor based on interwoven
capillary systems, which form an adhesion scaffold for the cells
residing in the extra-capillary space. The compared culture systems
differ in their culture characteristics, for example in the
bioreactor, cells are supplied with nutrients and oxygen via
perfusion, and mass exchange is mainly influenced by perfusion
rates and substance properties. In contrast, microspheroids are
maintained under static conditions and their size influences the
supply of oxygen and nutrients to the center of the 3D cell
aggregates. However, both 3D cultivation systems may support the
formation of an in vivo like cell-cell interaction and
microenvironment as already shown for PHH (41,42).
To evaluate the differentiation process and the
maturation state of the obtained cells, metabolic parameters,
secretion and expression of stage-specific markers, as well as CYP
enzyme activities, were analyzed in comparison to 2D cultures,
using freshly isolated or cultured PHH as controls.
Since the bioreactors and the microspheroids do not
allow microscopic analysis on a cellular level of the cells during
culture, alternative read outs were used to monitor the
differentiation process. Therefore, glucose consumption and lactate
production were analyzed to evaluate the metabolic activity of
hiPSCs in the different culture systems over time. The high rate of
glucose consumption and lactate production at the beginning of
differentiation in all systems is in consistence with the known
glycolytic state of undifferentiated hiPSCs (43). With increasing length of culture,
both rates decreased in all culture systems which could be
explained either by a decreased cell proliferation or by a shift to
oxidative phosphorylation as an energy source in association with
cell differentiation (43).
The finding of cell differentiation is also
supported by the observed downregulation of the pluripotency marker
NANOG, a temporal peak of DE markers and an upregulation of
hepatic markers on mRNA level, although the extent of
downregulation and upregulation varied between the compared culture
systems. For example, the mRNA expression of the hepatic markers
ALB, CYP3A4 and HNF4A [important for the
activation of CYP3A4 (44)] was
significantly higher in bioreactors compared to 2D cultures,
indicating a more mature state of the HLCs in the bioreactors.
Moreover, expression of SOX9, which is weakly expressed in
hepatoblasts and strongly in cholangiocytes (45) was observed in 2D cultures and
bioreactors and was even higher than in freshly isolated PHH. This
finding can be explained by the generation of cholangiocytes in
these two differentiation systems in accordance to observations by
Freyer et al (39) and
Miki et al (46). Another
explanation for the detected SOX9 expression may be the
presence of bipotent progenitors (hepatoblasts) in the
differentiated cell populations. This assumption is supported by De
Assuncao et al (47), who
developed a protocol for cholangiocyte differentiation from hiPSCs
and observed an increase in cholangiocyte markers already during
the hepatic progenitor phase. AFP expression was still
significantly higher in HLCs than in PHH, although the 3D culture
systems demonstrated a significant downregulation compared to the
2D culture. The downregulation of AFP expression during
hepatic maturation has been challenging in hepatic differentiation
protocols (17,22). The significant decrease of mRNA
expression levels of hepatic markers such as ALB,
HNF4A and CYP3A4 in cultured PHH compared to freshly
isolated PHH underlines again the importance of appropriate culture
models to maintain the hepatic phenotype. Additionally, it
demonstrates the difficulty to get standardized controls for
hepatic differentiation approaches.
The finding of a more mature state of HLCs in
bioreactors as compared with the other culture systems under
investigation is supported by the detection of ALB and A1AT protein
secretion solely in the culture perfusate of the bioreactors.
However, it cannot be excluded that the absence of ALB and A1AT
detection in the microspheroids and in 2D cultures is due to the
low ratio of cell number to culture volume in 2D cultures
(0.4×106 cells/ml) and in microspheroids
(0.07×106 cells/ml) as compared with a ratio of
5×106 cells/ml in the bioreactors. Thus, the measured
parameters are much more diluted in 2D cultures and in
microspheroids than in the bioreactor system, and thus ALB and A1AT
may be under the detection limit. This underlines another
characteristic of the bioreactor system, which enables high-density
culture of the differentiated cells. An exception to this
observation was the high peak in urea secretion detected for
microspheroids on day 13. This may be explained by the fact that
the HLCs were subjected to cell stress during enzymatic detachment
and cell reseeding for microspheroid formation on day 11. This is
in line with findings from studies using PHH, which also showed a
peak for urea secretion at the first day in culture together with a
high enzyme release, which was attributed to cell stress during the
preceding isolation procedure (40,48).
