1. Introduction
Precursor proteins are not active, and thus, need
further processing to become functional mature proteins.
Post-translational protein modification is the chemical
modification of proteins prior to or following protein
biosynthesis, and includes phosphorylation, ubiquitylation,
methylation, acetylation, and modifications by ubiquitin-like
modifiers (Ubls).
Ubiquitin (Ub) is a globular protein consisting of
76 amino acid residues, and the UPS (ubiquitin-proteasome system)
is responsible for degrading 80–90% of intracellular proteins. Ub
binds the lysine residues of target proteins by a series of enzymes
including the ubiquitin-activating enzyme E1, ubiquitin-conjugating
enzyme E2 and the ubiquitin E3 ligases. Subsequently, these
ubiquitylated proteins are recognized and degraded by the 26S
proteasome. Modifications by Ubls are similar to ubiquitylation and
they compete for the lysine residues of certain substrates
(1).
Small ubiquitin-related modifier (SUMO) proteins are
one type of Ubls. Although they only have ~18% sequence identity
with ubiquitin, they share a common three-dimensional structure;
namely, a globular β-grasp fold and a characteristic C-terminal
diglycine (Gly-Gly) motif that is exposed following maturation
(2). SUMOs have a flexible, 20
amino acid N-terminus that seems to primarily serve as an acceptor
in the formation of poly-SUMO chains (2). Mammalian cells express four SUMO
isoforms: SUMO-1, SUMO-2, SUMO-3 and SUMO-4. SUMO1-3 is
ubiquitously expressed in human tissues, whereas SUMO4 is only
expressed in the kidney, lymph nodes and spleen (3). SUMO-2 and 3 are 95% identical to
each other and only 50% identical to SUMO-1 (4); it is not yet clear whether these
three isoforms are functionally distinct (6). Unlike SUMO-1, SUMO-2 and 3 have
internal SUMO modification sites at their N-termini (K11), which
can form poly-SUMO chains (5).
The expression level and activity of SUMO proteins affect their
conjugation to substrates, although it remains unknown if SUMO-4
can be conjugated to cellular proteins. Similar to the
ubiquitylation pathway, SUMO conjugation is reversible and
catalyzed by a three-step enzymatic reaction that includes
activation, conjugation, and ligation (Fig. 1), which does not result in protein
decomposition, but rather, in the regulation of normal cell
functions such as protein-protein interactions, subcellular
localization, DNA repair, and cell cycle and transcription factor
regulation (6).
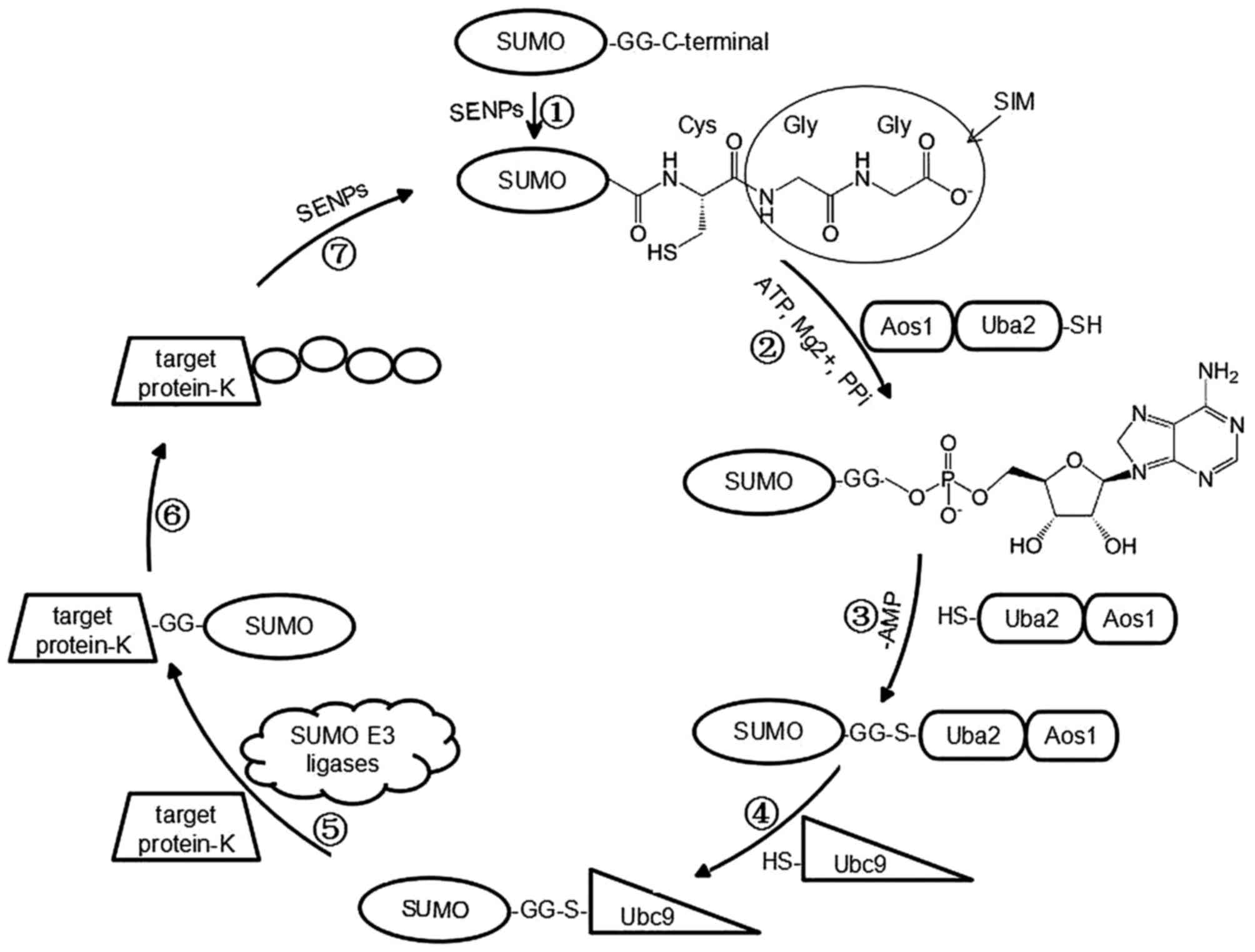 | Figure 1Major steps in the SUMOylation
pathway. SENPs cleave the C-terminus of SUMO to expose the Gly-Gly
motif required for conjugation, which is also called the
SUMO-interacting motif. SAE activates SUMO in a two-step reaction
that involves ATP hydrolysis. SAE1 (Aos1) promotes adenylation of
the SUMO C-terminus to form the SUMO-AMP intermediate, followed by
transfer of SUMO to SAE2 (Uba2) and thioester bond formation. Then
SUMO is transferred from SAE to Ubc9. Ubc9 catalyzes formation of
an isopeptide bond between the C-terminal glycine of SUMO and a
lysine residue in the substrate, usually together with a specific
SUMO E3 ligase. Finally, SUMO is removed from the lysine residues
of target proteins by SENPs using their isopeptidase activities.
SUMO, small ubiquitin-related modifier; SENPs, sentrin-specific
proteases; SAE, SUMO-activating enzyme; Aos1, SUMO1 activating
enzyme subunit 1; Uba2, ubiquitin like modifier activating enzyme
2; ATP, adenosine triphosphate; AMP, adenosine monophosphate; Ubc9,
ubiquitin conjugating enzyme E2. |
In SUMOylation, initially, sentrin-specific
proteases (SENPs) cleave the C-terminus of SUMO using their
endopeptidase activities to expose the Gly-Gly motif required for
conjugation, which is also termed the SUMO-interacting motif (SIM)
(7–9). SENPs have six isoforms (SENP1, 2, 3,
5, 6 and 7) in mammals, which are divided into three subfamilies
based on their sequence homology, substrate specificity, and
subcellular localization (10).
The catalytic cysteine protease domain at the C-terminus is ~250
amino acid residues in length, and controls the specificity and
function of SENP isoforms (11).
The catalytic domain crystal structures of SENP1, SENP2 and SENP7,
which are comprised of the typical catalytic triad (Cys-His-Asp),
are very similar (10). SENP1 and
SENP2 have SUMO maturation and deSUMOylation abilities, whereas the
other SENPs are unable to induce maturation and prefer SUMO-2/3
over SUMO-1 for deSUMOylation (10,12).
In the second step, SUMO-activating enzyme (SAE)
activates the C-terminus of SUMO in a two-step reaction that
involves ATP hydrolysis. There is only one heterodimeric E1 enzyme
[SAE1/SAE2, also termed activator of SUMO1 (Aos1)/ubiquitin like
modifier activating enzyme 2 (Uba2)] involved in the SUMOylation
pathway. SAE1 (Aos1) promotes adenylation of the SUMO C-terminus to
form the SUMO-adenosine monophosphate (AMP) intermediate, followed
by transfer of SUMO to the catalytic Cys173 residue on SAE2 (Uba2)
and thioester bond formation (13).
The third step is conjugation. Specifically, SUMO is
transferred from SAE to SUMO E2 (also termed ubiquitin conjugating
enzyme 2I, UbE2I or Ubc9), again resulting in formation of a
thioester linkage between the C-terminal glycine in SUMO and the
active site Cys93 residue in Ubc9. Ubc9 is the only known
SUMO-conjugating enzyme required for SUMOylation, and its deletion
abolishes SUMO conjugation (14).
