Introduction
Glioma is the most common primary brain tumor,
accounting for 80% of malignant brain tumors, and has a high rate
of recurrence, mortality and morbidity (1). The excessive proliferation,
resistance to apoptosis and high invasion rate of glioma cells lead
to the characteristic lack of boundaries with the surrounding brain
tissues, regardless of the differentiation of glioma cells
(2). It is thus challenging to
completely eradicate the tumor by surgery. With current standard
management by surgical resection and precise radiotherapy and
chemotherapy, the median survival time of patients with
glioblastoma multiforme is only 12–15 months, and the 5-year
survival rate remains <3% (3).
Therefore, elucidation of the mechanism underlying the
tumorigenesis, tumor development and clonal evolution of glioma is
essential for the identification of an effective target for gene
therapy.
MicroRNAs (miRNAs) are a class of small endogenous
non-coding RNAs that are ~22 nucleotides in length. These regulate
gene expression post-transcriptionally, primarily through complete
or incomplete integration with the mRNA 3′ untranslated region
(3′UTR) of the target gene. A single miRNA molecule may regulate
hundreds of target genes by forming a complex regulatory network
with the target genes. miRNAs serve broad roles in embryonic
development, cell proliferation, differentiation, apoptosis, cell
cycle progression, angiogenesis and a variety of other biological
processes (4). Studies have shown
that the aberrant expression of specific miRNAs is closely
associated with uncontrolled proliferation, apoptosis blockage,
invasion and migration, as well as resistance to chemotherapy
(5–7). MicroRNA-130b (miR-130b) is an
important member of the microRNA-130 family. A previous study
demonstrated that in breast tissues, miR-130b downregulates
p21Waf1/Cip1, a cell cycle inhibitor, which in turn
inhibits mammary epithelial cell apoptosis and promotes tumor
growth (8). In addition, Dong
et al (9) reported that a
mutant p53 gene induced epithelial-mesenchymal transition by
regulating the miR-130b-zinc finger E-box binding homeobox 1 axis
in endometrial carcinoma, resulting in the initiation of tumor
development. They also observed that in comparison with normal
tissues, miR-130b expression was significantly lower in endometrial
carcinoma tissues, and the long-term follow-up of patients with
endometrial carcinoma revealed a longer survival time for patients
with high levels of miR-130b expression (9). These studies suggest that miR-130b
is capable of functioning as either a tumor suppressor or a tumor
promoter, depending on the type of tumor involved. A study
conducted by Malzkorn et al (10) concluded that the expression of
miR-130b significantly increased in four patients with glioma as
they progressed from World Health Organization (WHO) stage II to
IV. However, the functions and mechanisms of miR-130b in glioma
require elucidation.
In the present study, reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) was
used to determine the expression pattern of miR-130b in 30 glioma
patients and 3 glioma cell lines. U87 cells were transfected with
an miR-130b inhibitor to downregulate the expression of miR-130b.
The biological characteristics of the proliferation, cell cycle,
cell invasion and migration of the transfected U87 cells were then
assessed. Western blotting and luciferase reporter gene technology
were used to verify the expression of phosphatase and tensin
homolog (PTEN), a downstream target gene of miR-130b, and its
underlying mechanism.
Materials and methods
Clinical samples and cell lines
Human glioma tumor tissues were obtained from the
surgical resection of glioma patients (52–73 years old; 21 males
and 9 females) in the Fuzhou General Hospital (Fuzhou, China)
between January 2015 and December 2016. A total of 30 glioma
samples were thoroughly reviewed by an experienced neuropathologist
according to the 2007 WHO classification (11), resulting in 10 glioma samples
being classified as grades I and II, 10 as grade III and 10 as
grade IV. In addition, 5 non-neoplastic brain specimens were
obtained from patients with traumatic brain injury during
decompression surgery. All tissue samples were frozen in liquid
nitrogen immediately following resection and stored at −80°C. Prior
patient consent was obtained for the use of these clinical
materials for research purposes. In addition, ethical approval for
the study was obtained from the Ethics Committee of Fuzhou General
Hospital. The normal human astrocyte (NHA) cell line was purchased
from ScienCell Research Laboratories, Inc. (Carlsbad, CA, USA). The
glioma LN229, U87 and U251 cell lines used in this study were
obtained in 2013 from the Institute of Biochemistry and Cell
Biology (Shanghai Institutes for Biological Sciences, Chinese
Academy of Science, Shanghai, China).