The results from protein secretion are in line with
the analysis of the marker expression by immunohistochemistry
showing the highest ratio of ALB to AFP positive cells in the
bioreactors. In contrast, the amount of HNF4A positive cells was
highest in 2D cultures whereas for CK18 positive cells no distinct
differences between the culture systems could be detected. In
addition, immunohistochemical analysis of the 3D cell aggregates in
microspheroids and bioreactors revealed a heterogeneous cell
population which may be a result of gradient formation of
differentiation promoting factors since it has been reported that
concentration gradients influence cell differentiation and tissue
formation (49,50).
The basal metabolic activity of CYP1A2 and CYP3A4 of
the in vitro generated cells was distinctly lower as
compared to PHH. However, cells in the 3D systems displayed a
higher CYP-functionality than 2D cultures, in line with previous
studies showing increased CYP activities of PSC-derived hepatocytes
in 3D models compared to 2D cultures (27,29,51). However, for pharmacological and
toxicological applications of hiPSC-derived HLCs, a further
increase of basal CYP activities would be desirable to approximate
the functionality of PHH as the current gold standard for in
vitro drug testing. The authors focused on basal CYP
activities, since application for pharmacological and toxicological
studies may require the opportunity to detect an induction by the
test-compound, which may be masked by the routine use of an inducer
(e.g. rifampin). To further confirm the present results, both basal
and induced levels of CYP-activities, should be measured in future
studies to judge the metabolic capabilities of HLC derivatives.
Beside the 3D cultivation, an extended culture duration may
increase the activity of CYP enzymes as described by Gieseck et
al (29). In addition,
repeated exposure to xenobiotics, mimicking the process of in
vivo drug metabolizing maturation during the first years of
childhood (52), may be another
approach to further increase CYP activity in future models
(17).
Normalization of the results
The cells in the different culture systems may
behave differently, thereby influencing the parameters used for
normalization. Hence, to allow a detailed comparison of different
culture systems the method of normalization needs to be considered
carefully. Therefore, in the present study, the authors used two
different methods for normalization. The initial cell number
reflects the hiPSC number plated on 2D culture plates or injected
into the bioreactor. In both systems the hiPSCs were subsequently
cultured for one or two days in mTeSR™1 medium, leading to the
proliferation of the hiPSCs and therefore to an unknown cell number
at the start of differentiation. In addition, the cells still
proliferated within the early differentiation phase as indicated by
glucose consumption and lactate production. In contrast, for the
microspheroid system the cell number reflects the number of
differentiated cells at the start of the spheroid formation at day
11. At this differentiation stage, the cells have a low
proliferation rate or have ceased to proliferate. Therefore, the
values of energy metabolism and protein secretion obtained for the
2D cultures and bioreactors are probably overestimated, whereas for
the microspheroid culture the values may represent more closely
what is really expressed per cell. For the analysis of the CYP
activities, which was performed only on day 18 of differentiation,
the authors determined the DNA content at day 18 and used these
values for normalization of CYP activities. This method is more
accurate for day 18, but it does not reflect the cell number at the
beginning of culture and during the differentiation. In conclusion,
both methods have their valid reasons for being used.
To conclude, the HLCs generated in the described
culture systems still demonstrate an immature hepatic phenotype
when compared to PHH and there have been recent expert workshops
(53), which attempt to move the
field forward. However, the 3D systems, particularly the
bioreactor, seem to increase the developmental state of the cells
as compared with 2D cultures, which may be due to the more
physiological environment in complex 3D culture systems. It can be
concluded that the choice of the culture system for HLCs depends
primarily on the intended application: The bioreactor system
provides an in vitro instrument for complex analyses, e.g.,
studies on the physiology and pathophysiology of cell
differentiation, or on complex effects of xenobiotics. In contrast,
microspheroids, due to their easy and cost-efficient handling,
could be used for investigation of a higher number of drug
candidates where a certain culture complexity is desired. Thus,
both culture systems open the perspective for the development of
improved in vitro hepatocyte models.
