Introduction
Medical advances in recent years have m\ade kidney
transplantation one of the most effective treatments for patients
with end-stage kidney diseases (1). Optimized surgical techniques and the
development of immunosuppressive therapy have resulted in improved
long-term outcomes for patients following transplantation (1–3).
However, these therapies may be impeded by a variety of different
factors, including drug doses, adverse side effects and the unique
and dynamic immune status of the individual, which leads to certain
patients suffering from acute allograft rejection (4,5).
Acute rejection, which is caused by the development of cellular
immunity following transplantation, has a strong negative
prognostic effect on the chances of allograft survival (6,7).
Multiple processes are involved in acute rejection, including
innate T-cell mediated rejection and adaptive antibody mediated
rejection (8). In addition, the
etiology of acute rejection is complicated and involves
interactions between renal hemodynamics, inflammatory responses and
molecular regulatory factors (9).
The specifics of these interactions and the molecular mechanisms by
which they occur are largely unknown.
MicroRNAs (miRNAs/miRs) are a class of ubiquitously
distributed, endogenous, non-coding single-stranded RNAs (10). They are very short 19–25
nucleotide sequences that regulate gene expression by
sequence-specific base pairing with the 3′-untranslated region
(UTR) of their mRNA target, causing the degradation or
translational repression of the mRNA (11). It has been determined that miRNAs
are associated with a variety of cellular processes, including
proliferation, differentiation, migration, growth, apoptosis and
development (12) The aberrant
expression of certain miRNAs is associated with multiple
pathological conditions, including heart disease (13), kidney disease (14) and various types of cancer
(15). Increasing evidence
suggests that miRNAs serve critical roles in the regulation of
innate and adaptive immune responses (16,17). In addition, previous studies have
reported that miRNA regulation is associated with organ
transplantation and acute allograft rejection, and that miRNAs may
be candidate biomarkers for the diagnosis of acute allograft
rejection (18–21). An improved understanding of the
multifunctional roles of miRNAs in the pathogenesis of acute
rejection is crucial for the development of novel diagnostic tools
and therapeutic strategies.
miR-650 has been reported to be upregulated in
different types of cancer, including gastric cancer (22), lung adenocarcinoma (23) and hepatocellular carcinoma
(24). miR-650 has been revealed
to contribute to tumor cell proliferation and apoptosis suppression
through interactions with upstream regulatory proteins. Zuo et
al (25) determined that
miR-650 activity was significantly correlated with the
downregulation of the tumor suppressor phosphatidylinositol
transfer protein CSR1. It has also been demonstrated that p16INK4a
induces the expression of miR-650 in MCF7 cells and that miR-650
may downregulate cyclin dependent kinase 1 (CDK1) by pairing with
the CDK1 3′-UTR (26). However,
to the best of our knowledge miR-650 has not yet been investigated
in acute allograft rejection.
The present study revealed that an increasing level
of miR-650 was associated with the downregulation of B-cell
CLL/lymphoma 11B (BCL11B) gene expression in acute renal allograft
rejection. In vitro study using human renal glomerular
endothelial cells (HRGECs) identified key events in acute allograft
rejection following transfection with miR-650 mimics, whereas the
opposite effects were observed in HRGECs transfected with a miR-650
inhibitor. The existence of a conserved miR-650 binding site on the
3′-UTR of BCL11B mRNA was predicted by computational algorithms and
confirmed by a luciferase reporter assay. Furthermore, the
knockdown of BCL11B using BCL11B-specific small interfering RNA
(siRNA) significantly decreased the apoptosis rate of HRGECs. The
results of the present study highlight a potential novel diagnostic
marker for acute renal allograft rejection and provide novel
therapeutic strategies for its treatment.
Materials and methods
Patient recruitment and sample
collection
Serum samples were collected from 29 recipients of
renal transplants who underwent surgery at Xiangya Hospital
(Central South University, Changsha, China) between March, 2014 and
January, 2016. The serum samples were subsequently divided into two
groups as follows: i) Transplantation patients with acute rejection
(acute rejection group; n=19); and ii) transplantation patients
with continuous stable kidney function (control group; n=10). The
samples collected from patients with acute rejection were obtained
within 24 h of their admission to hospital, the serum creatinine
level was measured and they were subsequently confirmed as
suffering from acute allograft rejection. Interpretation of the
biopsy results was performed according to the Updated Banff 07
criteria by a qualified physician (27). All tissue samples were obtained
from anonymized excess tissue, which was not required for
diagnostic or clinical purposes. The present study was approved by
the Ethics Committee of Xiangya Hospital of Central South
University. Written informed consent was obtained from all
participants. The demographic and clinical characteristics of all
patients included in the present study are detailed in Table I.
 | Table IDemographic and clinical
characteristics of renal allograft recipients. |
Table I
Demographic and clinical
characteristics of renal allograft recipients.
| Characteristic | Group
|
|---|
| No rejection | Acute
rejection |
|---|
| No. of
patients | 19 | 10 |
| Male, no. (%) | 12 (63) | 8 (80) |
| Female, no.
(%) | 7 (37) | 2 (20) |
| Age, year
(range) | 41.5 (25–65) | 38.7 (22–54) |
| Type of
allograft | – | – |
| Deceased donor | 19 | 9 |
| Living donor | 0 | 1 |
| Type of donor | – | – |
| Related | 0 | 1 |
| Unrelated | 19 | 9 |
| Data from
ultrasound | – | – |
| Increased
interlobar arteries RI | 0 | 7 |
| Normal interlobar
arteries RI | 19 | 3 |
| Urine volume (ml/24
h), mean ± SD | 2124.8±22.2 | 1790.6±24.7 |
| Blood creatinine
level (μmol/l), mean ± SD | 223.6±16.3 | 568.2±20.3 |
Hematoxylin and eosin (H&E)
staining
Histological sections (5-μm-thick) were
perfused with 4% paraformaldehyde at 4°C for 6 h and stained with
H&E (Sigma-Aldrich; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) for 5 min at room temperature. The results were observed
under an inverted microscope (Ix81; Olympus Corp., Tokyo, Japan) at
a magnification of ×20.
Immunohistochemistry
The sections were embedded in paraffin and sectioned
(5-μm-thick). Subsequently, the sections were perfused with
4% paraformaldehyde at 4°C for 6 h, then placed onto Poly-Prep
Slides (Sigma-Aldrich; Thermo Fisher Scientific, Inc.) and dried at
60°C for 1 h. The sections were deparaffinized in xylene (three
times, 5 min each) at room temperature and hydrated in 100% ethanol
followed by 95% ethanol. Then, the sections were treated with
sodium citrate (10 mM) at 95°C for 10 min and placed at room
temperature for 30 min. Expression levels were then detected using
a Vectastain Universal elite ABC kit (1:50; cat. no. PK-6200;
Vector Laboratories, Inc., Burlingame, CA, USA). The sections were
blocked with 5% normal horse serum (Dako; Agilent Technologies,
Inc., Santa Clara, CA, USA) for 30 min at room temperature and
incubated with anti-BCL11B antibodies (1:500; cat. no. ab18465;
Abcam, Cambridge, UK) at 4°C overnight. Next, the sections were
incubated with goat anti-rat IgG H&L (phycoerythrin) secondary
antibodies (1:1,000; cat. no. ab7010; Abcam) for 15 min at room
temperature. Subsequently, the sections were treated with a
3,3′-diamino-benzidine kit (Signet Laboratories, Inc., Dedham, MA,
USA). The nuclei were stained with hematoxylin (cat. no. 790-2208;
Ventana Medical Systems, Inc., Tucson, AZ, USA). The results were
observed under an Ix81 inverted microscope (magnification,
×20).
Cell culture
HRGECs (cat. no. 4000) were cultured in endothelial
cell medium (ECM) (both from ScienCell Research Laboratories, Inc.,
San Diego, CA, USA) with 10% fetal bovine serum (FBS; cat. no.
SH30071.03; HyClone; GE Healthcare Life Sciences, Logan, UT, USA),
100 U/ml penicillin, 100 mg/ml streptomycin and 2 mM glutamine
(cat. no. 11090-081) (both from Thermo Fisher Scientific, Inc.).
The HRGECs were incubated in an atmosphere with 5% CO2
at 37°C. Once they reached 70–80% confluence, the cells were split
according to standard procedures. Following the experimental
procedures, the cells were harvested in Dulbecco's modified Eagle's
medium (cat. no. 08459-35; Nacalai Tesque, Inc., Kyoto, Japan) with
1% FBS in preparation for biochemical analyses.
Immunofluorescence staining
Cells were fixed with 4% paraformaldehyde for 10 min
at room temperature, and subsequently blocked with 5% FBS
containing 0.5% Triton x-100 for 5 min at room temperature. The
HRGECs were then incubated overnight at 4°C with primary antibodies
directed against BCL11B (1:500; cat. no. ab18465; Abcam) and then
with Alexa Fluor® 488-conjugated goat anti-rabbit IgG
(cat. no. A-11034; Thermo Fisher Scientific, Inc.) for 1 h at room
temperature. Cell slides were mounted with mounting buffer
containing DAPI and immunofluorescence was observed under a
fluorescence microscope (magnification, ×10).