The fourth step is ligation. Ubc9 catalyzes
formation of an isopeptide bond between the C-terminal glycine of
SUMO and a lysine residue in the substrate, usually together with a
specific SUMO E3 ligase, which increases the efficiency of this
reaction by associating with both the substrate protein and Ubc9
(2,15). Specific E3 ligases in mammals
include the protein inhibitor of activated STAT-1 (PIAS) protein
family (PIAS1, PIAS3, PIASxα, PIASxβ and PIASy), the nucleoporin
Ran binding protein 2 and the human polycomb protein Pc2 (2). The tripartite motif family was
recently described as a fourth group of SUMO E3 ligases (16).
In the last step, SUMO is removed from the lysine
residues of target proteins by SENPs using their isopeptidase
activities (10).
Ubiquitin-specific protease-like 1 was identified as a new SUMO
isopeptidase, with roles in Cajal body functions (17). DeSUMOylating isopeptidase 1 is
another SUMO isopeptidase, which recognizes different sets of
substrates to SENPs (18).
All SUMOylation proteins are associated with cancer,
as elevated levels of SAE, Ubc9, SUMO E3s and SENPs have been
observed in various types of cancer (12,14–16). As target proteins of SUMOylation,
the Ras-related nuclear protein GTPase-activating protein
(RanGAPl), the nuclear factor-κB regulatory inhibitor-α, the tumor
suppressor gene p53, the androgen receptor, the progesterone
receptor and certain other substrates have been reported (19). The abnormal SUMOylation of these
target proteins is also associated with heart disease, diabetes,
arthritis, degenerative diseases, and brain ischemia/stroke
(6). Thus, there has been growing
appreciation for the potential importance of the SUMO conjugation
pathway as a target for treating these diseases (20–25).
This review discusses the compounds that inhibit the
SUMOylation pathway referred to in patents written in English or
Chinese (Table I). These
compounds can be divided into four categories according to their
mechanisms of action. Category 1 compounds are SUMO mimics that can
affect the entire pathway. Category 2 comprises SAE and Ubc9
inhibitors, because all SUMO isoforms are activated and conjugated
by SAE and Ubc9. Category 3 compounds are ligation inhibitors
because of the specificity of SUMO E3 ligases to target proteins.
Category 4 compounds are SENP inhibitors that may inhibit
maturation and deSUMOylation.
 | Table ISummary of compounds included in the
patents reviewed. |
Table I
Summary of compounds included in the
patents reviewed.
| No. | Patent ID | Invention | Mechanism |
|---|
| 2.1 | US0302815 | AuNP-ligand
conjugates | SIM mimic |
| 3.1 | EP2402334 | MLN4924 | AMP mimic |
| 3.2 | WO002994 | Heteroaryl
compounds | SAE inhibitor |
| 3.3 | US0317101
WO064898 | MLS-0437113
etc. | Singleton SAE/Ubc9
inhibitor |
| 3.4 | WO064897
US0245032
US9045483 | MLS-0207587
etc. | Tricyclic SAE/Ubc9
inhibitor |
| 3.5 | EP2545935
US0330738 | HLS5
polypeptide | Degenerating
Ubc9/PIAS1 |
| 4.1 | WO064887
US0302525 | NSC5068 etc. | SENP1, 2 and 7
inhibitor |
| 4.2 | CN104436196 | shRNA | SENP1
inhibitor |
| 4.3 | CN103961348 | Agents for treating
prostate cancer | SENP1
inhibitor |
| 4.4 | CN103877078 | Agents for treating
breast cancer | SENP2
inhibitor |
2. Category 1: SUMO mimics
Multivalent poly-Ubl chain inhibitors
(US0302815) (26)
Structure
Multivalent poly-SUMO chain inhibitors comprise gold
nanoparticle (AuNP)-ligand conjugates and include at least two
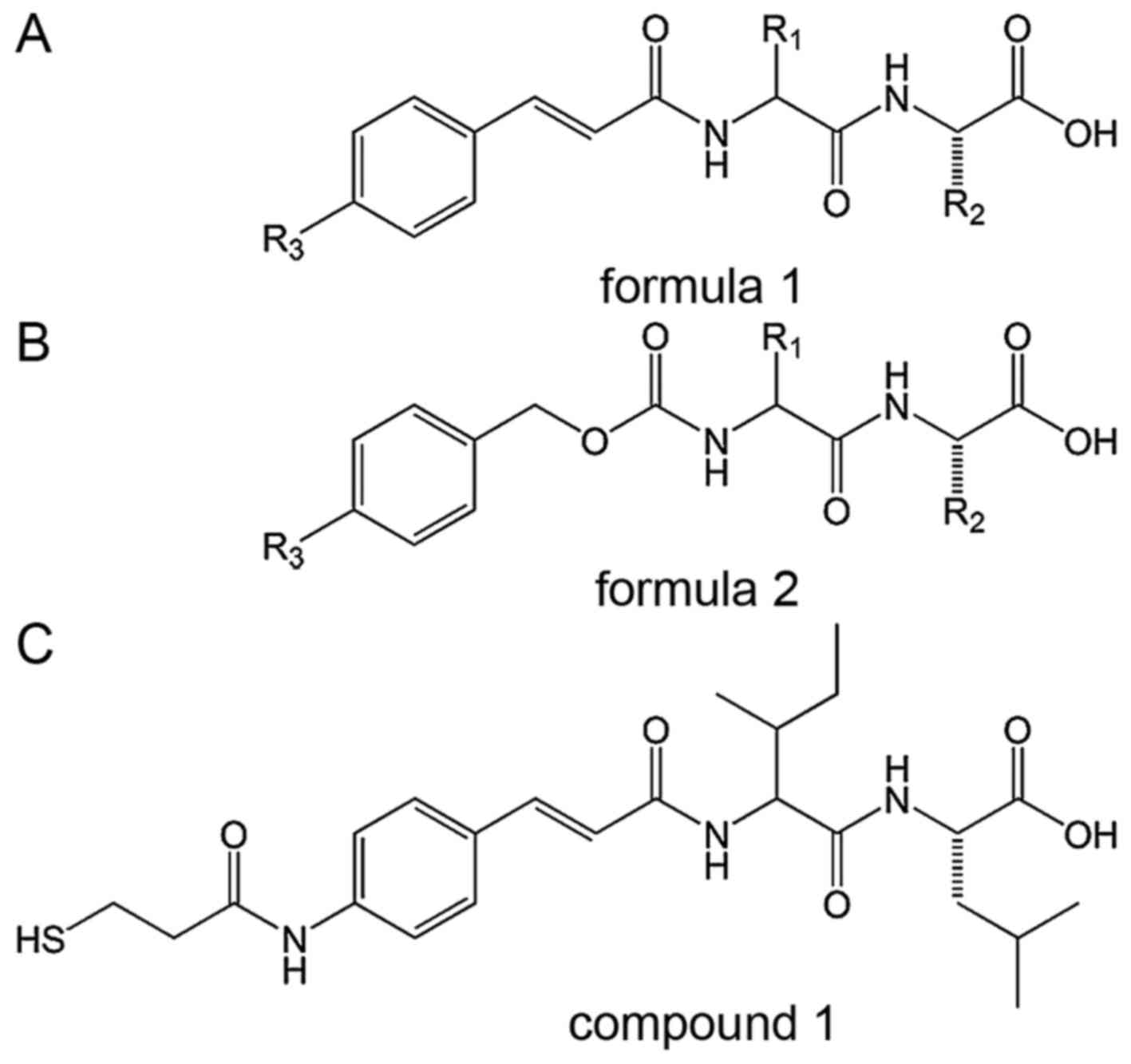
components: AuNPs and modified SIM mimics (26). The basic structures (formula 1 and
2) of SIM mimics and potential compound 1 are shown in Fig. 2 (26). The thiol tail [R3= -SH
or SH-(CH2)2-CO-NH-] allows the modified SIM
mimic to be conjugated to an AuNP by disulfide bonds (26).
Mechanism
The SIM mimic may recapitulate the action of SUMO in
protein modification to inhibit the SUMOylation pathway by: i)
Forming thioester conjugates with SAE; ii) being transferred from
SAE to Ubc9; and iii) being further transferred to the SUMOylation
target protein. Once the mimics are conjugated to SAE and Ubc9,
they block full-length SUMO from entering the cascade composed of
SAE and Ubc9. Thus, SUMO mimics can function as mechanism-based
inhibitors of the protein SUMOylation reaction (27). SUMO-2 and 3 are distinct from
SUMO-1 in that they harbor internal SUMO modification sites at
their N-termini to form poly-SUMO chains (5). Therefore, the conjugation of a weak
SUMO-2/3 ligand to AuNPs promotes selective multivalent
interactions with poly-SUMO-2/3 chains, resulting in the efficient
inhibition of poly-SUMO-chain-mediated protein-protein interactions
(5). Metals with high atomic
numbers, such as gold, preferentially absorb more X-ray energy than
soft tissues, thus augmenting the effects of ionizing radiation
when delivered to cells (28,29).
Application
Multivalent poly-SUMO chain inhibitors can kill
cancer cells by increasing radiation sensitivity, while sparing
normal cells (26). Thus, these
inhibitors provide a viable approach for treating cancer by
enhancing the effects of ionizing radiation therapy, which is
commonly used in cancer treatment. Combining the properties of
nanomaterials with nanoparticles as platforms for multivalent
interactions creates important potential therapeutic applications
(30).