Acknowledgments
The study leading to these results has received
support from the Innovative Medicines Initiative Joint Undertaking
under (grant no. 115439), resources of which are composed of
financial contribution from the European Union's Seventh Framework
Programme (FP7/2007-2013) and EFPIA companies. This publication
reflects only the author's views and neither the IMI JU nor EFPIA
nor the European Commission are liable for any use that may be made
of the information contained therein. L. Armstrong and M. Lako were
additionally funded by European Research Council (grant no. 614620)
and BBSRC UK (grant no. BB/I020209/1).
References
|
1
|
Lauschke VM and Ingelman-Sundberg M: The
importance of patient-specific factors for hepatic drug response
and toxicity. Int J Mol Sci. 17:17142016. View Article : Google Scholar :
|
|
2
|
Mueller SO, Guillouzo A, Hewitt PG and
Richert L: Drug biokinetic and toxicity assessments in rat and
human primary hepatocytes and HepaRG cells within the EU-funded
Predict-IV project. Toxicol In Vitro. 30:19–26. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors. Cell. 126:663–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Takahashi K, Tanabe K, Ohnuki M, Narita M,
Ichisaka T, Tomoda K and Yamanaka S: Induction of pluripotent stem
cells from adult human fibroblasts by defined factors. Cell.
131:861–872. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rashid ST, Corbineau S, Hannan N,
Marciniak SJ, Miranda E, Alexander G, Huang-Doran I, Griffin J,
Ahrlund-Richter L, Skepper J, et al: Modeling inherited metabolic
disorders of the liver using human induced pluripotent stem cells.
J Clin Invest. 120:3127–3136. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tafaleng EN, Chakraborty S, Han B, Hale P,
Wu W, Soto-Gutierrez A, Feghali-Bostwick CA, Wilson AA, Kotton DN,
Nagaya M, et al: Induced pluripotent stem cells model personalized
variations in liver disease resulting from α1-antitrypsin
deficiency. Hepatology. 62:147–157. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Si-Tayeb K, Lemaigre FP and Duncan SA:
Organogenesis and development of the liver. Dev Cell. 18:175–189.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hay DC, Fletcher J, Payne C, Terrace JD,
Gallagher RC, Snoeys J, Black JR, Wojtacha D, Samuel K, Hannoun Z,
et al: Highly efficient differentiation of hESCs to functional
hepatic endoderm requires ActivinA and Wnt3a signaling. Proc Natl
Acad Sci USA. 105:12301–12306. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hannan NRF, Segeritz CP, Touboul T and
Vallier L: Production of hepatocyte-like cells from human
pluripotent stem cells. Nat Protoc. 8:430–437. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Czysz K, Minger S and Thomas N: DMSO
efficiently down regulates pluripotency genes in human embryonic
stem cells during definitive endoderm derivation and increases the
proficiency of hepatic differentiation. PLoS One. 10:e01176892015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Szkolnicka D, Farnworth SL,
Lucendo-Villarin B and Hay DC: Deriving functional hepatocytes from
pluripotent stem cells. Curr Protoc Stem Cell Biol. 30:1G.5.1–12.
2014. View Article : Google Scholar
|
|
12
|
Tasnim F, Phan D, Toh YC and Yu H:
Cost-effective differentiation of hepatocyte-like cells from human
pluripotent stem cells using small molecules. Biomaterials.