Cell culture and transfection
Cells were grown in Dulbecco's modified Eagle's
medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco,
Grand Island, NY, USA) and 1% penicillin/streptomycin in a 5%
CO2 atmosphere at 37°C. U87 cells (1×105)
were seeded into 6-well plates and transfected with negative
control (NC), miR-130b inhibitor and PTEN-targeted small
interfering RNA (siPTEN; sequence, 5′-GACUUGAAGGCGUAUACAGtt-3′)
purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China)
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. The medium
was removed following a 24 h transfection, and the cells were
placed in the complete medium and maintained at 37°C in 5%
CO2.
RNA extraction and RT-qPCR
Total RNAs were extracted from cultured cells and
fresh glioma tissues using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) or total miRNAs using mirVana (Ambion;
Thermo Fisher Scientific, Inc.) kits according to the
manufacturer's protocols. Reverse transcription was then performed
by using a Reverse Transcription kit (Promega Corporation, Madison,
WI, USA) in accordance with the instructions of the manufacturer.
Gene-specific primers were used to synthesize miR-130b cDNA from
total RNA, in accordance with the recommended protocol for the
miRNA-specific TaqMan miRNA Assay kit (Applied Biosystems; Thermo
Fisher Scientific, Inc.); U6 small nuclear RNA (snRNA) was used as
an internal control. The expression level of miR-130b and PTEN was
examined by qPCR with a SYBR-Green PCR Master Mix kit in
conjunction with an ABI-Prism 7300 system (both Applied Biosystems;
Thermo Fisher Scientific, Inc.). The primer sequences were as
follows: PTEN forward, 5′-TTTGAAGACCATAACCCACCAC-3′ and reverse,
5′-ATTACACCAGTTCGTCCCTTTC-3′; GAPDH forward,
5′-TCGGAGTCAACGGATTTGG-3′ and reverse, 5′-CATGGGTGGAATCATATTGGA-3′;
U6 RT-primer, 5′-CGCTTCACGAATTTGCGTGTCAT-3′, forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
The conditions for PCR were as follows: stage 1, 95°C for 10 min;
stage 2, 40 cycles at 95°C for 15 sec, 57°C for 1 min; stage 3
(dissociation stage), 95°C for 15 sec, 60°C for 15 sec, 95°C for 15
sec. The relative expression levels of mature miR-130b and PTEN
mRNA were calculated by the 2−ΔΔCq method (12) and normalized to U6 snRNA and GAPDH
mRNA levels, respectively.
Western blotting
Total cell lysates from different experiments were
obtained by lysing the cells in radioimmunoprecipitation assay
buffer (Sigma, St. Louis, MO, USA). Protein concentrations were
determined using a BCA protein assay (Pierce; Thermo Fisher
Scientific, Inc.). Total protein from each sample (40 μg)
was resolved by 10% SDS-PAGE and transferred to PVDF membranes
(both from EMD Millipore, Billerica, MA, USA). The membranes were
incubated with primary antibodies PTEN (1:1,000 dilution; Abcam,
Cambridge, MA, USA), phosphorylated (p)-AKT (1:500 dilution;
sc-7985-R), p27 (1:500 dilution; sc-528) and cyc1in D1 (1:500
dilution; sc-70899) (all from Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA), anti-matrix metalloproteinase (MMP)-2 (1:500
dilution) and anti-MMP-9 (1:500 dilution) (both from Abcam)
overnight at 4°C, followed by incubation with a horseradish
peroxidase-conjugated donkey anti-rabbit secondary antibody
(sc-2317; 1:5,000 dilution; Santa Cruz Biotechnology, Inc.) or
HRP-conjugated donkey anti-mouse antibody (sc-2318;1:5,000
dilution; Santa Cruz Biotechnology, Inc.) at room temperature for
30 min. Membranes were stripped and then reprobed with a primary
antibody against GAPDH (1:1;000 dilution; Santa Cruz Biotechnology,
Inc.). The signal intensity was determined with ImageJ gel analysis
software version 1.48u (National Institutes of Health, Bethesda,
MD, USA). GAPDH was used as an endogenous protein for
normalization. Specific bands were visualized using enhanced
chemiluminescence detection (Thermo Fisher Scientific, Inc.).