3. Category 2: SUMO enzyme inhibitors
Activating E1 inhibitors (EP2402334)
(31)
Structure
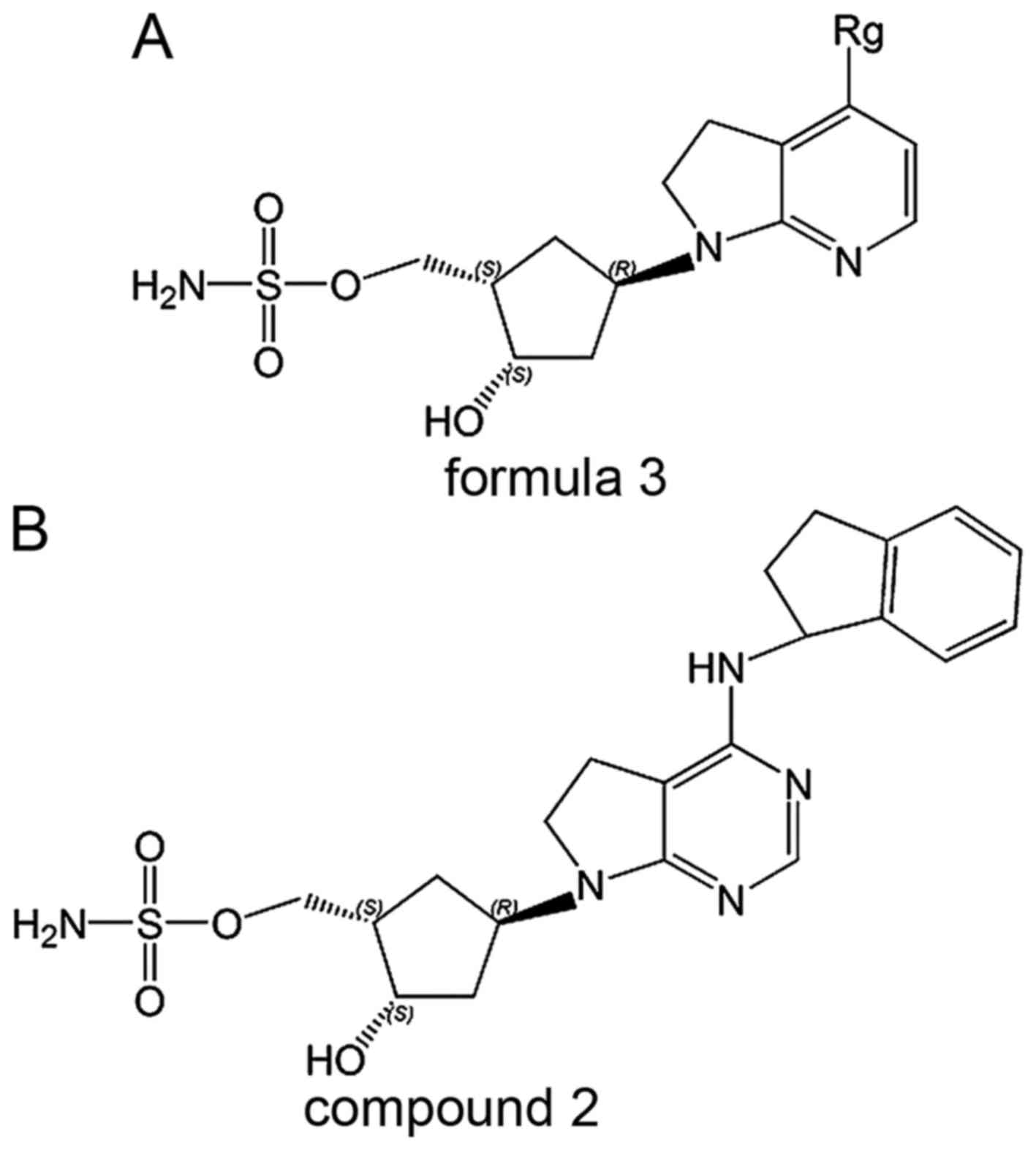
There are 86 compounds disclosed in EP2402334, of
which most are 4-substituted
[(1S,2S,4R)-2-hydroxy-4-{7H-pyrrolo(2,3-d)pyrimidin-7-yl}cyclopentyl)]
methyl sulfamates (formula 3; Fig.
3) (31).
Mechanism
The E1 inhibitors described herein are selective and
preferably target human NEDD8-activating enzyme (NAE), SAE, or
ubiquitin-activating enzyme, with the most preference for NAE
(31). The disclosed SAE
inhibitors can also be referred to as AMP mimics; they directly
bind to SAE1 and inhibit formation of the SAE-SUMO intermediate.
MLS 4924 (compound 3; Fig. 3), an
AMP mimic, is a selective and highly potent inhibitor of NAE, and
also inhibits SAE with a half maximal inhibitory concentration
(IC50) value of 8.2 (32).
Application
SAE inhibitors are useful for treating disorders,
particularly cell proliferation disorders, including cancer (not
limited to solid tumors and hematological tumors), inflammatory and
neurodegenerative disorders, and inflammation associated with
infection and cachexia (31).
Heteroaryl compounds (WO002994) (33)
Structure
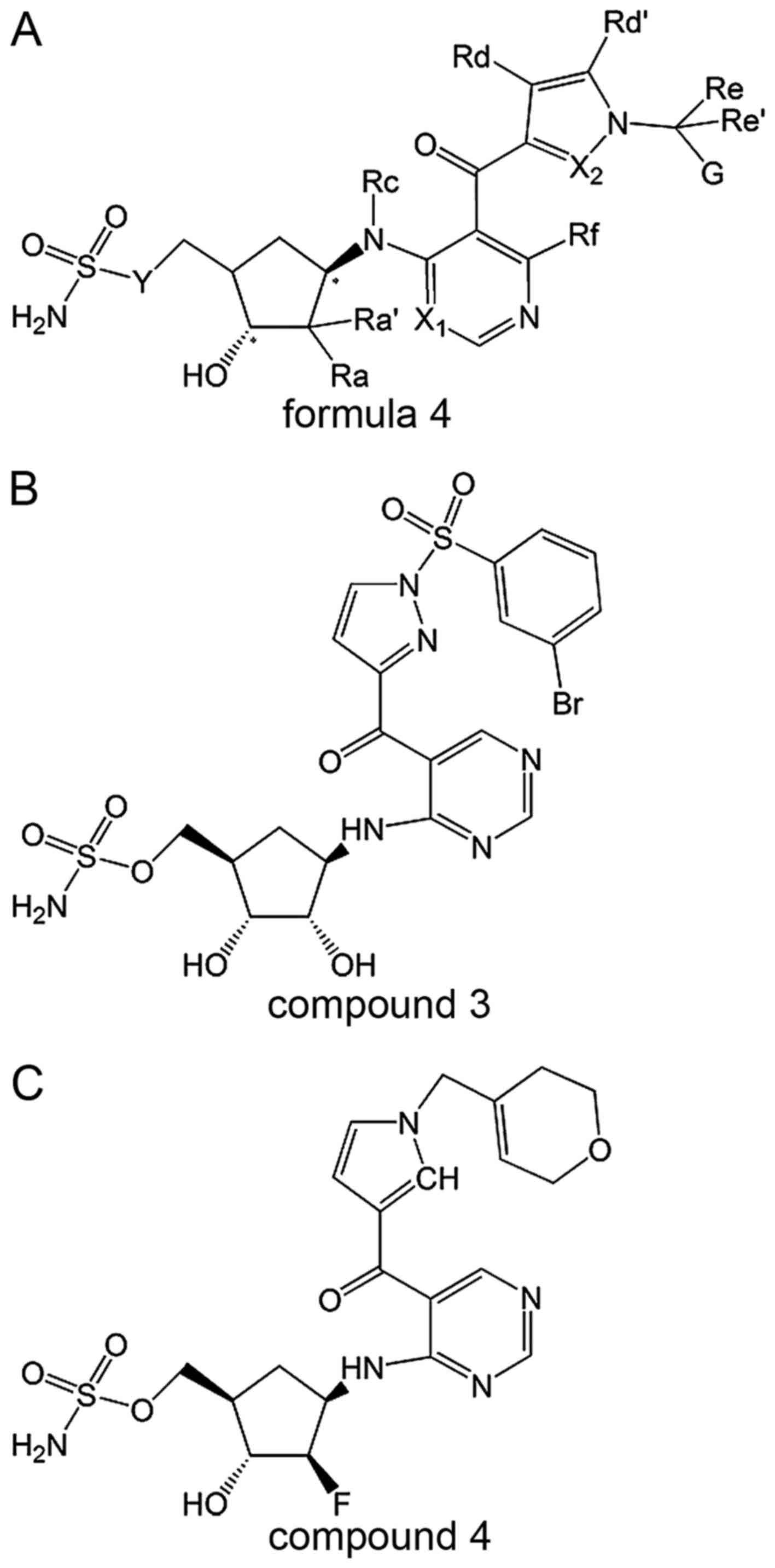
The basic structure (formula 4) of heteroaryl
compounds is shown in Fig. 4. The
-Y- is often replaced by -O-, -X1- is often replaced by -N-, -X2-
is often replaced by -N- or -CH-, Ra can be -OH or -F or -H, and Rc
is often replaced by -H. I-1 (compound 4; Fig. 4) and I-5 (compound 5; Fig. 4) are two examples of these
heteroaryl compounds (33).