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol reagent (cat.
no. 15596-026; Invitrogen; Thermo Fisher Scientific, Inc.) from
acute rejection and normal allograft tissues, and the transfected
HRGECs, according to the manufacturer's protocol. First-strand cDNA
was synthesized from 1 μg of total RNA using the RevertAid
First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. qPCR was then performed
with the KAPA SYBR FAST qPCR kit (cat. no. KK4601; Kapa Biosystems,
Inc., Wilmington, MA, USA) on a real-time PCR system. The reaction
was performed in a 20 μl volume and the thermocycling
conditions were as follows: 95°C for 10 min; 38 cycles of 95°C for
10 sec, 60°C for 2 min and 72°C for 2 min; and then 72°C for 10
min. Each assay was performed in triplicate and β-actin or U6 was
used as the endogenous control gene. The primer sequences used were
as follows: BCL11B forward, 5′-TGCCAGTGTCAGTTGTCAGG-3′ and reverse,
5′-CCAGGTAGATGCGGAAGC-3′; miR-650 forward,
5′-ACACTCCAGCTGGGAGGAGGCAGCGCTCT-3′ and reverse,
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGTCCTG-3′; U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′;
and β-actin forward, 5′-ATCGTGCGTGACATTAAGGAGAAG-3′ and reverse,
5′-AGGAAGGAAGGCTGGAAGAGTG-3′. The relative amount of miR-650 and
BCL11B was calculated using the 2−ΔΔCq method with U6
and β-actin as the controls, respectively (28).
miR-650 mimic, inhibitor and mock
transfection
An miR-650 mimic, inhibitor and mock (scrambled
control) were chemically synthesized by Shanghai GenePharma Co.,
Ltd. (Shanghai, China). The sequences for the miR-650 mimic,
inhibitor and mock were 5′-AGGAGGCAGCGCUCUCAGGAC-3′,
5′-GUCCUGAGAGCGCUGCCUCCU-3′ and 5′-UUCUCCGAACGU GUCACGUTT-3′,
respectively. Cells (5×104 cells/well) were seeded in
6-well plates in ECM without antibiotics. The miR-650 mimic,
inhibitor and mock (50 nM) were transfected into HRGECs at 80%
confluence using Lipofectamine™ 3000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
protocol.
siRNA transfection
The BCL11B-specific siRNA and a non-silencing
negative control (NC) siRNA were chemically synthesized by Shanghai
GenePharma Co., Ltd. The BCL11B targeting sequence was
5′-CCUGGAGAAACACAUGAA ATT-3′ (sense). The sequence of the control
siRNA was 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense). HRGECs
(2×104 cells/well) were transfected with BCL11B-sepcific
siRNA (50 μM) or NC siRNA (50 μM) using Lipofectamine
3000 reagent for 24 h according to the manufacturer's
protocols.
Determination of cell proliferation using
the cell counting kit-8 (CCK-8) assay
The proliferation assay was performed using a CCK-8
assay (cat. no. CK04; Dojindo Molecular Technologies, Inc.,
Shanghai, China) according to the manufacturer's protocol to
evaluate cell proliferation. Cells were seeded at a density of
5×104 cells/well in a 96-well plate. At each time point
(0, 24, 48 and 72 h) CCK-8 solution (15 μl) was added to
each well, and the cells were further incubated for 2 h at 37°C.
The absorbance of samples at 450 nm was then determined using a
microplate plate reader.
Determination of apoptosis by flow
cytometry
The rate of cell apoptosis was determined using an
Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI)
apoptosis detection kit (BioVision, Inc., Milpitas, CA, USA).
HRGECs were detached with trypsin-EDTA and washed with
phosphate-buffered saline (PBS). They were subsequently resuspended
in a binding buffer [10 mM HEPES (pH 7.4); 150 mM NaCl; 5 mM KCl; 1
mM MgCl2; 1.8 mM CaCl2] containing Annexin
V-FITC (1 g/ml) and further incubated for 20 min at the room
temperature. At 10 min prior to the end of incubation, PI (10 g/ml)
was added to the cell suspension to stain necrotic cells. The cells
were then analyzed via fluorescence-activated cell sorting using a
FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA)
equipped with a 488 nm excitation laser, and analyzed using ModFit
LT 2.0 software (Verity Software House, Topsham, ME, USA).
Cell cycle analysis using flow
cytometry
Cells were harvested and washed with cold PBS, and
subsequently fixed with 70% ethanol overnight at −20°C. The fixed
cells were then washed with cold PBS, centrifuged (1,000 × g for 10
min at 4°C) and the supernatant was discarded. The cells were
stained with a PI solution (10 μg/ml RNase A; 50
μg/ml PI) at 37°C for 30 min in the dark. The cell cycle
distribution was then analyzed using a flow cytometer with
CellQuest 3.0 software (BD Biosciences).
Cytokine and chemokine enzyme-linked
immunosorbent assay (ELISA)
The quantification of cytokines and chemokines were
evaluated using ELISA kits, as described below (eBioscience; Thermo
Fisher Scientific, Inc.). The levels of pro-inflammatory cytokines
and chemokines were analyzed 48 h post-transfection. The
homogenates were sonicated for 20 sec at 400 W at 4°C and
centrifuged at 13,000 × g for 10 min at 4°C. The total protein
concentration was measured using a DC™ protein assay (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) prior to quantification of
interleukin-2 (IL-2) (cat. no. BMS221-2), IL-9 (cat. no.
88-7958-88), IL-13 (cat. no. 88-7439-88), interferon-γ (IFN-γ)
(cat. no. KHC3014) and chemokine (C-C motif) ligand 5 (cat. no.
BMS287-2INST) by ELISA according to the manufacturer's protocol.
Data was expressed as pg/mg of total protein.
Macrophage chemotactic assay
A macrophage chemotactic assay was performed using
Transwell cell migration chambers with 5 μm-pore inserts
(Cell Biolabs, Inc., San Diego, CA, USA) in a 24-well plate. The
lower chambers contained ECM that the HRGECs were cultured in for
48 h after transfection with the miR-650 mimic, inhibitor or mock
as a chemotactic stimulus. A total of 3×105 (10,000
cells/insert) THP-1 macrophages (cat. no. AA-CELL-63; American Type
Culture Collection, Manassas, VA, USA) were placed into the upper
chambers in a serum-free RPMI-1640 (cat. no. 430-1800EG; Gibco;
Thermo Fisher Scientific, Inc.). The percentage of cells that
migrated to the lower chamber was determined by Invitrogen CyQuant™
GR (Thermo Fisher Scientific, Inc.) staining for 20 min at 37°C.
Fluorescence was measured with a Victor 1420 Multilabel Counter
microplate reader (Wallac, Turku, Finland) at 480 and 520 nm, and
analyzed by ImageJ (version 1.45s; National Institutes of Health,
Bethesda, MD, USA) and 3D Slicer (version 4.3; slicer.org) software.
Western blot analysis
For western blot analysis, acute rejection and
normal allograft samples were sonicated for 5 sec on ice twice at
100 W in 50 mM lysis buffer [3.1 mM sucrose (pH 7.4); 1 mM
dithiothreitol, 10 μg/ml leupeptin; 10 μg/ml soybean
trypsin inhibitor; 2 μg/ml aprotinin; 0.1% Triton x-100].
Homogenates were centrifuged at 10,000 × g at 4°C for 20 min and
the supernatants were collected. The total protein concentration
was measured using the Bradford protein assay (Bio-Rad
Laboratories, Inc.). Protein lysates (30 μg/lane) were
separated using 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and transferred to polyvinylidene
difluoride membranes. Following blocking with 1% bovine serum
albumin (BSA; cat. no. EQBAH62; Europa Bioproducts, Ltd.,
Cambridge, UK) for 1.5 h at room temperature, the membranes were
incubated overnight with monoclonal primary antibodies directed
against caspase-8 (cat. no. ab32397; Abcam) and BCL11B (cat. no.
ab28448) [both Abcam and diluted 1:1,000 in PBS-Tween-20 (PBS-T)
with 1% BSA] at 4°C overnight. The membranes were then washed in
PBS-T three times (10 min/wash) and probed with the appropriate
secondary antibody (1:2,000; cat. no. ab6721; Abcam) for 1 h at
room temperature. The membranes were developed using enhanced
chemiluminescence (Immun-Star HRP Chemiluminescent kit; Bio-Rad
Laboratories, Inc.), and the band densities were measured with a
Versa Doc™ MP 5000 molecular imager and Quantity One software
(version 4.6) (both Bio-Rad Laboratories, Inc.). Equal protein
loading was verified by measurement of the glyceraldehyde
3-phosphate dehydrogenase (GAPDH) level with a mouse monoclonal
antibody (1:2,000; cat. no. ab8245; Abcam).
miR-650 target prediction and
verification by luciferase assay
The putative targets of miR-650 were predicted using
TargetScan (version 6.2; targetscan.org), miRBase (version 21; miRBase.org), PicTar (version 4.0.24;
pictar.mdcberlin.de) and miRanda (version 3.3a; microrna.org). The human BCL11B wild-type and mutant
3′-UTR reporter vectors were constructed by inserting annealed
oligonucleotides with flanking restriction sites into a pmirGLO
Dual-Luciferase miRNA Target Expression Vector (Promega Corp.,
Madison, WI, USA). A total of 3×105 cells were seeded in
a 24-well plate and co-transfected with the wild-type or mutant
BCL11B vectors and the miR-650 mimic using Lipofectamine 3000
(Thermo Fisher Scientific, Inc.). Firefly luciferase and
Renilla luciferase signals were measured after 48 h using
the Dual-luciferase assay reporter kit (cat. no. E1910; Promega
Corp.) and quantified using a Lumat LB 9501 luminator. A
Renilla plasmid was used as an internal reference and
Firefly luciferase activity was normalized to Renilla
luciferase.