70:115–125. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schwartz RE, Trehan K, Andrus L, Sheahan
TP, Ploss A, Duncan SA, Rice CM and Bhatia SN: Modeling hepatitis C
virus infection using human induced pluripotent stem cells. Proc
Natl Acad Sci USA. 109:2544–2548. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bukong TN, Lo T, Szabo G and Dolganiuc A:
Novel developmental biology-based protocol of embryonic stem cell
differentiation to morphologically sound and functional yet
immature hepatocytes. Liver Int. 32:732–741. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Siller R, Greenhough S, Naumovska E and
Sullivan GJ: Small-molecule-driven hepatocyte differentiation of
human pluripotent stem cells. Stem Cell Reports. 4:939–952. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Asplund A, Pradip A, van Giezen M,
Aspegren A, Choukair H, Rehnström M, Jacobsson S, Ghosheh N, El
Hajjam D, Holmgren S, et al: One standardized differentiation
procedure robustly generates homogenous hepatocyte cultures
displaying metabolic diversity from a large panel of human
pluripotent stem cells. Stem Cell Rev. 12:90–104. 2016. View Article : Google Scholar
|
|
17
|
Kim JH, Jang YJ, An SY, Son J, Lee J, Lee
G, Park JY, Park HJ, Hwang DY, Kim JH, et al: Enhanced metabolizing
activity of human ES cell-derived hepatocytes using a 3D culture
system with repeated exposures to xenobiotics. Toxicol Sci.
147:190–206. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Park H-J, Choi Y-J, Kim JW, Chun HS, Im I,
Yoon S, Han YM, Song CW and Kim H: Differences in the epigenetic
regulation of cytochrome P450 genes between human embryonic stem
cell-derived hepatocytes and primary hepatocytes. PLoS One.
10:e01329922015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Baxter M, Withey S, Harrison S, Segeritz
CP, Zhang F, Atkinson-Dell R, Rowe C, Gerrard DT, Sison-Young R,
Jenkins R, et al: Phenotypic and functional analyses show stem
cell-derived hepatocyte-like cells better mimic fetal rather than
adult hepatocytes. J Hepatol. 62:581–589. 2015. View Article : Google Scholar :
|
|
20
|
Godoy P, Schmidt-Heck W, Natarajan K,
Lucendo-Villarin B, Szkolnicka D, Asplund A, Björquist P, Widera A,
Stöber R, Campos G, et al: Gene networks and transcription factor
motifs defining the differentiation of stem cells into
hepatocyte-like cells. J Hepatol. 63:934–942. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kondo Y, Iwao T, Nakamura K, Sasaki T,
Takahashi S, Kamada N, Matsubara T, Gonzalez FJ, Akutsu H, Miyagawa
Y, et al: An efficient method for differentiation of human induced
pluripotent stem cells into hepatocyte-like cells retaining drug
metabolizing activity. Drug Metab Pharmacokinet. 29:237–243. 2014.
View Article : Google Scholar
|
|
22
|
Cameron K, Tan R, Schmidt-Heck W, Campos
G, Lyall MJ, Wang Y, Lucendo-Villarin B, Szkolnicka D, Bates N,
Kimber SJ, et al: Recombinant laminins drive the differentiation
and self-organization of hESC-derived hepatocytes. Stem Cell
Reports. 5:1250–1262. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Takayama K, Inamura M, Kawabata K,
Katayama K, Higuchi M, Tashiro K, Nonaka A, Sakurai F, Hayakawa T,
Furue MK, et al: Efficient generation of functional hepatocytes
from human embryonic stem cells and induced pluripotent stem cells
by HNF4alpha transduction. Mol Ther. 20:127–137. 2012. View Article : Google Scholar
|
|
24
|
Watanabe H, Takayama K, Inamura M,
Tachibana M, Mimura N, Katayama K, Tashiro K, Nagamoto Y, Sakurai
F, Kawabata K, et al: HHEX promotes hepatic-lineage specification
through the negative regulation of eomesodermin. PLoS One.
9:e907912014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tomizawa M, Shinozaki F, Motoyoshi Y,
Sugiyama T, Yamamoto S and Ishige N: Transcription factors and
medium suitable for initiating the differentiation of human induced
pluripotent stem cells to the hepatocyte lineage. J Cell Biochem.