Mechanism
These heteroaryl compounds may be useful as SAE
inhibitors (33).
Application
Heteroaryl compounds can be used for the treatment
of proliferative, inflammatory, cardiovascular and
neurodegenerative disorders (33). The altered expression of proteins
in the SAE pathway have been noted in a variety of cancer types
including multiple myeloma (MM) (34) and breast cancer (21). In addition, preclinical studies
have indicated that Myc-driven cancers may be especially sensitive
to SAE inhibition (35).
Singleton inhibitors [US0317101 (36) and WO064898 (37)]
Structure
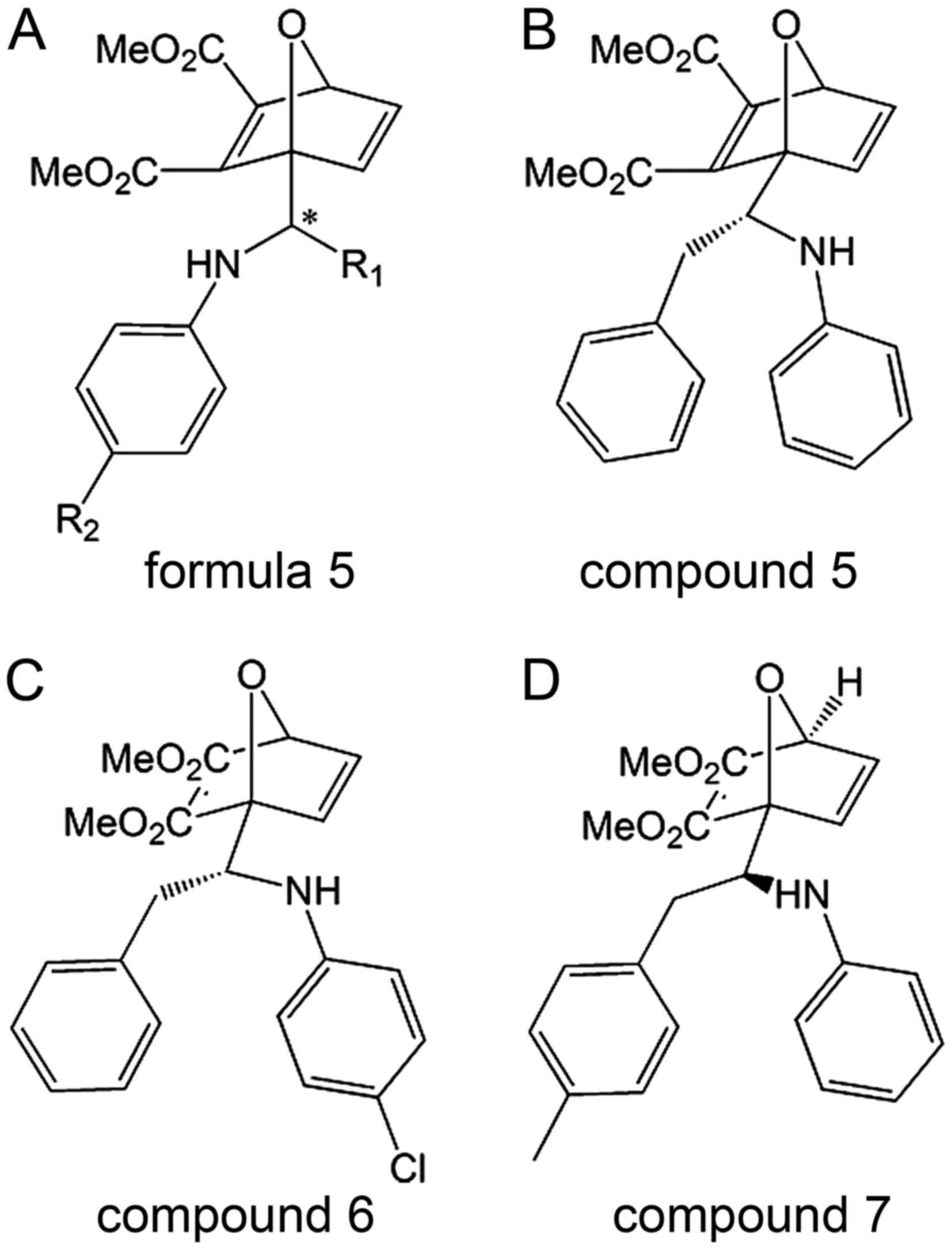
The structure (formula 5) of singleton inhibitors is
presented in Fig. 5 (36,37).
Mechanism
Singleton inhibitors inhibit the function of SAE
and/or Ubc9. MLS-0437113 (compound 5; Fig. 4) is the lead and most potent
compound in this series, and strongly inhibits SUMO-RanGap1 and
Ubc9 conjugation (36,37). MLS-0417120 (compound 6; Fig. 4) is another potential inhibitor
(36,37). There are 16 SAE-specific
inhibitors in these two patents. MLS-0437317 (compound 7; Fig. 5) is a representative SAE-specific
inhibitor that can bind to SAE with high affinity and specificity,
and inhibits global SUMOylation in a dose-dependent manner
(36,37).
Application
Singleton inhibitors have toxicity in cancer and
other diseases by sensitizing cells to genotoxic treatments.
Chemoradiotherapy (CRT) is frequently used as a preoperative
treatment for colorectal cancer to facilitate surgical intervention
and improve long-term survival (38,39). Novel SAE inhibitors can enhance
the effects of CRT and/or impair tumor viability, improving
treatment outcomes, preserving quality of life and reducing
healthcare costs (37). Such
inhibitors may be similarly useful in other cancer types (16), diseases, and conditions associated
with the overexpression of SAE or Ubc9 (36,37).
Bicyclic and tricyclic inhibitors
[WO064897 (40), US0245032
(41) and US9045483 (42)]
Structure
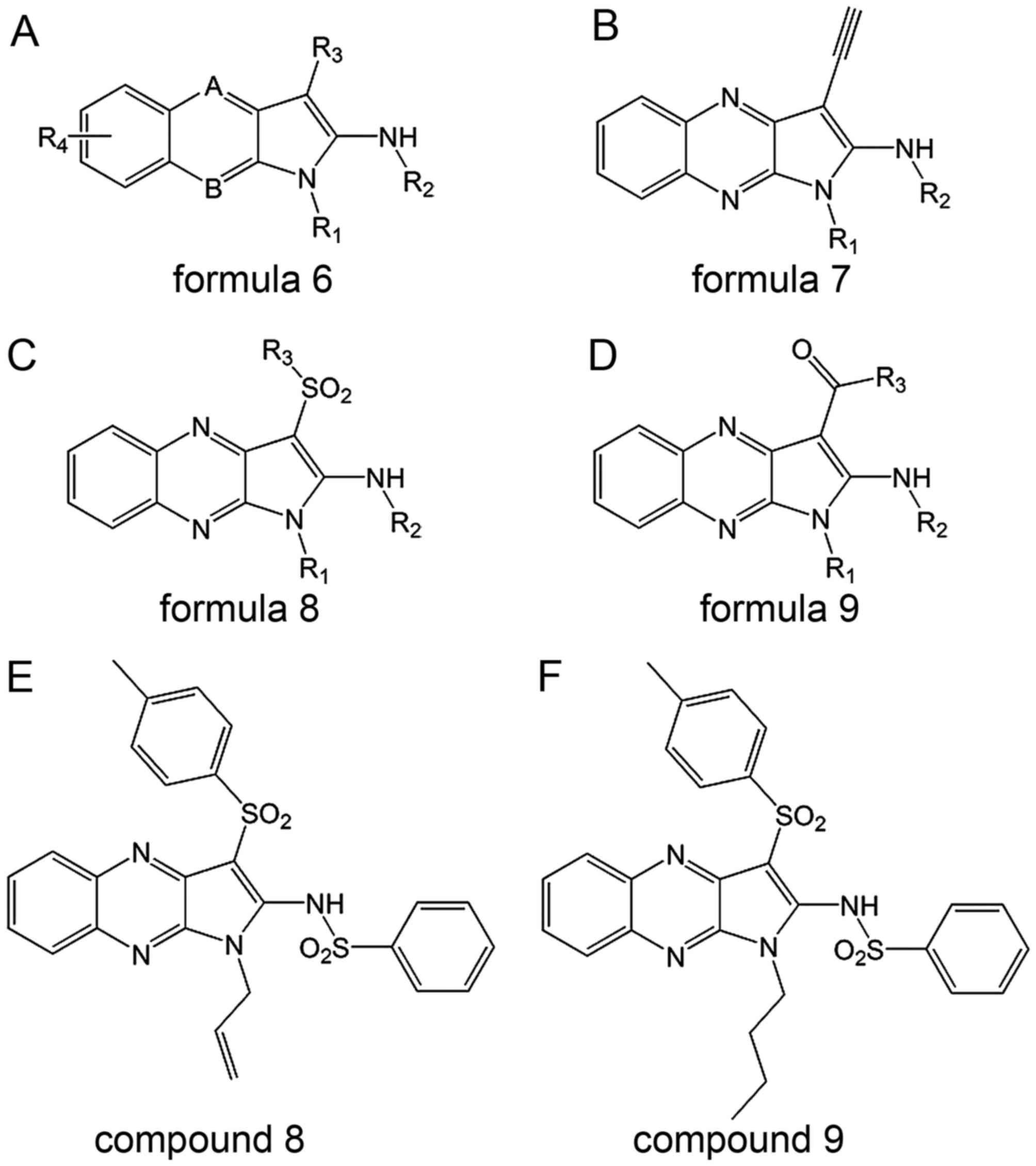
The basic structure in these three patents is
formula 6 (Fig. 6). Most
tricyclic inhibitors disclosed herein are derived from formulas 7,
8 and 9 (Fig. 6). There are 13
tricyclic SAE inhibitors disclosed in these patents. Compound 8
(Fig. 5) is similar to
MLS-0207587 (compound 9; Fig. 6),
which also showed an IC50 of 0.5 µM (40–42).