Statistical analysis
Statistical calculations were performed using
GraphPad Prism software (version 6; GraphPad Software, Inc., La
Jolla, CA, USA). Data are presented as the mean ± standard error of
the mean. A Student's t-test was used for comparisons between two
groups and multiple-factor analysis was used was used for the
comparisons of multiple groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
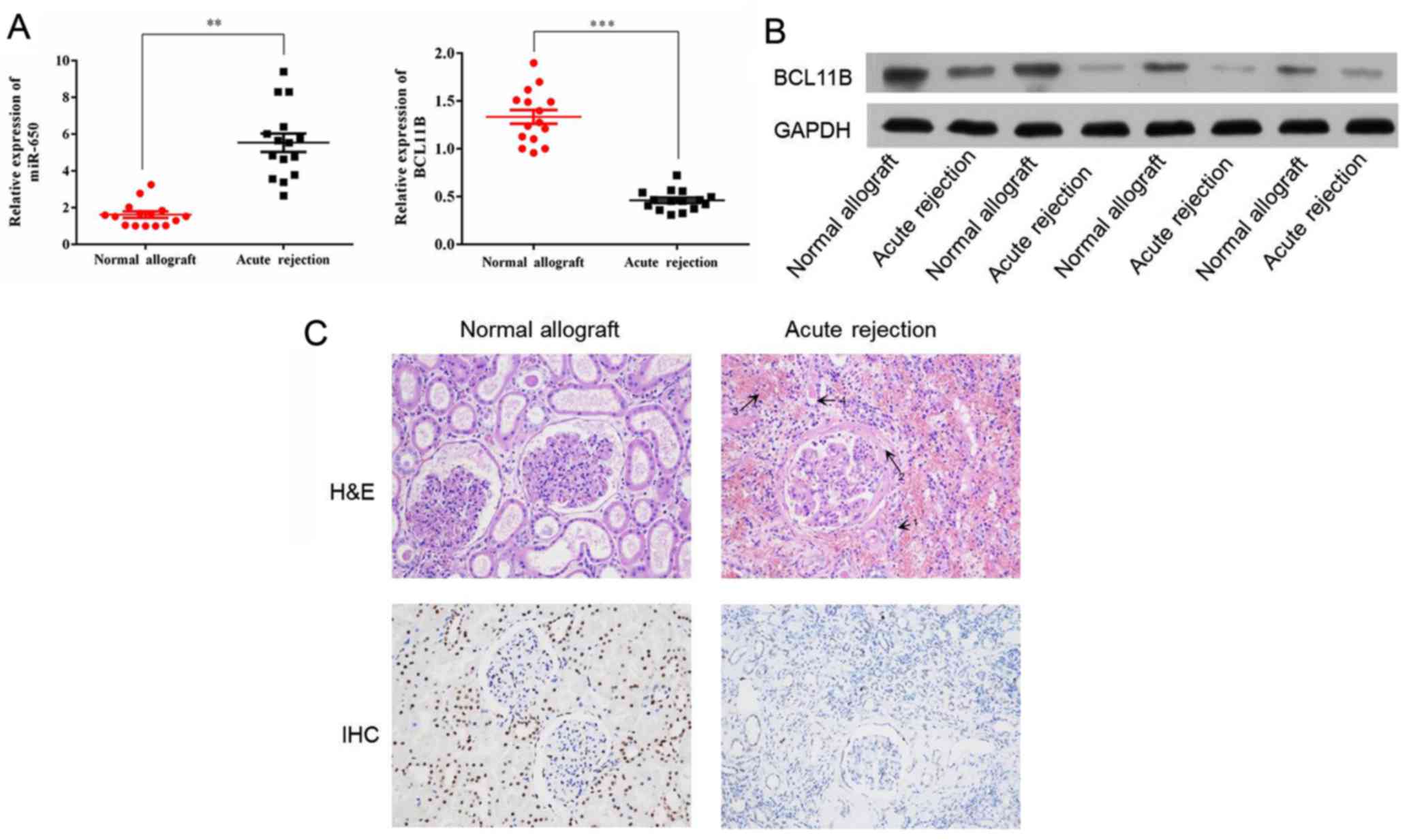
miR-650 expression is increased and
BCL11B expression is decreased in acute kidney rejection
The expression of miR-650 has been evaluated in
different types of cancer (29);
however, to the best of our knowledge, it has not yet been
investigated in acute allograft rejection after kidney
transplantation. The level of miR-650 expression in the acute
rejection group compared with the control group was determined
using RT-qPCR. Significant upregulation of miR-650 was identified
in the acute rejection group in comparison with the control group
(Fig. 1A). The protein expression
of BCL11B was evaluated by western blot analysis. This indicated
that BCL11B expression was notably lower in the acute rejection
group compared with the control group (Fig. 1B). RT-qPCR reported similar
results in regards to BCL11B mRNA expression, which was
significantly lower in the acute rejection group compared with the
control group (Fig. 1A).
Histological features of acute
rejection
Histological sections from the patients were stained
with H&E in order to determine the main histologic features in
the acute rejection group compared with the control group (Fig. 1C). These included margination of
neutrophils in peritubular capillaries, arterial fibrinoid
necrosis, thrombotic microangiopathy and acute tubular injury.
Furthermore, immunohistochemistry analysis revealed that BCL11B
expression was markedly increased in the acute rejection group in
comparison with the control (Fig.
1C).
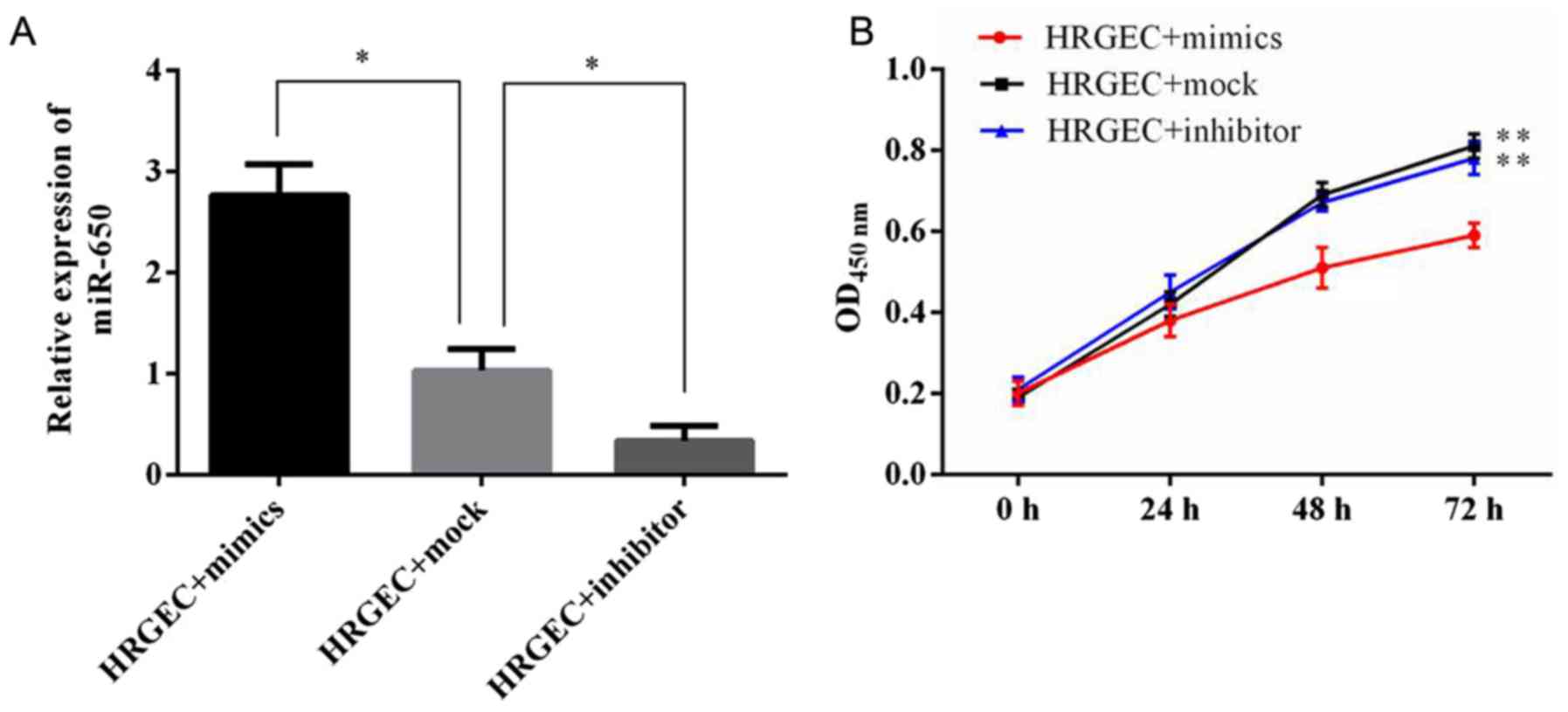
Transfection of the miR-650 inhibitor and
mimic into HRGECs
To explore the functional role of miR-650 in acute
renal rejection, an miR-650 mimic, inhibitor or mock were
transfected into HRGECs. The expression level of miR-650 was then
evaluated using RT-qPCR (Fig.
2A). It was revealed that the miR-650 mimic significantly
increased the level of miR-650, whereas the miR-650 inhibitor
significantly decreased the level of endogenous miR-650, in
comparison with cells transfected with the miR-650 mock.
Upregulation of miR-650 significantly
decreases HRGEC viability and increases apoptosis
The effect of miR-650 on cell viability and
proliferation were determined. The CCK-8 assay demonstrated that 48
h after transfection with the miR-650 mimic cell viability was
significantly decreased compared with cells transfected with the
miR-360 inhibitor or mock (Fig.
2B).