Luciferase reporter assay
The 3′UTR sequence of PTEN predicted to interact
with miR-130b or a mutated sequence with the predicted target sites
was synthesized and inserted into the XbaI and FseI
sites of a pGL3 control vector (Promega Corporation) by integrating
the search results of TargetScan and miRDB programs (13). These constructs were designated
pGL3-PTEN-3′-UTR and pGL3-PTEN-3′-UTR-mut, respectively. For the
reporter assay, U87 cells were plated onto 24-well plates and
transfected with pGL3, pGL3-PTEN-3′-UTR or pGL3-PTEN-3′-UTR-mut
vector and miR-130b inhibitor or NC vectors using the FuGENE HD
transfection reagent (Promega Corporation). A Renilla
luciferase vector pRL-SV50 (Promega Corporation) was co-transfected
in order to normalize the differences in transfection efficiency.
Subsequent to 48 h transfection, the cells were harvested and
assayed using a Dual-Luciferase Reporter Assay system (Promega
Corporation), according to the manufacturer's protocol.
Transfection was repeated in triplicate in three independent
experiments.
Proliferation assays
Cells (5×103/well) were plated in 96-well
plates and grown for 24, 48, 72 and 96 h following transfection.
Cell proliferation was documented every 24 h for 4 days using a
Cell Counting kit-8 (CCK-8) assay (Genview, Carlsbad, CA, USA). The
absorbance at a wavelength of 570 nm was detected using a
microplate reader.
Colony formation assay
The stably-transfected glioma cells (300 cells/well)
were seeded into 6-well plates and cultured in cell culture medium
for 2 weeks to allow colony formation. The culture medium was
refreshed every third day. The colonies were then fixed in 100%
methanol for 30 min and stained with crystal violet solution for 30
min. Subsequently, the number of macroscopically observable
colonies was recorded.
Cell cycle and apoptosis assays
The effects of miR-310b on the cell cycle and
apoptosis of U87 glioma cells were examined by flow cytometry. In
brief, the cells were harvested by trypsinization 48 h following
transfection. They were then washed three times with ice-cold
phosphate-buffered saline (PBS) and fixed with 70% ethanol
overnight at 4°C. The cell cycle and apoptotic rate were measured
by flow cytometry (BD Biosciences, San Jose, CA, USA) and the data
were analyzed using BD CellQuest™ PRO software (BD Biosciences).
Annexin V-APC/7-AAD Apoptosis Detection kit (KaiJi, Nanjing, China)
was used.
Transwell and scratch-wound assays
Cell invasion was determined using Transwell and
scratch-wound assays. For the Transwell assay, U87 cells were
transfected with miR-130b inhibitor or NC. Subsequent to incubation
for 48 h, 3×104 cells were transferred to the top of a
Matrigel-coated invasion chamber (BD Biosciences) in serum-free
DMEM. DMEM containing 10% FBS was added to the lower chamber. After
24 h, the non-invading cells were removed, and the invading cells
were fixed using 95% ethanol for 30 min, stained with 0.1% crystal
violet for 30 min and images were captured under a magnification of
×100. For the scratch-wound assay, miR-130b inhibitor or NC was
transfected into the cells in 6-well plates. The cell layers were
then scratched using a 20-μl sterile pipette tip to form
wound gaps. The wound location in the 6-well plates was marked and
the cells were then allowed to migrate into the cell-free wound in
serum-free DMEM for 24 h. The tests were repeated in three
independent experiments. The wound widths were measured and the
relative wound widths were calculated. Data are shown as means ±
standard deviation (SD) of 3 independent experiments.