Mechanism
The tricyclic inhibitors competitively inhibit ATP
binding, sensitizing cells to genotoxic stress and inhibiting HIV
infection. Some compounds also inhibit Ubc9 (40–42).
Application
These inhibitors can be used to treat cancer,
degenerative diseases, and viral infections (e.g. HIV). The
characterized tricyclic SAE inhibitors may also induce significant
sensitivity to radiation in various cancer cell lines (40–42).
Hemopoietic lineage switch 5 polypeptide
[EP2545935 (43) and US0330738
(44)]
Structure
Hemopoietic lineage switch 5 (HLS5) is a member of
the RING finger B-box coiled-coil protein family. The gene is
located on chromosome 8p21, a region implicated in numerous
leukemias and solid tumors (45).
The overexpression of HLS5 in HeLa cells inhibited cell growth,
clonogenicity, and tumorigenicity; thus, it is conceivable that
HLS5 is a tumor suppressor protein (46).
Mechanism
HLS5 binds SUMO-1 and is SUMOylated in vivo.
It also binds Ubc9 and PIAS1, and has a global effect on
SUMOylation by causing the degradation of PIAS1 and Ubc9 through
its coiled-coil domain (43,44). In addition, HLS5 also increases
the SUMOylation of some proteins. It is possible that the effects
of HLS5 are concentration-dependent, in that it may promote greater
cell death at higher concentrations (47).
Application
Ligation inhibitors can be used to treat diseases
such as Parkinson's disease, diabetes, Huntington's disease,
familial neuronal intranuclear inclusion disease, Alzheimer's
disease, neuronal intranuclear inclusion disease, cancer (48–52), polyglutamine disease and human
immunodeficiency virus (HIV) infection (43,44).
4. Category 3: SENP inhibitors
Inhibitors of deSUMOylation enzymes
[WO064887 (53) and US0302525
(54)]
Structure
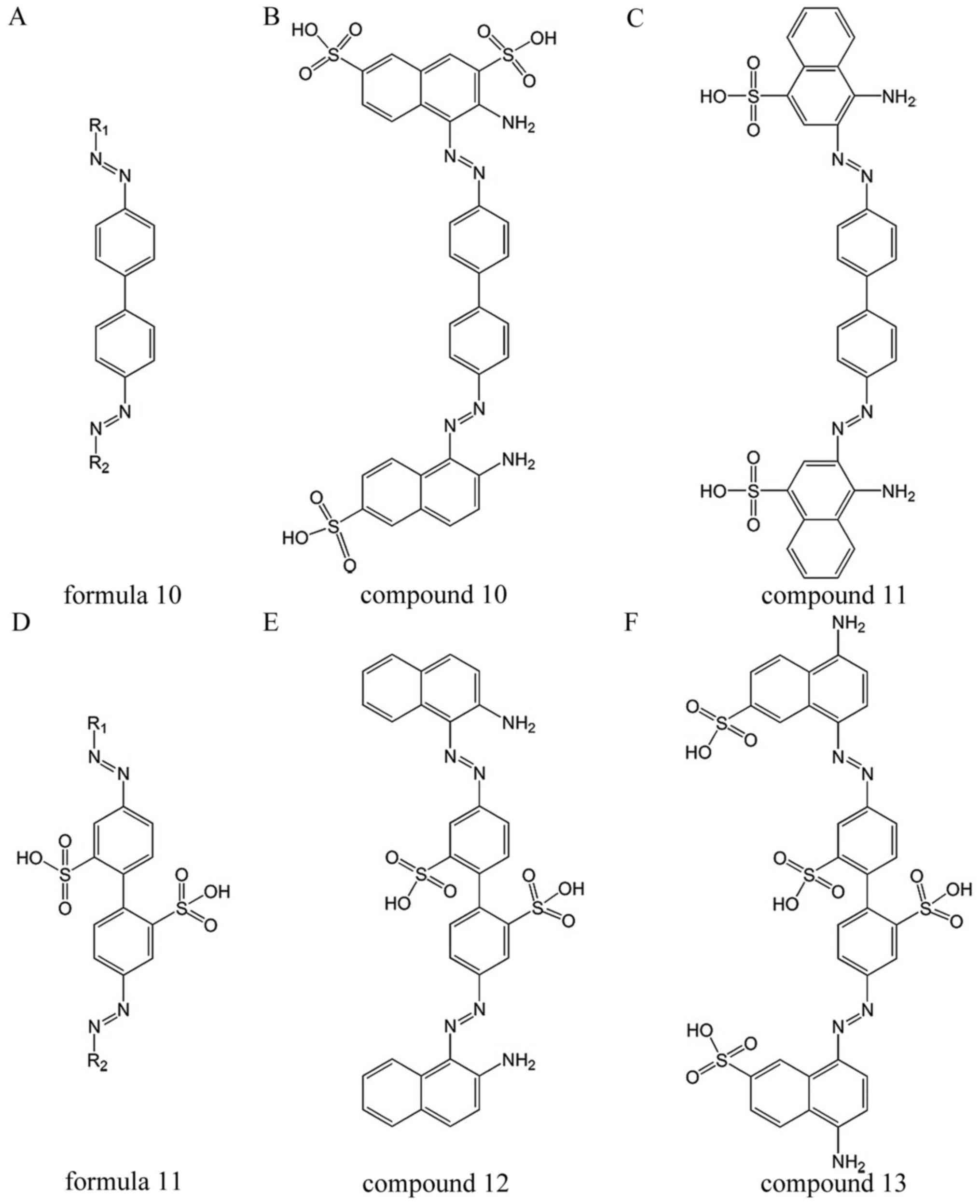
Compounds 10–11 in Fig. 7 are derived from formula 10 and
compounds 12–13 are derived from formula 11 (53,54). They inhibit SENPs and also inhibit
HIV infection (55).
Mechanism
SENPs share a conserved catalytic domain that is a
cysteine protease of ~225 amino acids (11). Nuclear magnetic resonance and
enzyme kinetics data indicate that SENP inhibitors bind in the
vicinity of the catalytic center and inhibit the enzyme by a mixed
inhibitory mechanism (56). The
catalytic domain crystal structures of SENP1, SENP2 and SENP7 are
very similar (10). As a result,
the inhibitors disclosed herein mostly inhibit SENP1, SENP2 and
SENP7.
Application
The SENP inhibitors may sensitize target
cells/cancer cells to DNA-damaging therapy, and then minimize or
eliminate harm to healthy cells at reduced doses. Inhibitors of
deSUMOylation significantly reduce viral infectivity, and limit
maintenance of the latent HIV reservoir, which may be valuable to
future HIV eradication strategies (57). The SENP inhibitors described
herein may also be used to treat cardiovascular disease,
neurodegenerative diseases and diabetes in a preventative manner
(53,54).
An SENP1 inhibitor: small hairpin RNA
[shRNA; CN 104436196 (58)]
Structure
The sequence of the shRNA described herein is
5′-CCGGGCGCCAGAUUGAAGAACAGAACUCGAGUUCUGUUCUUCAAUCUGGCGCUUUUU-3′
(SEQ ID no:1) (58).
Mechanism
The SENP1 inhibitor described herein is an shRNA,
including the shRNA recombinant vector and host cells containing
the recombinant vector (58). As
a precursor of small interfering RNA, shRNA can inhibit expression
of the SENP1 protein via the RNA interference mechanism, leading to
inhibition of cell proliferation and induction of apoptosis of
malignant tumor cells (58).
Application
The SENP1 inhibitors described herein can be used
for the preparation of certain special agents that can prevent or
treat malignant hematological tumors, and those that inhibit the
proliferation of malignant hematological tumor cells and/or promote
apoptosis in malignant hematological tumor cells (58).