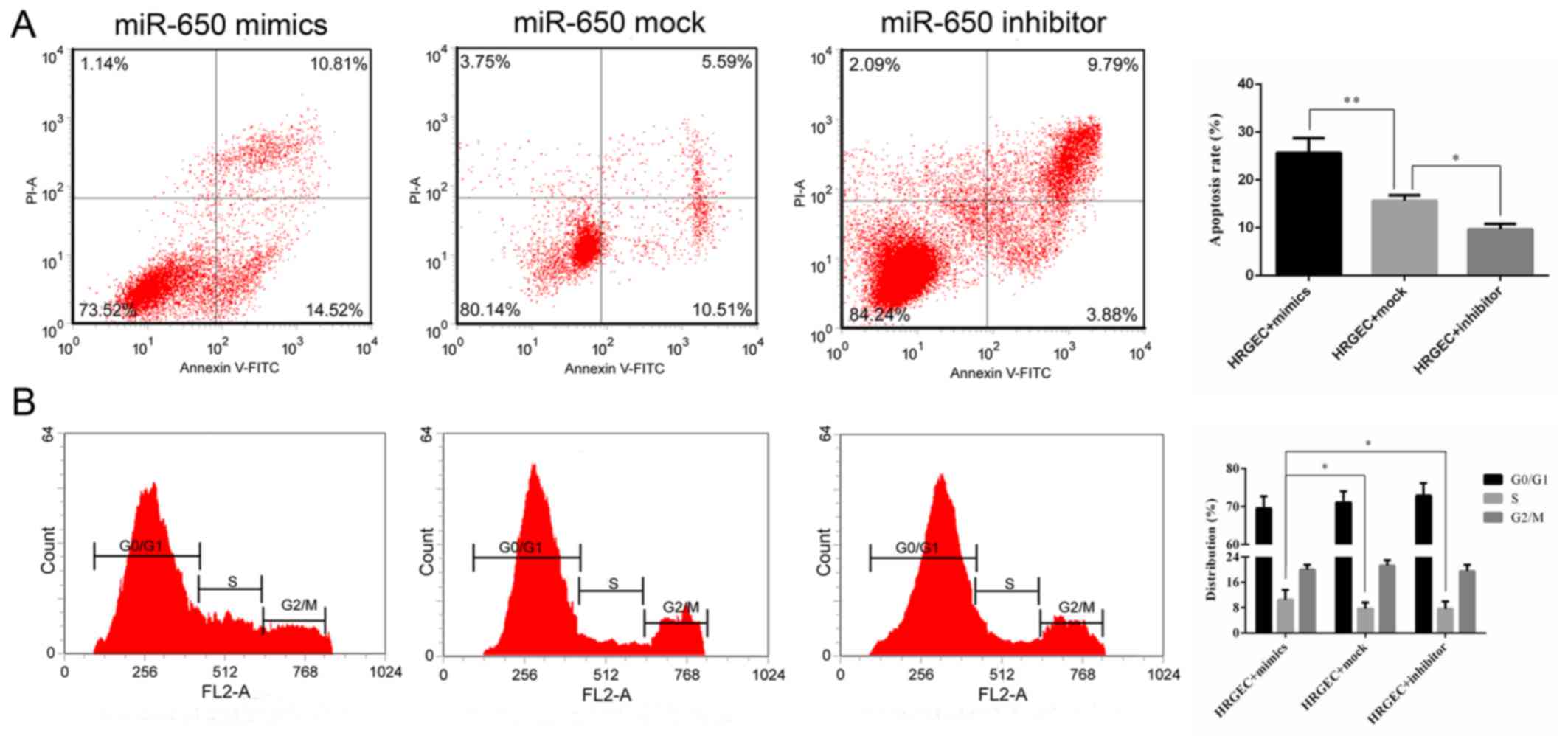
The rate of apoptosis was determined by flow
cytometry. The cell apoptosis rate was significantly increased in
cells transfected with miR-650 mimics compared with the cells
transfected with the miR-650 mock (Fig. 3A). Additionally, the cells
transfected with the miR-650 inhibitor had a significantly lower
apoptosis rate compared with the mock group (Fig. 3A). Cell cycle distribution was
measured by flow cytometry, and it was revealed that the proportion
of cells in S phase was significantly increased in the miR-650
mimic group compared with the mock and inhibitor groups, indicating
that miR-650 inhibited G1 and G2/M phase
arrest to promote cell proliferation (Fig. 3B).
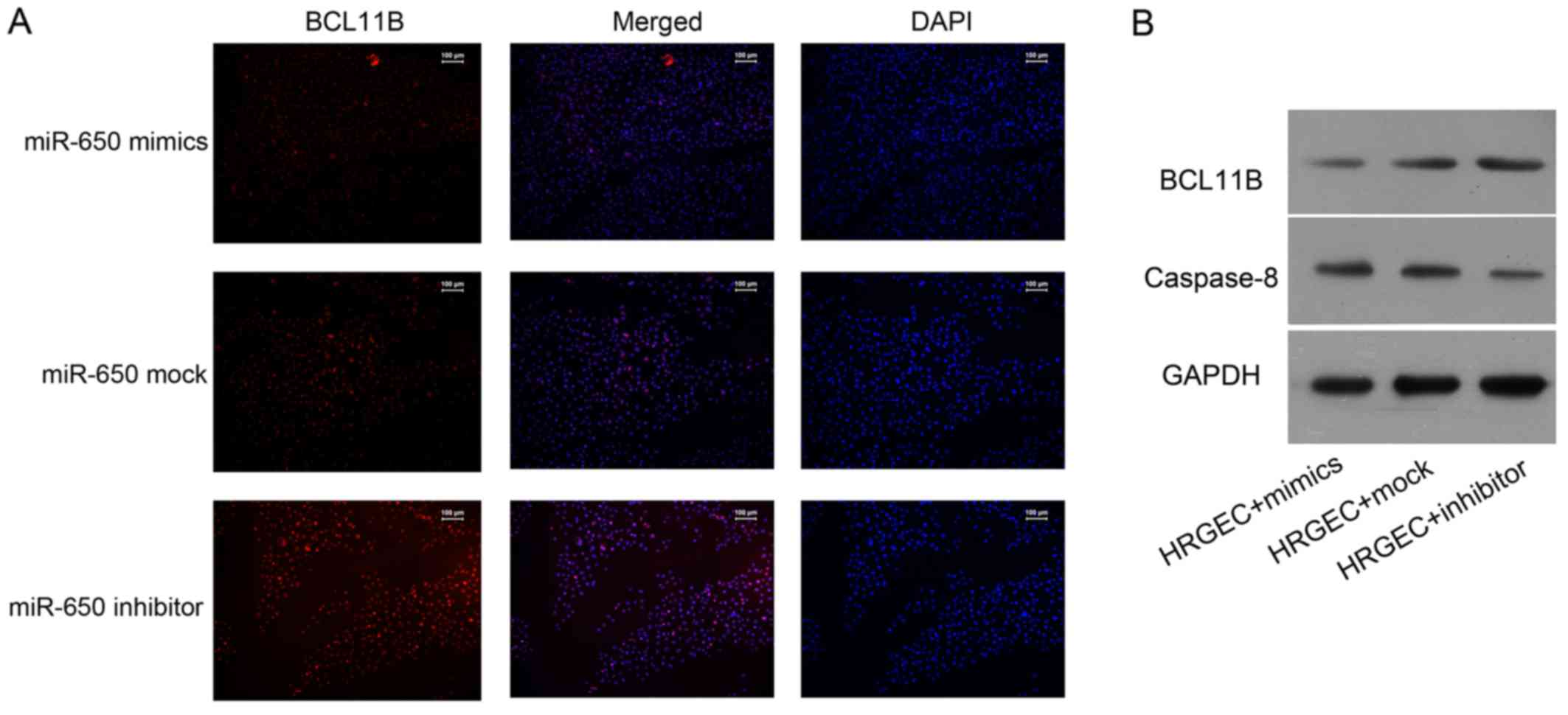
Immunofluorescence analysis revealed that the level
of BCL11B was notably reduced in cells transfected with the miR-650
mimic, whereas it was notably increased in cells transfected with
the miR-650 inhibitor (Fig. 4A).
Western blot analysis demonstrated that the BCL11B protein
expression was markedly higher in cells transfected with the
miR-650 inhibitor compared with cells transfected with the miR-650
mimic (Fig. 4B). By contrast, the
expression of caspase-8 was enhanced in cells transfected with the
miR-650 mimic, while an opposite effect was observed in miR-650
inhibitor-transfected cells (Fig.
4B).
miR-650 regulates the release of
allograft rejection-associated cytokines from HRGECs
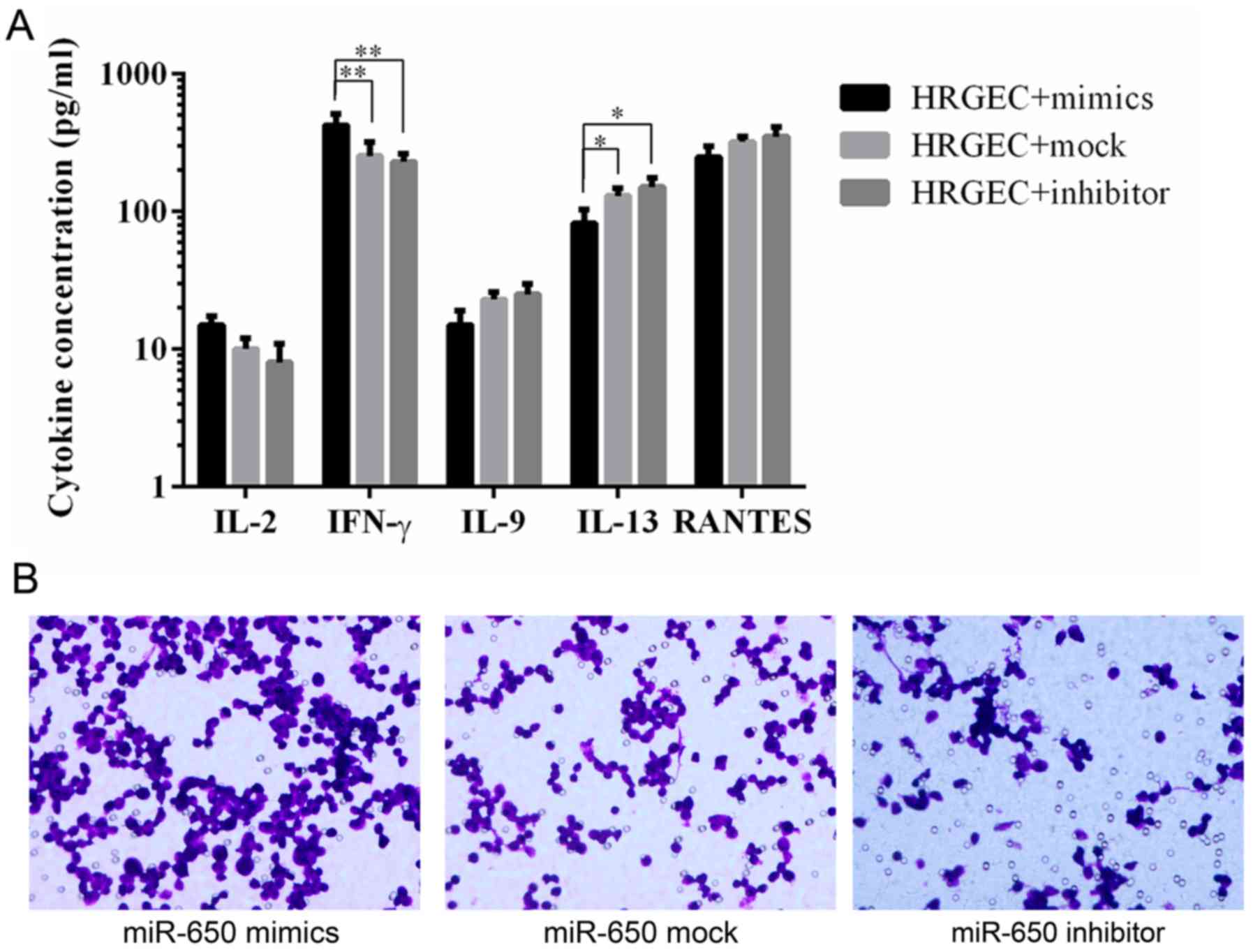
ELISA was used to determine the levels of
pro-inflammatory cytokines and chemokines. The results revealed
that HRGECs transfected with the miR-650 mimic released a
significantly higher amount of IFN-γ and a significantly lower
amount of IL-13 compared with HRGECs transfected with the miR-650
inhibitor and mock (Fig. 5A).