Nude mouse tumorigenicity
U87 cells stably-transfected with miR-130b inhibitor
or NC were resuspended in PBS and 1.5×106 cells were
injected subcutaneously into the right flanks of male BALB/c
athymic (nude) mice (6–8 weeks old; weighing 20 ± 2 g obtained from
Shanghai SLAC Experimental Animal Co., Ltd., Shanghai, China). Mice
were maintained in a pathogen-free environment. The experiment was
conducted using 10 mice (n=5/group). Tumor volumes were measured at
different time-points using the formula: Tumor volume = length (mm)
× width2 (mm2)/2.
Statistical analysis
All data are shown as mean ± standard deviation, and
the experiments were repeated three times. Statistical analyses
were performed using a two-tailed Student's t-test with SPSS
version 12.0 software (SPSS, Inc., Chicago, IL, USA) and
multiple-group comparisons were analyzed using one-way ANOVA for
the subsequent individual group comparisons. P<0.05 was
considered to indicate a statistically significant difference.
Results
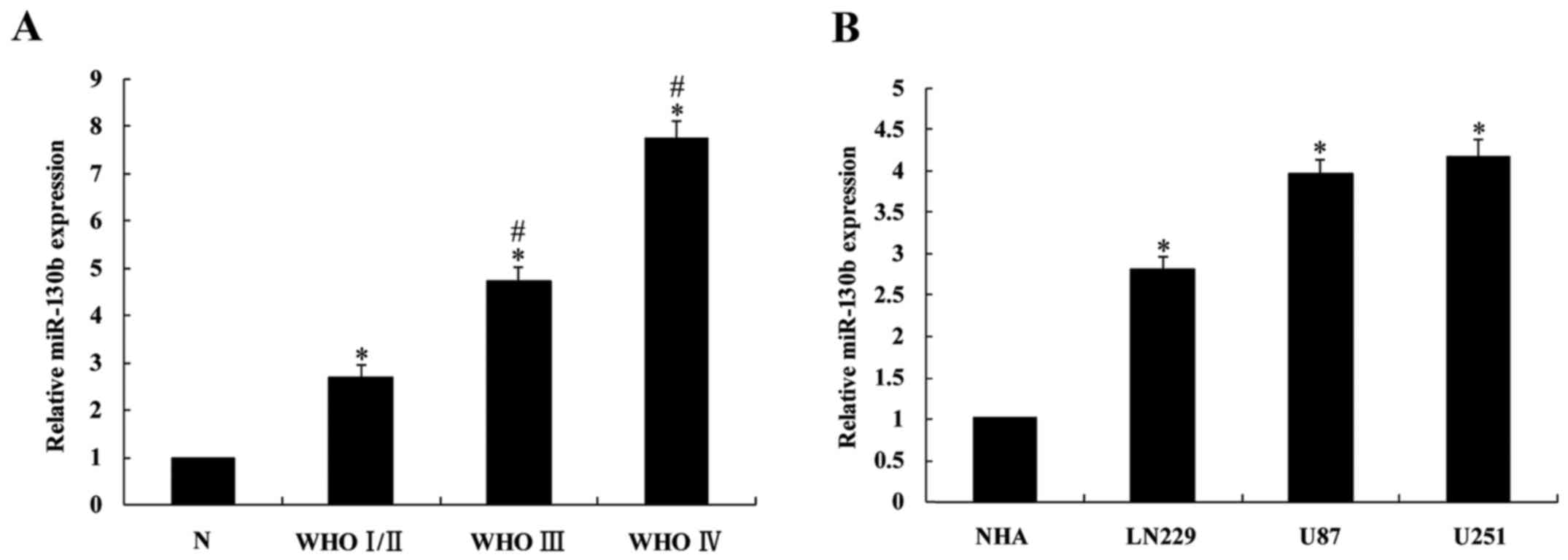
Upregulation of miR-130b in glioma
tissues and cells
The expression levels of miR-130b in 30 glioma
cases, 5 normal brain tissues and 3 glioma cell lines were detected
using RT-qPCR. The results demonstrated that miR-130b expression in
glioma tissues was significantly higher than that in normal brain
tissues (P<0.05; Fig. 1A), and
its expression in high level glioma tissues (III and IV) was
significantly higher (P<0.05) than in low grade glioma (I/II).