SENP1 inhibitors [CN 103961348 (59)]
Structure
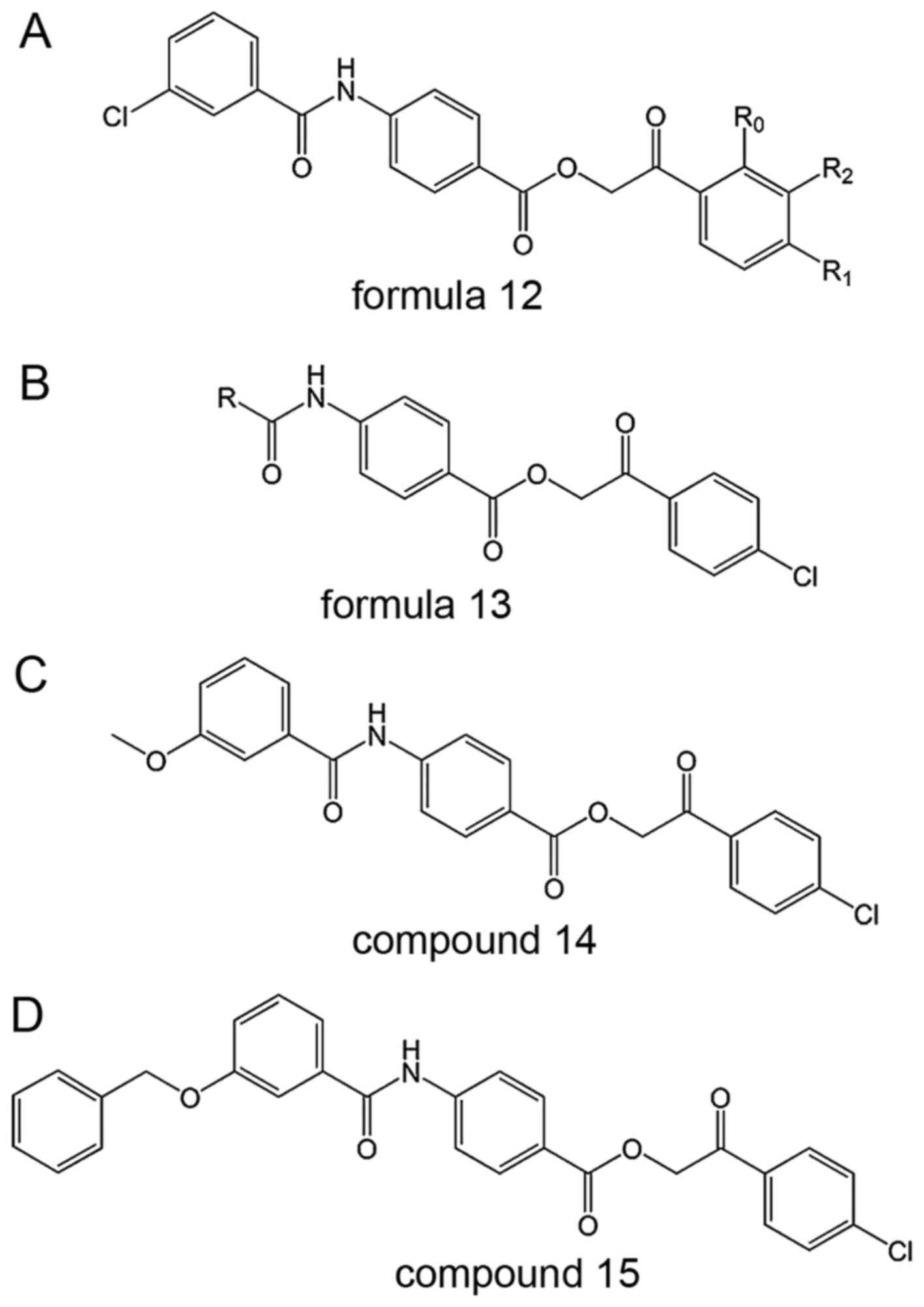
These SENP1 inhibitors were derived from formula 12
and 13 in Fig. 8 (59), of which the representatives are
2-(4-chlorophenyl)-2-oxoethyl 4-benzamidobenzoate derivatives
(60).
Mechanism
The compounds in this patent inhibited the activity
of SENP1 in vitro and certain cloned prostate cancer strains
(59). Compounds 14 and 15
(Fig. 8) have been reported to
have IC50 values of 2.38 and 1.08 µM,
respectively (10).
Application
SENP1 affects the function of hypoxia-inducible
factor 1α in tumor angiogenesis, thus promoting the development of
various tumors (61). The SENP1
inhibitors described herein can inhibit the growth of prostate
cancer cells stimulated by androgen (62–64). Thus, these inhibitors may be
useful candidate anticancer drugs.
SENP 2 inhibitors [CN 103877078 (65)]
Structure
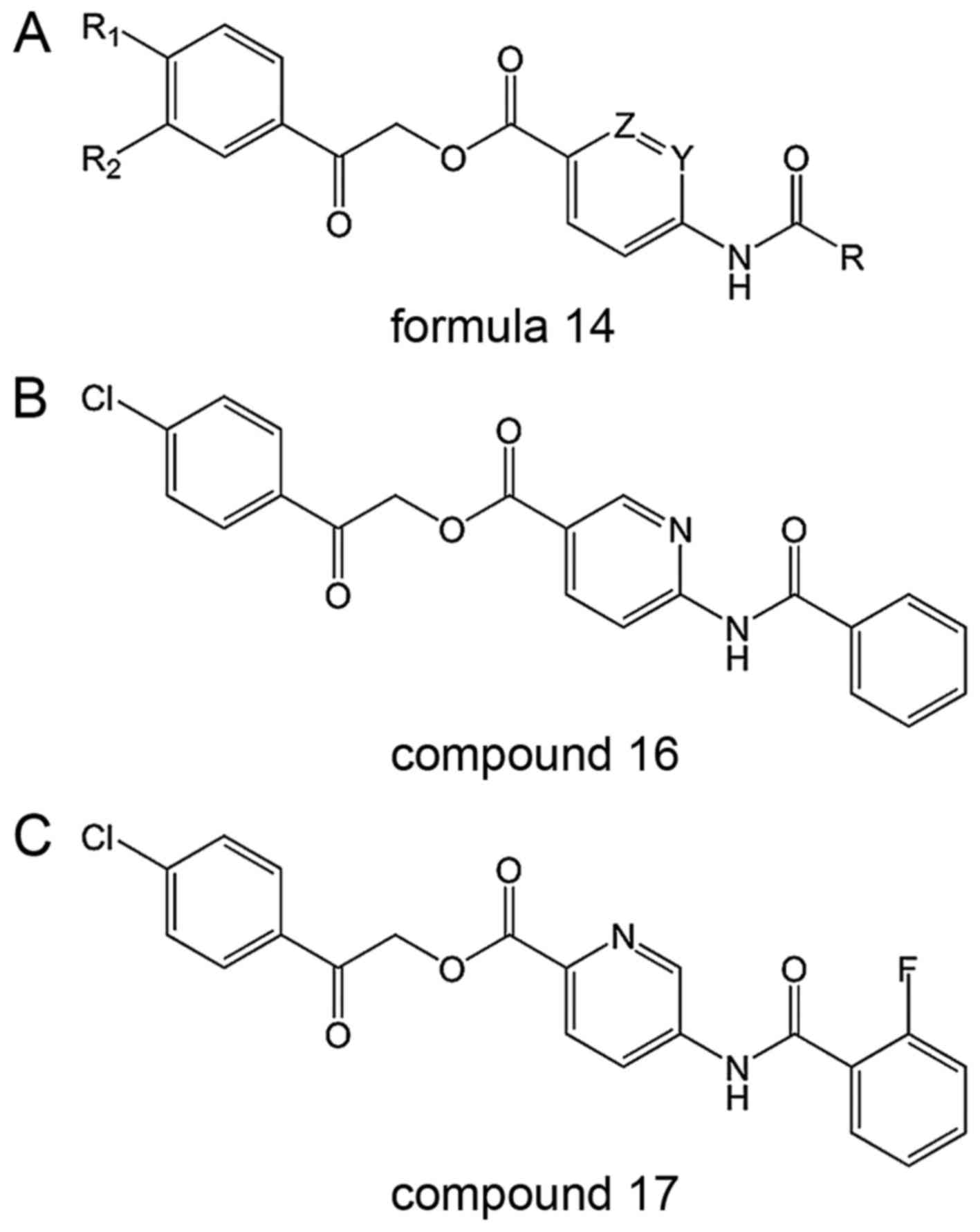
These inhibitors comprise SENP2 inhibitors
containing formula 14 (Fig. 9) as
an active ingredient, including its pharmacological derivatives or
acceptable salts. This patent has 864 compounds derived from
formula 14, differing in Z, Y, R, R1 and R2, compounds 16 and 17
are used here as two examples (65).
Mechanism
The SENP2 inhibitors described have high efficiency
and low toxicity, and significantly inhibit SENP2 activity in
vitro (65).
Application
The inhibitors of SENP2 described herein can be used
to treat breast cancer. By removing the SUMO modification of Pc2 in
the protein regulator of cytokinesis 1 protein, the regulation of
polycomb group target gene activity is achieved. SENP2 is involved
in the regulation of cell differentiation and development, and is
highly expressed in a large number of patients with breast cancer
(65). Thus, SENP2 inhibitors may
be novel drug candidates for anticancer therapy.
5. Discussion
The proteasome inhibitors, bortezomib and
carfizomib, which target the ubiquitin pathway, have been used for
the treatment of MM and hematologic malignancies (66). The second-generation proteasome
inhibitor, carfizomib, induces responses in a minority of patients
with MM that relapsed from or were refractory to bortezomib
treatment, and experienced dose-limiting peripheral neuropathy.
However, carfizomib still results in some adverse reactions that
are commonly shared with other antineoplastic agents. To identify
more effective agents with less toxicity, inhibitors targeting the
SUMOylation pathway have been investigated.
This review summarizes compounds inhibiting the
SUMOylation pathway that were described in patents written in
English or Chinese between 2012 and 2015. Among these compounds,
SAE and SENP1/2 inhibitors appear to have the greatest therapeutic
potential, as once protein maturation or activation is blocked,
downstream biological events will be inhibited. However, SAE, Ubc9
and SENP1/2 inhibitors do not have target selectivity, whereas SUMO
E3 ligases inhibitors may be more selective with fewer side
effects.