A chemotaxis assay was used to evaluate macrophage
chemotaxis towards a conditioned medium from cells transfected with
the miR-650 mimic, inhibitor or mock. The results revealed that the
conditioned medium from miR-650 inhibitor-transfected cells notably
decreased macrophage migration compared with the miR-650
mock-transfected cells, whereas the medium from miR-650 mimic
transfected-cells had a notably increased level of macrophage
migration compared with the mock transfected cells (Fig. 5B).
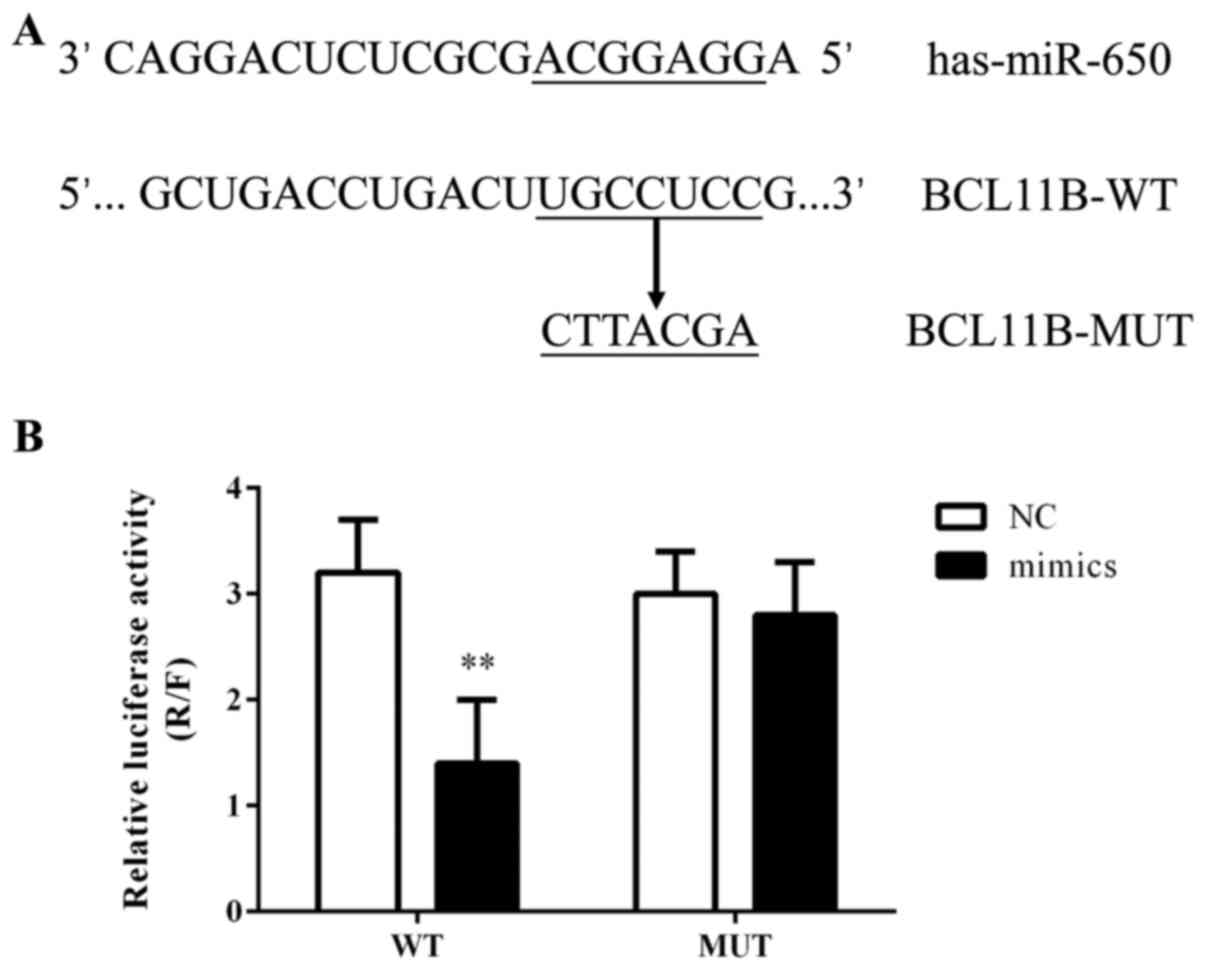
BCL11B as a target of miR-650
The computational prediction software packages
miRBase, TargetScan, PicTar and miRanda were used to identify the
potential binding sites for BCL11B mRNA on miR-650. It was revealed
that the 3′-UTR of BCL11B mRNA possessed the miR-650 binding site
(Fig. 6A) and that this binding
site was conserved in mammals (data not shown). The effect of
miR-650 on the translation of BCL11B mRNA into protein was
evaluated using a luciferase reporter assay. The miR-650 mimic
significantly decreased the luciferase activity of the reporter
gene with a wild-type BCL11B 3′-UTR compared with the NC (Fig. 6B). However, the regulatory effect
of miR-650 was suppressed when the predicted miR-650 binding site
in BCL11B mRNA was mutated.
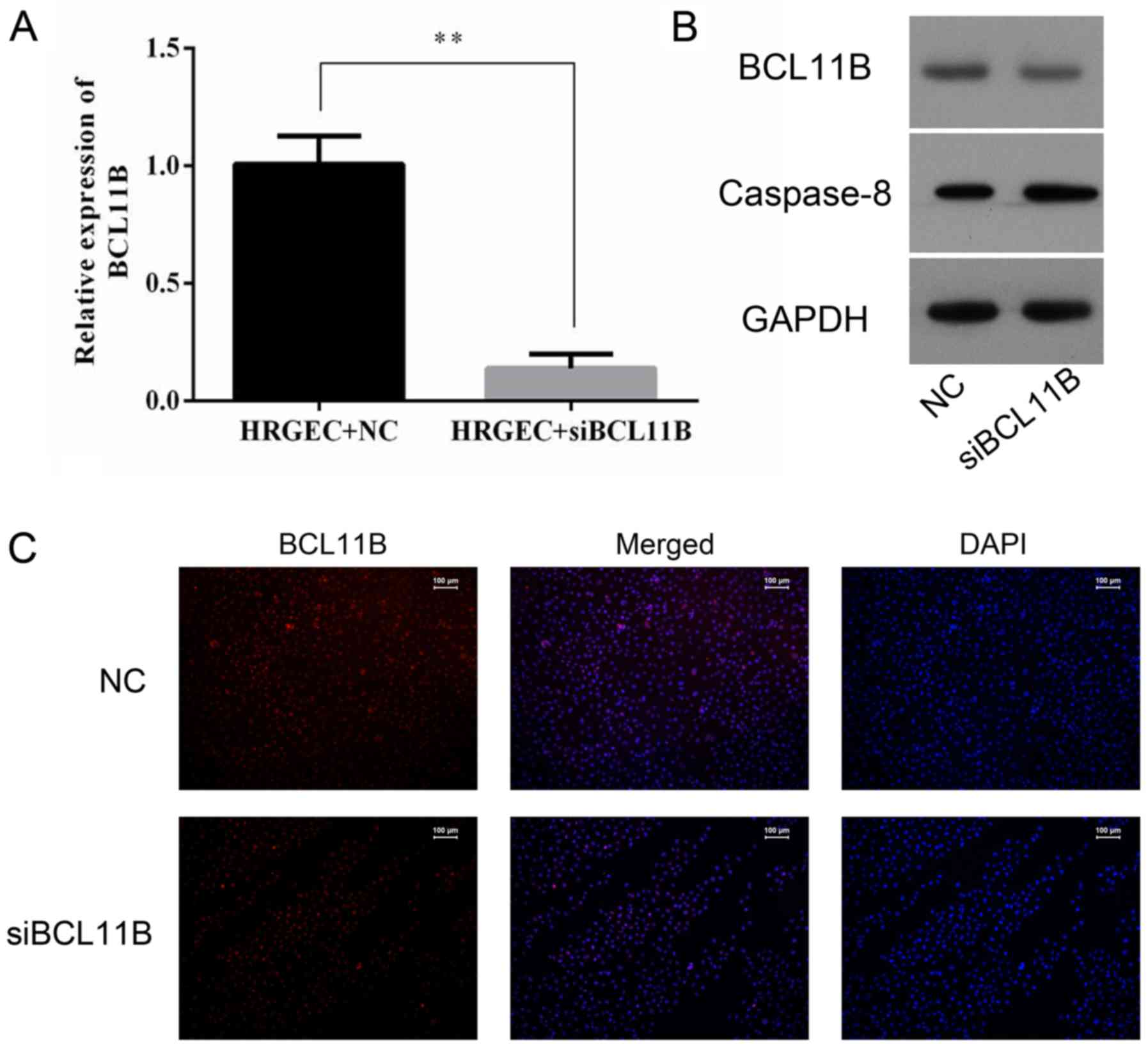
Transfection of BCL11B siRNA
The NC siRNA-transfected cells did not differ in any
evaluation in all experimental groups (data not shown); therefore,
the NC siRNA-transfected cells were used as a control for the
BCL11B siRNA-transfected cells in all siRNA experiments. Knockdown
of BCL11B expression by BCL11B siRNA was evaluated by RT-qPCR,
western blot analysis and immunofluorescence. The level of BCL11B
in the BCL11B siRNA-transfected cells was significantly reduced
compared with the NC cells (Fig.