117:2001–2009. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vinken M, Papeleu P, Snykers S, De Rop E,
Henkens T, Chipman JK, Rogiers V and Vanhaecke T: Involvement of
cell junctions in hepatocyte culture functionality. Crit Rev
Toxicol. 36:299–318. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ramasamy TS, Yu JS, Selden C, Hodgson H
and Cui W: Application of three-dimensional culture conditions to
human embryonic stem cell-derived definitive endoderm cells
enhances hepatocyte differentiation and functionality. Tissue Eng
Part A. 19:360–367. 2013. View Article : Google Scholar
|
|
28
|
Sivertsson L, Synnergren J, Jensen J,
Björquist P and Ingelman-Sundberg M: Hepatic differentiation and
maturation of human embryonic stem cells cultured in a perfused
three-dimensional bioreactor. Stem Cells Dev. 22:581–594. 2013.
View Article : Google Scholar :
|
|
29
|
Gieseck RL III, Hannan NR, Bort R, Hanley
NA, Drake RA, Cameron GW, Wynn TA and Vallier L: Maturation of
induced pluripotent stem cell derived hepatocytes by 3D-culture.
PLoS One. 9:e863722014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Takebe T, Sekine K, Enomura M, Koike H,
Kimura M, Ogaeri T, Zhang RR, Ueno Y, Zheng YW, Koike N, et al:
Vascularized and functional human liver from an iPSC-derived organ
bud transplant. Nature. 499:481–484. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zeilinger K, Schreiter T, Darnell M,
Söderdahl T, Lübberstedt M, Dillner B, Knobeloch D, Nüssler AK,
Gerlach JC and Andersson TB: Scaling down of a clinical
three-dimensional perfusion multicompartment hollow fiber liver
bioreactor developed for extracorporeal liver support to an
analytical scale device useful for hepatic pharmacological in vitro
studies. Tissue Eng Part C Methods. 17:549–556. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
van de Bunt M, Lako M, Barrett A, Gloyn
AL, Hansson M, McCarthy MI, Beer NL and Honoré C: Insights into
islet development and biology through characterization of a human
iPSC-derived endocrine pancreas model. Islets. 8:83–95. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Friedrich J, Seidel C, Ebner R and
Kunz-Schughart LA: Spheroid- based drug screen: Considerations and
practical approach. Nat Protoc. 4:309–324. 2009. View Article : Google Scholar
|
|
34
|
Pfeiffer E, Kegel V, Zeilinger K,
Hengstler JG, Nüssler AK, Seehofer D and Damm G: Featured article:
Isolation, characterization, and cultivation of human hepatocytes
and non-parenchymal liver cells. Exp Biol Med (Maywood).
240:645–656. 2015. View Article : Google Scholar
|
|
35
|
Brzeszczyńska J, Johns N, Schilb A, Degen
S, Degen M, Langen R, Schols A, Glass DJ, Roubenoff R, Greig CA, et
al: Loss of oxidative defense and potential blockade of satellite
cell maturation in the skeletal muscle of patients with cancer but
not in the healthy elderly. Aging (Albany NY). 8:1690–1702. 2016.
View Article : Google Scholar
|
|
36
|
Vandesompele J, De Preter K, Pattyn F,
Poppe B, Van Roy N, De Paepe A and Speleman F: Accurate
normalization of real-time quantitative RT-PCR data by geometric
averaging of multiple internal control genes. Genome Biol.
3:RESEARCH00342002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
38
|
Liu J, Brzeszczynska J, Samuel K, Black J,
Palakkan A, Anderson RA, Gallagher R and Ross JA: Efficient
episomal reprogramming of blood mononuclear cells and
differentiation to hepatocytes with functional drug metabolism. Exp
Cell Res. 338:203–213. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Freyer N, Knöspel F, Strahl N, Amini L,
Schrade P, Bachmann S, Damm G, Seehofer D, Jacobs F, Monshouwer M,
et al: Hepatic differentiation of human induced pluripotent stem
cells in a perfused three-dimensional multicompartment bioreactor.