Research studies should not just focus on SUMO
inhibitors, but should also take SUMO activators, such as cysteine
protease polypeptides, into consideration (67–70). These proteins function as SENPs,
with the ability to cleave SUMO from a target protein and/or cleave
the precursor form of SUMO to release its active form. These SENP
analogs (cysteine protease poly-peptides) may be useful in the
treatment of various cancers and some other diseases related to
cysteine family members (71).
PIAS1 is a breast cancer suppressor protein (72). An artificial SUMO ligase
reportedly promotes the process of ligation (73).
The application of SUMO inhibitors for treating
cancer is of utmost importance. Thus, additional studies should
focus on identifying the roles of SUMO enzymes in different cancer
types. In addition, an understanding of the basic biology and
specificity/selectivity of SUMO inhibitors is required before they
can be identified and developed for use in the clinic. Although, to
date, no SUMO inhibitors have been tested in humans, and further
experiments are required in cells and animals, compounds targeting
the SUMOylation pathway may represent a new direction for the
treatment of cancer and other diseases.
Acknowledgments
This study was sponsored by the grants from the
National Natural Science Foundation of China (grant no. 81571568);
the Jiangsu Province's Key Provincial Talents Program (grant no.
RC201170); the Priority Academic Program Development of Jiangsu
Higher Education Institutions (PAPD); the Six Talents Peak projects
of Jiangsu Province (to J.-F.W. and H.Y.).
References
|
1
|
Herrmann J, Lerman LO and Lerman A:
Ubiquitin and ubiquitin-like proteins in protein regulation. Circ
Res. 100:1276–1291. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Flotho A and Melchior F: Sumoylation: A
regulatory protein modification in health and disease. Annu Rev
Biochem. 82:357–385. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bohren KM, Nadkarni V, Song JH, Gabbay KH
and Owerbach D: A M55V polymorphism in a novel SUMO gene (SUMO-4)
differentially activates heat shock transcription factors and is
associated with susceptibility to type I diabetes mellitus. J Biol
Chem. 279:27233–27238. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Geiss-Friedlander R and Melchior F:
Concepts in sumoylation: A decade on. Nat Rev Mol Cell Biol.
8:947–956. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tatham MH, Jaffray E, Vaughan OA, Desterro
JM, botting CH, Naismith JH and Hay RT: Polymeric chains of SUMO-2
and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and
Ubc9. J Biol Chem. 276:35368–35374. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang W and Paschen W: SUMO proteomics to
decipher the SUMO-modified proteome regulated by various diseases.
Proteomics. 15:1181–1191. 2015. View Article : Google Scholar :
|
|
7
|
Hannich JT, Lewis A, Kroetz MB, Li SJ,
Heide H, Emili A and Hochstrasser M: Defining the SUMO-modified
proteome by multiple approaches in Saccharomyces cerevisiae. J Biol
Chem. 280:4102–4110. 2005. View Article : Google Scholar
|
|
8
|
Hecker CM, Rabiller M, Haglund K, Bayer P
and Dikic I: Specification of SUMO1- and SUMO2-interacting motifs.
J Biol Chem. 281:16117–16127. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song J, Durrin LK, Wilkinson TA, Krontiris
TG and Chen Y: Identification of a SUMO-binding motif that
recognizes SUMO-modified proteins. Proc Natl Acad Sci USA.
101:14373–14378. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kumar A and Zhang KY: Advances in the
development of SUMO specific protease (SENP) inhibitors. Comput
Struct Biotechnol J. 13:204–211. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hickey CM, Wilson NR and Hochstrasser M:
Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol.
13:755–766. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bettermann K, Benesch M, Weis S and
Haybaeck J: SUMOylation in carcinogenesis. Cancer Lett.
316:113–125. 2012. View Article : Google Scholar
|
|
13
|
Olsen SK, Capili AD, Lu X, Tan DS and Lima
CD: Active site remodelling accompanies thioester bond formation in
the SUMO E1. Nature. 463:906–912. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kumar A, Ito A, Hirohama M, Yoshida M and
Zhang KY: Identification of sumoylation inhibitors targeting a
predicted pocket in Ubc9. J Chem Inf Model. 54:2784–2793. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kumar A, Ito A, Hirohama M, Yoshida M and
Zhang KY: Identification of sumoylation activating enzyme 1
inhibitors by structure-based virtual screening. J Chem Inf Model.
53:809–820. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mattoscio D and Chiocca S: SUMO pathway
components as possible cancer biomarkers. Future Oncol.
11:1599–1610. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schulz S, Chachami G, Kozaczkiewicz L,
Winter U, Stankovic-Valentin N, Haas P, Hofmann K, Urlaub H, Ovaa
H, Wittbrodt J, et al: Ubiquitin-specific protease-like 1 (USPL1)
is a SUMO isopeptidase with essential, non-catalytic functions.
EMbO Rep. 13:930–938. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shin EJ, Shin HM, Nam E, Kim WS, Kim JH,
Oh BH and Yun Y: DeSUMOylating isopeptidase: A second class of SUMO
protease. EMBO Rep. 13:339–346. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shirong L and Wei H: Sumoylation - a
muitffunctional post-translational protein modification. J Med Mol
Biol. 3:212–215. 2006.
|
|
20
|
Liu X, Xu Y, Pang Z, Guo F, Qin Q, Yin T,
Sang Y, Feng C, Li X, Jiang L, et al: Knockdown of SUMO-activating
enzyme subunit 2 (SAE2) suppresses cancer malignancy and enhances
chemotherapy sensitivity in small cell lung cancer. J Hematol
Oncol. 8:672015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen SF, Gong C, Luo M, Yao HR, Zeng YJ
and Su FX: Ubc9 expression predicts chemoresistance in breast
cancer. Chin J Cancer. 30:638–644. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rabellino A, Carter B, Konstantinidou G,
Wu SY, Rimessi A, byers LA, Heymach JV, Girard L, Chiang CM,
Teruya-Feldstein J, et al: The SUMO E3-ligase PIAS1 regulates the
tumor suppressor PML and its oncogenic counterpart PML-RARA. Cancer
Res. 72:2275–2284. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li T, Huang S, Dong M, Gui Y and Wu D:
Prognostic impact of SUMO-specific protease 1 (SENP1) in prostate
cancer patients undergoing radical prostatectomy. Urol Oncol.
31:1539–1545. 2013. View Article : Google Scholar
|
|
24
|
Bossis G, Sarry JE, Kifagi C, Ristic M,
Saland E, Vergez F, Salem T, Boutzen H, Baik H, Brockly F, et al:
The ROS/SUMO axis contributes to the response of acute myeloid
leukemia cells to chemotherapeutic drugs. Cell Rep. 7:1815–1823.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dassouki Z, Sahin U, El Hajj H, Jollivet
F, Kfoury Y, Lallemand-Breitenbach V, Hermine O, de Thé H and
Bazarbachi A: ATL response to arsenic/interferon therapy is
triggered by SUMO/PML/RNF4-dependent Tax degradation. Blood.
125:474–482. 2015. View Article : Google Scholar
|
|
26
|
Chen Y, Horne D, Li YJ, Ma LY,
Perkins-Harki AL and Su Y: Multi-valent poly-ubl chain inhibitors
and methods of use. US Patent 0302815. Filed February 21, 2012;
issued August 5, 2014.
|
|
27
|
Zhao B, Villhauer EB, Bhuripanyo K,
Kiyokawa H, Schindelin H and Yin J: SUMO-mimicking peptides
inhibiting protein SUMOylation. Chembiochem. 15:2662–2666. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Butterworth KT, Coulter JA, Jain S, Forker
J, McMahon SJ, Schettino G, Prise KM, Currell FJ and Hirst DG:
Evaluation of cytotoxicity and radiation enhancement using 1.9 nm
gold particles: Potential application for cancer therapy.
Nanotechnology. 21:2951012010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li YJ, Perkins AL, Su Y, Ma Y, Colson L,
Horne DA and Chen Y: Gold nanoparticles as a platform for creating
a multivalent poly-SUMO chain inhibitor that also augments ionizing
radiation. Proc Natl Acad Sci USA. 109:4092–4097. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chithrani DB, Jelveh S, Jalali F, van
Prooijen M, Allen C, Bristow RG, Hill RP and Jaffray DA: Gold
nanoparticles as radiation sensitizers in cancer therapy. Radiat
Res. 173:719–728. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Langston SP, Olhava EJ and Vyskocil S:
Inhibitors of E1 activating enzymes. EP Patent 2402334 A1. Filed
February 2, 2006; issued Junuary 26, 2012.
|
|
32
|
Soucy TA, Smith PG, Milhollen MA, Berger
AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP,
Critchley S, et al: An inhibitor of NEDD8-activating enzyme as a
new approach to treat cancer. Nature. 458:732–736. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Diffey MO, England Db, Hu ZG, Ito M,
Langston SP, Mcintyre C, Mizutani H and Xu H: Heteroaryl compounds
useful as inhibitors of SUMO activating enzyme. WO Patent
2015002994 A2. Filed July 1, 2014; issued January 8, 2015.
|
|
34
|
Driscoll JJ, Pelluru D, Lefkimmiatis K,
Fulciniti M, Prabhala RH, Greipp PR, Barlogie B, Tai YT, Anderson
KC, Shaughnessy JD Jr, et al: The sumoylation pathway is
dysregulated in multiple myeloma and is associated with adverse
patient outcome. Blood. 115:2827–2834. 2010. View Article : Google Scholar :
|
|
35
|
Kessler JD, Kahle KT, Sun T, Meerbrey KL,
Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, et
al: A SUMOylation-dependent transcriptional subprogram is required
for Myc-driven tumorigenesis. Science. 335:348–353. 2012.