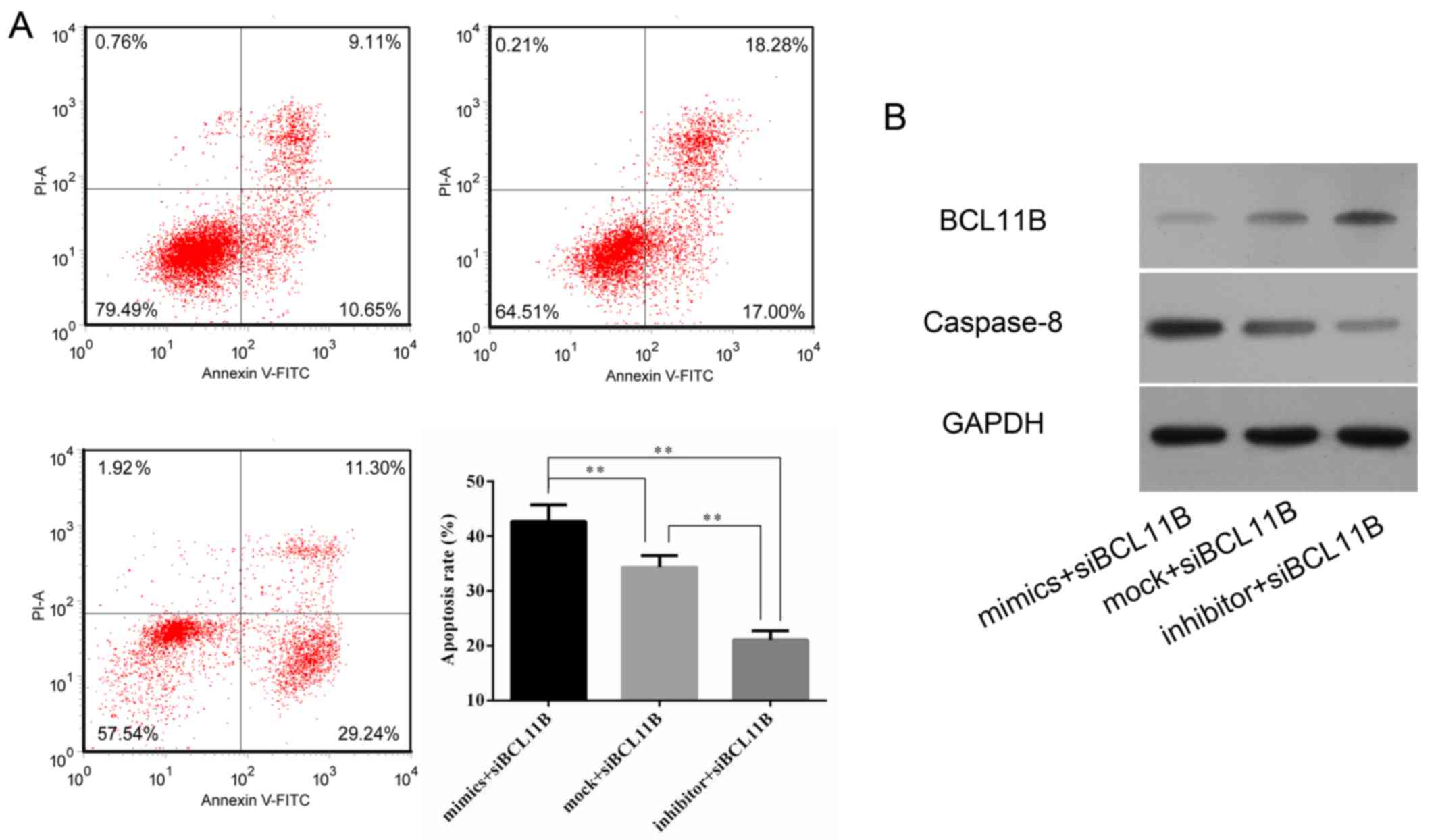
7). A lower level of caspase-8 protein expression was also
observed in the BCL11B siRNA-transfected group (Fig. 7B).
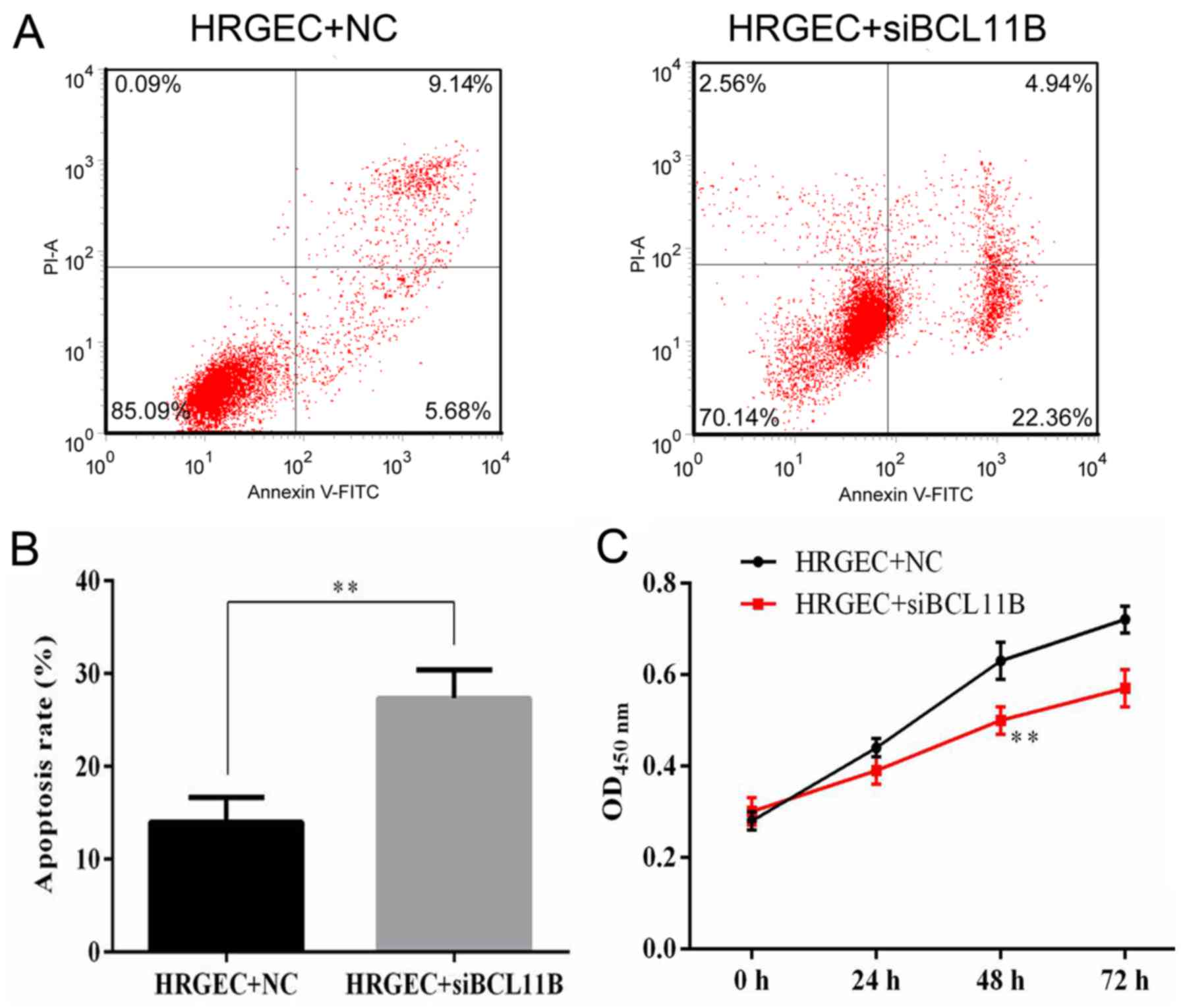
Cell viability and apoptosis rates were measured by
flow cytometry and a CCK8 assay. It was revealed that BCL11B siRNA
transfection significantly increased the apoptotic rate of HRGECs
in comparison with the NC (Fig. 8A
and B). The survival of HRGECs was identified as being
significantly reduced in cells transfected with BCL11B siRNA
compared with the NC at 48 h after transfection (Fig. 8C).
To explore the combined effect of the miR-650 mimic
and inhibition of BCL11B, BCL11B siRNA was transfected into HRGECs
in combination with mock, inhibitor or mimic miR-650. Flow
cytometry revealed that the apoptotic rate was significantly
increased in cells transfected with BCL11B siRNA and the miR-650
mimic compared with the other combinations (Fig. 9A). Western blot analysis indicated
that transfection of BCL11B siRNA combined with the miR-650 mimic
further enhanced the expression of caspase-8 in comparison with the
other groups. (Fig. 9B)
Discussion
Acute renal rejection is a major cause of allograft
dysfunction and the most common type of kidney transplant rejection
(30). Acute renal rejection has
a rapid onset and can occur just weeks after transplantation. The
risk of developing acute renal rejection can be reduced by the use
of prophylactic immunosuppressive drugs (31), which has dramatically decreased
the incidence rate of acute renal rejection over the past three
decades (32,33). However, the optimization of
immunosuppressive methods to prevent allograft rejection and
minimize drug toxicity, infection and malignancy remains
challenging. The early diagnosis of acute renal rejection is
important for effective treatment with immunosuppressants and
corticosteroids. Increasing evidence has demonstrated that miRNAs
are differentially expressed in acute renal rejection and serve
essential roles in its progression (34–36). Previous studies have reported that
miRNAs may be used as a liquid biopsy tool, for applications
including distinguishing squamous cell carcinoma in lung cancer
biopsies (37), and predicting
the pathological response (38)
of patients with and without acute rejection (39,40). This suggests that the abnormal
expression of miRNAs is highly correlated with the progression of
acute renal rejection where they serve an essential regulatory
role. Therefore, the identification of miRNA markers may be
critical for the early diagnosis of acute renal rejection.