Biores Open Access. 5:235–248. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hoffmann SA, Müller-Vieira U, Biemel K,
Knobeloch D, Heydel S, Lübberstedt M, Nüssler AK, Andersson TB,
Gerlach JC and Zeilinger K: Analysis of drug metabolism activities
in a miniaturized liver cell bioreactor for use in pharmacological
studies. Biotechnol Bioeng. 109:3172–3181. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schyschka L, Sánchez JJ, Wang Z, Burkhardt
B, Müller-Vieira U, Zeilinger K, Bachmann A, Nadalin S, Damm G and
Nussler AK: Hepatic 3D cultures but not 2D cultures preserve
specific transporter activity for acetaminophen-induced
hepatotoxicity. Arch Toxicol. 87:1581–1593. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rennert K, Steinborn S, Gröger M,
Ungerböck B, Jank AM, Ehgartner J, Nietzsche S, Dinger J, Kiehntopf
M, Funke H, et al: A microfluidically perfused three dimensional
human liver model. Biomaterials. 71:119–131. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Varum S, Rodrigues AS, Moura MB,
Momcilovic O, Easley CA IV, Ramalho-Santos J, Van Houten B and
Schatten G: Energy metabolism in human pluripotent stem cells and
their differentiated counterparts. PLoS One. 6:e209142011.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tirona RG, Lee W, Leake BF, Lan LB, Cline
CB, Lamba V, Parviz F, Duncan SA, Inoue Y, Gonzalez FJ, et al: The
orphan nuclear receptor HNF4α determines PXR- and CAR-mediated
xenobiotic induction of CYP3A4. Nat Med. 9:220–224. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Miki T, Ring A and Gerlach J: Hepatic
differentiation of human embryonic stem cells is promoted by
three-dimensional dynamic perfusion culture conditions. Tissue Eng
Part C Methods. 17:557–568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
De Assuncao TM, Sun Y, Jalan-Sakrikar N,
Drinane MC, Huang BQ, Li Y, Davila JI, Wang R, O'Hara SP, Lomberk
GA, et al: Development and characterization of human-induced
pluripotent stem cell-derived cholangiocytes. Lab Invest.
95:684–696. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Knöspel F, Jacobs F, Freyer N, Damm G, De
Bondt A, van den Wyngaert I, Snoeys J, Monshouwer M, Richter M,
Strahl N, et al: In vitro model for hepatotoxicity studies based on
primary human hepatocyte cultivation in a perfused 3D bioreactor
system. Int J Mol Sci. 17:5842016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Singh M, Berkland C and Detamore MS:
Strategies and applications for incorporating physical and chemical
signal gradients in tissue engineering. Tissue Eng Part B Rev.
14:341–366. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Uzel SG, Amadi OC, Pearl TM, Lee RT, So PT
and Kamm RD: Simultaneous or sequential orthogonal gradient
formation in a 3D cell culture microfluidic platform. Small.
12:612–622. 2016. View Article : Google Scholar :
|
|
51
|
Takayama K, Kawabata K, Nagamoto Y,
Kishimoto K, Tashiro K, Sakurai F, Tachibana M, Kanda K, Hayakawa
T, Furue MK, et al: 3D spheroid culture of hESC/hiPSC-derived
hepatocyte-like cells for drug toxicity testing. Biomaterials.
34:1781–1789. 2013. View Article : Google Scholar
|
|
52
|
Ginsberg G, Hattis D, Sonawane B, Russ A,
Banati P, Kozlak M, Smolenski S and Goble R: Evaluation of
child/adult pharmacokinetic differences from a database derived
from the therapeutic drug literature. Toxicol Sci. 66:185–200.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Goldring C, Antoine DJ, Bonner F, Crozier
J, Denning C, Fontana RJ, Hanley NA, Hay DC, Ingelman-Sundberg M,
Juhila S, et al: Stem cell-derived models to improve mechanistic
understanding and prediction of human drug-induced liver injury.
Hepatology. 65:710–721. 2017. View Article : Google Scholar
|