View Article : Google Scholar
|
|
36
|
Chen Y, Li YJ, Divlianska D, Bobkova E and
Roth G: Singleton inhibitors of SUMOylation enzymes and methods for
their use. US Patent 20130317101 A1. Filed May 9, 2013; issued
November 28, 2013.
|
|
37
|
Chen Y and Li YJ: Singleton inhibitors of
SUMOylation enzymes and methods for their use. WO Patent 064898 A1.
Filed May 9, 2013; issued January 19, 2016.
|
|
38
|
Maas M, Nelemans PJ, Valentini V, Crane
CH, Capirci C, Rödel C, Nash GM, Kuo LJ, Glynne-Jones R,
García-Aguilar J, et al: Adjuvant chemotherapy in rectal cancer:
Defining subgroups who may benefit after neoadjuvant chemoradiation
and resection: A pooled analysis of 3,313 patients. Int J Cancer.
137:212–220. 2015. View Article : Google Scholar
|
|
39
|
García-Aguilar J, Hernandez de Anda E,
Sirivongs P, Lee SH, Madoff RD and Rothenberger DA: A pathologic
complete response to preoperative chemoradiation is associated with
lower local recurrence and improved survival in rectal cancer
patients treated by mesorectal excision. Dis Colon Rectum.
46:298–304. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen Y and Li YJ: Bicyclic and tricyclic
inhibitors of SUMOylation enzymes and methods for their use. WO
Patent 064897 A2. Filed November 9, 2011; issued May 18, 2012.
|
|
41
|
Chen Y, Li YJ, Divlianska D, Bobkova E,
Roth G, Jun PU and Khan P: Bicyclic and tricyclic inhibitors of
SUMOylation enzymes and methods for their use. US Patent
20130245032 A1. Filed May 9, 2013; issued September 19, 2013.
|
|
42
|
Chen Y, Li YJ, Divlianska D, bobkova E,
Roth G, Jun PU and Khan P: Inhibitors of small ubiquitin-like
modifier enzymes with substituted pyrrolo(2,3-B)quinoxalines. US
Patent 9045483 B2. Filed May 9, 2013; issued June 2, 2015.
|
|
43
|
Lalonde JP, Lim R, Scaife R, Gallagher S
and Klinken SP: HLS-5 a control SUMOylation agent. EP Patent
2545935 A1. Filed October 6, 2006; issued January 16, 2013.
|
|
44
|
Klinken SP, Lalonde JP, Lim R, Scaife R
and Gallagher S: SUMOylation control agent and uses thereof. US
Patent 20130330738 A1. Filed Junuary 4, 2013; issued December 12,
2013.
|
|
45
|
Knuutila S, Aalto Y, Autio K, Björkqvist
AM, El-Rifai W, Hemmer S, Huhta T, Kettunen E, Kiuru-Kuhlefelt S,
Larramendy ML, et al: DNA copy number losses in human neoplasms. Am
J Pathol. 155:683–694. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lalonde JP, Lim R, Ingley E, Tilbrook PA,
Thompson MJ, McCulloch R, Beaumont JG, Wicking C, Eyre HJ,
Sutherland GR, et al: HLS5, a novel RBCC (ring finger, B box,
coiled-coil) family member isolated from a hemopoietic lineage
switch, is a candidate tumor suppressor. J Biol Chem.
279:8181–8189. 2004. View Article : Google Scholar
|
|
47
|
Kimura F, Suzu S, Nakamura Y, Nakata Y,
Yamada M, Kuwada N, Matsumura T, Yamashita T, Ikeda T, Sato K, et
al: Cloning and characterization of a novel RING-B-box-coiled-coil
protein with apoptotic function. J Biol Chem. 278:25046–25054.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Alshareeda AT, Negm OH, Green AR, Nolan C,
Tighe P, Albarakati N, Sultana R, Madhusudan S, Ellis IO and Rakha
EA: SUMOylation proteins in breast cancer. Breast Cancer Res Treat.
144:519–530. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Coppola D, Parikh V, boulware D and Blanck
G: Substantially reduced expression of PIAS1 is associated with
colon cancer development. J Cancer Res Clin Oncol. 135:1287–1291.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen P, Zhao D, Sun Y, Huang L, Zhang S
and Yuan Y: Protein inhibitor of activated STAT-1 is downregulated
in gastric cancer tissue and involved in cell metastasis. Oncol
Rep. 28:2149–2155. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wei J, Costa C, Ding Y, Zou Z, Yu L,
Sanchez JJ, Qian X, Chen H, Gimenez-Capitan A, Meng F, et al: mRNA
expression of bRCA1, PIAS1, and PIAS4 and survival after
second-line docetaxel in advanced gastric cancer. J Natl Cancer
Inst. 103:1552–1556. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hoefer J, Schäfer G, Klocker H, Erb HH,
Mills IG, Hengst L, Puhr M and Culig Z: PIAS1 is increased in human
prostate cancer and enhances proliferation through inhibition of
p21. Am J Pathol. 180:2097–2107. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen Y, Li S, Li S, Li YJ, Su Y, Wong S
and Zaia J: Inhibitors of deSUMOylation enzymes and methods for
their use. WO Patent 2012064887 A1. Filed November 9, 2011; issued
May 18, 2012.
|
|
54
|
Chen Y: Methods of identifying SENP1
inhibitors. US Patent 20140302525 A1. Filed April 7, 2014; issued
October 9, 2014.
|
|
55
|
Madu IG, Namanja AT, Su Y, Wong S, Li YJ
and Chen Y: Identification and characterization of a new chemotype
of noncovalent SENP inhibitors. ACS Chem Biol. 8:1435–1441. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chen CH, Namanja AT and Chen Y:
Conformational flexibility and changes underlying activation of the
SUMO-specific protease SENP1 by remote substrate binding. Nat
Commun. 5:49682014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Madu IG, Li S, Li B, Li H, Chang T, Li YJ,
Vega R, Rossi J, Yee JK, Zaia J, et al: A Novel Class of HIV-1
Antiviral Agents Targeting HIV via a SUMOylation-Dependent
Mechanism. Sci Rep. 5:178082015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang LS, Sun HY, Wu ZZ, Xiao FJ, Wang H
and Yang YF: SENP1 protein inhibitors and uses thereof. CN Patent
104436196 A. Filed September 23, 2013; issued March 25, 2015.
|
|
59
|
Zhang J, Cheng JK, Lu SY, Cheng YY, Zhang
JM and Huang M: Senp1 small molecule inhibitors and their
applications. CN Patent 103961348 B. Filed February 5, 2013; issued
August 17, 2016.
|
|
60
|
Chen Y, Wen D, Huang Z, Huang M, Luo Y,
Liu B, Lu H, Wu Y, Peng Y and Zhang J:
2-(4-Chlorophenyl)-2-oxoethyl 4-benzamidobenzoate derivatives, a
novel class of SENP1 inhibitors: Virtual screening, synthesis and
biological evaluation. Bioorg Med Chem Lett. 22:6867–6870. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Cheng J, Kang X, Zhang S and Yeh ET:
SUMO-specific protease 1 is essential for stabilization of
HIF1alpha during hypoxia. Cell. 131:584–595. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Bawa-Khalfe T, Cheng J, Lin SH, Ittmann MM
and Yeh ET: SENP1 induces prostatic intraepithelial neoplasia
through multiple mechanisms. J Biol Chem. 285:25859–25866. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yamaguchi T, Sharma P, Athanasiou M, Kumar
A, Yamada S and Kuehn MR: Mutation of SENP1/SuPr-2 reveals an
essential role for desumoylation in mouse development. Mol Cell
Biol. 25:5171–5182. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Cheng J, Bawa T, Lee P, Gong L and Yeh ET:
Role of desumoylation in the development of prostate cancer.
Neoplasia. 8:667–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhang J, Cheng JK, Huang M, Chen YY, Huang
ZM, Lu SY and Shi T: SENP2 small molecule inhibitor and
applications thereof. CN Patent 103877078 A. Filed December 20,
2012; issued June 25, 2014.
|
|
66
|
Dou QP and Zonder JA: Overview of
proteasome inhibitor-based anticancer therapies: Perspective on
bortezomib and second generation proteasome inhibitors versus
future generation inhibitors of ubiquitin-proteasome system. Curr
Cancer Drug Targets. 14:517–536. 2014. View Article : Google Scholar
|
|
67
|
Chen JW, Ink BS and Lewis AP: Cysteine
protease polypeptides GB2371801. 2002
|
|
68
|
Ink BS and Lewis AP: Cysteine protease
polypeptide GB2372504. 2002
|
|
69
|
Chen JW, Ink BS and Lewis AP: Cysteine
protease polypeptide GB2372994. 2002
|
|
70
|
Ink BS: Protease polypeptide GB2382078.
2003
|
|
71
|
Góra J and Latajka R: Involvement of
cysteine proteases in cancer. Curr Med Chem. 22:944–957. 2015.
View Article : Google Scholar
|
|
72
|
Wu HJ, Liu WD, Hong YD and Guo QP:
Identification of a breast cancer suppressor and methods for use
CN101923095. 2010
|
|
73
|
German RA and Sangita P: Modulation of
cellular protein function by artifical SUMO ligases. US Patent
20120246757 A1. Filed March 22, 2012; issued September 27,
2012.
|