Previous studies have revealed that specific miRNAs
are favorable for the prediction of acute renal rejection,
including miR-142-5q (41),
miR-155 (42), miR-142-3q
(43) and miR-223-3q (44). These miRNAs were identified to be
upregulated in transplanted renal tissue and peripheral blood
lymphocytes. In addition, urinary levels of specifics miRNAs have
been considered as diagnostic markers for acute renal rejection;
Lorenzen et al (45)
identified a significantly decreased level of miR-210 in the urine
samples from patients with acute renal rejection compared with
those without rejection. In the present study, the expression and
role of miR-650 in acute renal rejection was investigated. An
increased expression of miR-650 was identified in patients with
acute renal rejection compared with patients with normal
allografts, which suggests that miR-650 may serve a functional role
in the development of acute renal rejection. Furthermore, the in
vitro model using HRGECs transfected with a miR-650 mimic
revealed that the upregulation of miR-650 significantly suppressed
cell proliferation and induced apoptosis. Transfection with the
miR-650 mimic also significantly enhanced the release of
inflammatory cytokines and chemokines, and promoted macrophage
chemotaxis. These results suggest that miR-650 serves an essential
regulatory role in acute renal rejection and may be considered as a
potential diagnostic marker for acute renal rejection.
Previous studies have demonstrated that miR-650
contributes to the development of cancer by targeting tumor
suppressor and apoptotic factors during cancer progression
(46,47). Huang et al (23) revealed that miR-650 is a
prognostic factor in human lung adenocarcinoma and regulates the B
cell lymphoma-2/Bax signaling pathway by targeting tumor suppressor
inhibitor of growth protein 4. In the present study, it was
revealed that the upregulation of miR-650 contributed to the
apoptosis of HRGECs by targeting BCL11B. BCL11B, also known as
CTIP2, is a transcriptional factor expressed in several different
types of cancer, including lymphoma, melanoma and prostate cancer
(48–50). BCL11B is implicated in multiple
biological processes, including cell proliferation (51), apoptosis (52) and the immune response (53), as well as different pathological
conditions, including cancer (54), inflammation (55), cardiac hypertrophy (56) and human immunodeficiency virus
latency (57). Previous studies
have demonstrated that BCL11B also contributes to T-cell
development and T-cell identity (53,58,59). Additionally, BCL11B serves a
critical role in the production and release of cytokines, including
IFN-γ, IL-5 and IL-13 (60–62). T-cells and cytokines are important
modulators of acute renal injury (61). In the present study, it was
revealed that BCL11B expression was negatively associated with the
level of miR-650 and was significantly decreased in patients with
acute renal rejection in comparison to patients with normal
allografts. The potential binding sites for miR-650 on BCL11B mRNA
were identified by computational prediction. The results of the
present study suggest that miR-650 targets BCL11B mRNA, and that
its upregulation reduces the expression of BCL11B, resulting in
apoptosis, the release of cytokines and chemokines, and macrophage
chemotaxis. The expression of caspase-8 was also negatively
associated with the level of BCL11B, indicating that apoptotic
signaling was upregulated by the suppression of BCL11B in acute
renal rejection. The effect of BCL11B demonstrated in the present
study is different from its previously demonstrated effect as a
tumor suppressor (63) and immune
response inducer (64). The
results of the present study also revealed mechanistic insights
into the functioning of miR-650, as well as the novel regulatory
role of BCL11B.
To the best of our knowledge, the present study is
the first to demonstrate that the upregulation of miR-650
contributes to the progression of acute renal rejection, which
occurs via promoting apoptosis and immune responses through the
targeting of BCL11B. In addition, the inhibition of miR-650 was
identified to provide an efficient protective effect by reducing
the expression of BCL11B, resulting in a lower level of apoptosis
and immune responses, and increased cell migration. These results
indicate that miR-650 is a potential novel diagnostic factor for
acute renal rejection and may provide novel therapeutic strategies
for its treatment.
References
|
1
|
Mas VR, Le TH and Maluf DG: Epigenetics in
kidney transplantation: Current evidence, predictions, and future
research directions. Transplantation. 100:23–38. 2016. View Article : Google Scholar
|
|
2
|
Li PK, Burdmann EA and Mehta RL; World
Kidney Day Steering Committee 2013: Acute kidney injury: Global
health alert. Transplantation. 95:653–657. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Van Sandwijk MS, Ten Berge IJ, Majoie CB,
Caan MW, De Sonneville LM, Van Gool WA and Bemelman FJ: Cognitive
changes in chronic kidney disease and after transplantation.
Transplantation. 100:734–742. 2016. View Article : Google Scholar
|
|
4
|
Hakenberg O: Kidney transplantation.
Urologe. 54:13552015.In German. View Article : Google Scholar
|
|
5
|
Verhave J, Boucher A, Dandavino R,
Collette S, Senécal L, Hebert MJ, Girardin C and Cardinal H: The
incidence, management, and evolution of rapamycin-related side
effects in kidney transplant recipients. Clin Transplant.
28:616–622. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
El Ters M, Grande JP, Keddis MT, Rodrigo
E, Chopra B, Dean PG, Stegall MD and Cosio FG: Kidney allograft
survival after acute rejection, the value of follow-up biopsies. Am
J Transplant. 13:2334–2341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vlachopanos G and Georgalis A: On the
value and pitfalls of follow-up biopsies after acute rejection. Am
J Transplant. 14:2372014. View Article : Google Scholar
|
|
8
|
Goldberg RJ, Weng FL and Kandula P: Acute
and chronic allograft dysfunction in kidney transplant recipients.
Med Clin North Am. 100:487–503. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Becker LE, Morath C and Suesal C: Immune
mechanisms of acute and chronic rejection. Clin Biochem.
49:320–323. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hammond SM: An overview of microRNAs. Adv
Drug Deliv Rev. 87:3–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Iwakawa HO and Tomari Y: The Functions of
MicroRNAs: mRNA decay and translational repression. Trends Cell
Biol. 25:651–665. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Small EM and Olson EN: Pervasive roles of
microRNAs in cardiovascular biology. Nature. 469:336–342. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fan PC, Chen CC, Chen YC, Chang YS and Chu
PH: MicroRNAs in acute kidney injury. Hum Genomics. 10:292016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lujambio A and Lowe SW: The microcosmos of
cancer. Nature. 482:347–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim JK, Kim TS, Basu J and Jo EK: MicroRNA
in innate immunity and autophagy during mycobacterial infection.
Cell Microbiol. 19:e126872017. View Article : Google Scholar
|
|
17
|
Zawada AM, Zhang L, Emrich IE, Rogacev KS,
Krezdorn N, Rotter B, Fliser D, Devaux Y, et al: MicroRNA profiling
of human intermediate monocytes. Immunobiology. 222:587–596. 2017.
View Article : Google Scholar
|
|
18
|
Oda H, Ikeguchi R, Yurie H, Kaizawa Y,
Ohta S, Yamamoto K, Aoyama T and Matsuda S: Plasma microRNAs are
potential biomarkers of acute rejection after hindlimb
transplantation in rats. Transplantation Direct. 2:e1082016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fujino M, Zhu P, Cai S, Nishio Y, Zhuang J
and Li XK: MicroRNAs involved in acute rejection and tolerance in
murine cardiac allografts. Exp Clin Transplant. 14:424–430.
2016.PubMed/NCBI
|
|
20
|
Soltaninejad E, Nicknam MH, Nafar M,
Ahmadpoor P, Pourrezagholi F, Sharbafi MH, Hosseinzadeh M, Foroughi
F, Yekaninejad MS, Bahrami T, et al: Differential expression of
microRNAs in renal transplant patients with acute T-cell mediated
rejection. Transpl Immunol. 33:1–6. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wilflingseder J, Reindl-Schwaighofer R,
Sunzenauer J, Kainz A, Heinzel A, Mayer B and Oberbauer R:
MicroRNAs in kidney transplantation. Nephrol Dial Transplant.
30:910–917. 2015. View Article : Google Scholar :
|
|
22
|
Zhang X, Zhu W, Zhang J, Huo S, Zhou L, Gu
Z and Zhang M: MicroRNA-650 targets ING4 to promote gastric cancer
tumorigenicity. Biochem Biophys Res Commun. 395:275–280. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang JY, Cui SY, Chen YT, Song HZ, Huang
GC, Feng B, Sun M, De W, Wang R and Chen LB: MicroRNA-650 was a
prognostic factor in human lung adenocarcinoma and confers the
docetaxel chemoresistance of lung adenocarcinoma cells via
regulating Bcl-2/Bax expression. PLoS One. 8:e726152013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Feng L, Xie Y, Zhang H and Wu Y:
Down-regulation of NDRG2 gene expression in human colorectal cancer
involves promoter methylation and microRNA-650. Biochem Biophys Res
Commun. 406:534–538. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zuo ZH, Yu YP, Ding Y, Liu S, Martin A,
Tseng G and Luo JH: Oncogenic activity of miR-650 in prostate
cancer is mediated by suppression of CSR1 expression. Am J Pathol.
185:1991–1999. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chien WW, Domenech C, Catallo R, Kaddar T,
Magaud JP, Salles G and Ffrench M: Cyclin-dependent kinase 1
expression is inhibited by p16(INK4a) at the post-transcriptional
level through the microRNA pathway. Oncogene. 30:1880–1891. 2011.
View Article : Google Scholar
|
|
27
|
Mengel M, Sis B and Halloran PF: SWOT
analysis of Banff: Strengths, weaknesses, opportunities and threats
of the international Banff consensus process and classification
system for renal allograft pathology. Am J Transplant. 7:2221–2226.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
29
|
Farooqi AA, Qureshi MZ, Coskunpinar E,
Naqvi SK, Yaylim I and Ismail M: MiR-421, miR-155 and miR-650:
Emerging trends of regulation of cancer and apoptosis. Asian Pac J
Cancer Prev. 15:1909–1912. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Aktaş A: Transplanted kidney function
evaluation. Semin Nucl Med. 44:129–145. 2014. View Article : Google Scholar
|
|
31
|
Montero N, Pérez-Sáez MJ, Pascual J,
Abramowicz D, Budde K, Dudley C, Hazzan M, Klinger M, Maggiore U,
Oberbauer R, et al; DESCARTES Working Group. DESCARTES ERA-EDTA
Board: Immunosuppression in the elderly renal allograft recipient:
A systematic review. Transplant Rev (Orlando). 30:144–153. 2016.
View Article : Google Scholar
|
|
32
|
O'Leary JG, Samaniego M, Barrio MC, Potena
L, Zeevi A, Djamali A and Cozzi E: The influence of
immunosuppressive agents on the risk of de novo donor-specific HLA
antibody production in solid organ transplant recipients.
Transplantation. 100:39–53. 2016. View Article : Google Scholar
|
|
33
|
Watson CJ and Dark JH: Organ
transplantation: Historical perspective and current practice. Br J
Anaesth. 108(Suppl 1): i29–i42. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu Z, Yang W, Steward N, Sweet SC,
Danziger-Isakov L, Heeger PS and Mohanakumar T: Role of circulating
microRNAs in the immunopathogenesis of rejection following
pediatric lung transplantation. Transplantation. 101:2461–2468.
2017. View Article : Google Scholar
|
|
35
|
Matz M, Lorkowski C, Fabritius K, Durek P,
Wu K, Rudolph B, Neumayer HH, Mashreghi MF and Budde K: Free
microRNA levels in plasma distinguish T-cell mediated rejection
from stable graft function after kidney transplantation. Transpl
Immunol. 39:52–59. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schmuck RB, Reutzel-Selke A, Raschzok N,
Morgul HM, Struecker B, Lippert S, de Carvalho Fischer C, Schmelzle
M, Boas-Knoop S, Bahra M, et al: Bile: miRNA pattern and
protein-based biomarkers may predict acute cellular rejection after
liver transplantation. Biomarkers. 22:19–27. 2017. View Article : Google Scholar
|
|
37
|
Patnaik S, Mallick R, Kannisto E, Sharma
R, Bshara W, Yendamuri S and Dhillon SS: MiR-205 and MiR-375
microRNA assays to distinguish squamous cell carcinoma from
adenocarcinoma in lung cancer biopsies. J Thorac Oncol. 10:446–453.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wen J, Luo K, Liu H, Liu S, Lin G, Hu Y,
Zhang X, Wang G, Chen Y, Chen Z, et al: MiRNA expression analysis
of pretreatment biopsies predicts the pathological response of
esophageal squamous cell carcinomas to neoadjuvant
chemoradiotherapy. Ann Surg. 263:942–948. 2016. View Article : Google Scholar
|
|
39
|
Gharib SA, Edelman JD, Ge L and Chen P:
Acute cellular rejection elicits distinct microRNA signatures in
airway epithelium of lung transplant patients. Transplant Direct.
1:e442015. View Article : Google Scholar
|
|
40
|
Merhi B, Bayliss G and Gohh RY: Role for
urinary biomarkers in diagnosis of acute rejection in the
transplanted kidney. World J Transplant. 5:251–260. 2015.
View Article : Google Scholar
|
|
41
|
Iwasaki K, Yamamoto T, Inanaga Y,
Hiramitsu T, Miwa Y, Murotani K, Narumi S, Watarai Y, Katayama A,
Uchida K, et al: MiR-142-5p and miR-486-5p as biomarkers for early
detection of chronic antibody-mediated rejection in kidney
transplantation. Biomarkers. 22:45–54. 2017. View Article : Google Scholar
|
|
42
|
Anglicheau D, Sharma VK, Ding R, Hummel A,
Snopkowski C, Dadhania D, Seshan SV and Suthanthiran M: MicroRNA
expression profiles predictive of human renal allograft status.
Proc Natl Acad Sci USA. 106:5330–5335. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wei Q, Mi QS and Dong Z: The regulation
and function of microRNAs in kidney diseases. IUBMB Life.
65:602–614. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ulbing M, Kirsch AH, Leber B, Lemesch S,
Münzker J, Schweighofer N, Hofer D, Trummer O, Rosenkranz AR,
Müller H, et al: MicroRNAs 223-3p and 93-5p in patients with
chronic kidney disease before and after renal transplantation.
Bone. 95:115–123. 2017. View Article : Google Scholar
|
|
45
|
Lorenzen JM, Volkmann I, Fiedler J,
Schmidt M, Scheffner I, Haller H, Gwinner W and Thum T: Urinary
miR-210 as a mediator of acute T-cell mediated rejection in renal
allograft recipients. Am J Transplant. 11:2221–2227. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang IP, Tsai HL, Miao ZF, Huang CW, Kuo
CH, Wu JY, Wang WM, Juo SH and Wang JY: Development of a
deregulating microRNA panel for the detection of early relapse in
postoperative colorectal cancer patients. J Transl Med. 14:1082016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zeng ZL, Li FJ, Gao F, Sun DS and Yao L:
Upregulation of miR-650 is correlated with down-regulation of ING4
and progression of hepatocellular carcinoma. J Surg Oncol.
107:105–110. 2013. View Article : Google Scholar
|
|
48
|
Kominami R: Role of the transcription
factor Bcl11b in development and lymphomagenesis. Proc Jpn Acad Ser
B Phys Biol Sci. 88:72–87. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Uddin MN, Zhang Y, Harton JA, MacNamara KC
and Avram D: TNF-alpha-dependent hematopoiesis following Bcl11b
deletion in T cells restricts metastatic melanoma. J Immunol.
192:1946–1953. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Mahapatra S, Klee EW, Young CY, Sun Z,
Jimenez RE, Klee GG, Tindall DJ and Donkena KV: Global methylation
profiling for risk prediction of prostate cancer. Clin Cancer Res.
18:2882–2895. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li Z, Chen G, Yang Y, Guo W and Tian W:
Bcl11b regulates enamel matrix protein expression and dental
epithelial cell differentiation during rat tooth development. Mol
Med Rep. 15:297–304. 2017. View Article : Google Scholar
|
|
52
|
Huang X, Chen S, Shen Q, Chen S, Yang L,
Grabarczyk P, Przybylski GK, Schmidt CA and Li Y: Down regulation
of BCL11B expression inhibits proliferation and induces apoptosis
in malignant T cells by BCL11B-935-siRNA. Hematology. 16:236–242.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hirose S, Touma M, Go R, Katsuragi Y,
Sakuraba Y, Gondo Y, Abe M, Sakimura K, Mishima Y and Kominami R:
Bcl11b prevents the intrathymic development of innate CD8 T cells
in a cell intrinsic manner. Int Immunol. 27:205–215. 2015.
View Article : Google Scholar
|
|
54
|
Huang X, Du X and Li Y: The role of BCL11B
in hematological malignancy. Exp Hematol Oncol. 1:222012.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang Z, Zhang LJ, Guha G, Li S, Kyrylkova
K, Kioussi C, Leid M, Ganguli-Indra G and Indra AK: Selective
ablation of Ctip2/Bcl11b in epidermal keratinocytes triggers atopic
dermatitis-like skin inflammatory responses in adult mice. PLoS
One. 7:e512622012. View Article : Google Scholar
|
|
56
|
Cherrier T, Le Douce V, Eilebrecht S,
Riclet R, Marban C, Dequiedt F, Goumon Y, Paillart JC, Mericskay M,
Parlakian A, et al: CTIP2 is a negative regulator of P-TEFb. Proc
Natl Acad Sci USA. 110:12655–12660. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Cismasiu VB, Paskaleva E, Suman Daya S,
Canki M, Duus K and Avram D: BCL11B is a general transcriptional
repressor of the HIV-1 long terminal repeat in T lymphocytes
through recruitment of the NuRD complex. Virology. 380:173–181.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Takachi T, Takahashi M, Takahashi-Yoshita
M, Higuchi M, Obata M, Mishima Y, Okuda S, Tanaka Y, Matsuoka M,
Saitoh A, et al: Human T-cell leukemia virus type 1 Tax oncoprotein
represses the expression of the BCL11B tumor suppressor in T-cells.
Cancer Sci. 106:461–465. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li L, Zhang JA, Dose M, Kueh HY, Mosadeghi
R, Gounari F and Rothenberg EV: A far downstream enhancer for
murine Bcl11b controls its T-cell specific expression. Blood.
122:902–911. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Liu P, Li P and Burke S: Critical roles of
Bcl11b in T-cell development and maintenance of T-cell identity.
Immunol Rev. 238:138–149. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kinsey GR and Okusa MD: Expanding role of
T cells in acute kidney injury. Curr Opin Nephrol Hypertens.
23:9–16. 2014. View Article : Google Scholar :
|
|
62
|
Yu Y, Wang C, Clare S, Wang J, Lee SC,
Brandt C, Burke S, Lu L, He D, Jenkins NA, et al: The transcription
factor Bcl11b is specifically expressed in group 2 innate lymphoid
cells and is essential for their development. J Exp Med.
212:865–874. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Gutierrez A, Kentsis A, Sanda T, Holmfeldt
L, Chen SC, Zhang J, Protopopov A, Chin L, Dahlberg SE, Neuberg DS,
et al: The BCL11B tumor suppressor is mutated across the major
molecular subtypes of T-cell acute lymphoblastic leukemia. Blood.
118:4169–4173. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhong C and Zhu J: Bcl11b drives the birth
of ILC2 innate lymphocytes. J Exp Med. 212:8282015. View Article : Google Scholar : PubMed/NCBI
|