Furthermore, miR-130b expression in the 3 glioma cell lines was
significantly upregulated compared with that in the NHAs
(P<0.05; Fig. 1B).
PTEN is a potential downstream target of
miR-130b
Using the TargetScan bioinformatic database, the
target site for miR-130b interaction was detected in the 3′UTR of
the PTEN gene. Fig. 2A shows that
the target sequence was located at nucleotides 412–418 of the 3′UTR
region of the PTEN gene. Then, the 3′-UTR of a PTEN fragment
containing the miR-130b interaction site was ligated into a vector
at a region downstream of the luciferase reporter gene. The
luciferase reporter assay revealed that following transfection with
the miR-130b inhibitor, a significant increase in wild-type
luciferase activity was observed (P<0.05). By contrast, the
luciferase activity of the mutant showed no significant change
compared with that of the NC (P>0.05; Fig. 2B). The present study thus shows
that miR-130b directly binds the 3′UTR of the PTEN gene, thereby
inhibiting its expression.
Inhibition of the proliferation and
induction of apoptosis of U87 cells by miR-130b
To confirm that U87 glioma cell proliferation is
inhibited by miR-130b, U87 cells were transfected with miR-130b
inhibitor, and the CCK-8 colorimetric assay and colony formation
proliferation experiments were conducted to examine the inhibitory
effects of the miR-130b inhibitor on U87 cell proliferation. At ~24
h after transfection, the cells were harvested and seeded at a
density of 1×105 cells/well in 6-well plates. The cells
were collected at 24, 48, 72 and 96 h after plating, and cell
proliferation was examined. The results demonstrated that following
transfection of the miR-130b inhibitor, cell proliferation at 48,
72 and 96 h was significantly inhibited, with a lower proliferation
rate compared with that of the NC (P<0.05; Fig. 3A). Similarly, the results of the
colony formation assay suggested that the downregulation of
miR-130b significantly inhibited the colony formation of U87 cells,
compared with that of the NC (P<0.05; Fig. 3B and C). The cells transiently
transfected with the miR-130b inhibitor were subjected to cell
cycle analysis by flow cytometry. Compared with the NC, U87 cells
transfected with the miR-130b inhibitor underwent G0/G1 cell cycle
arrest (Fig. 3D and E).
Furthermore, transient transfection with the miR-130b inhibitor
significantly induced apoptosis of the U87 cells (Fig. 3F and G). These in vitro
experiments suggest that the reduced expression of miR-130b
suppressed glioblastoma cell proliferation via cell cycle arrest
and the promotion of cell apoptosis.
Inhibition of nude mouse tumorigenicity
by miR-130b
To substantiate the effect of the miR-130b inhibitor
in glioma carcinogenesis, U87 cells transfected with miR-130b
inhibitor or miR-130b NC were implanted into the right flanks of
BALB/c athymic mice by subcutaneous injection. At 28 days
post-injection, the mean volumes of tumors generated from the
miR-130b cells were significantly smaller than those originating
from NC cells (Fig. 4A–C).
Western blotting for the glioma xenograft tissues revealed that
expression of PTEN was higher in the miR-130b inhibitor group
compared with the NC group (Fig.
4D).
Inhibition of the invasion and migration
of U87 cells by the miR-130b inhibitor
Following the transfection of U87 cells with the
miR-130b inhibitor for 24 h, cell invasion ability was examined
using a Transwell assay. U87 cells transfected with the miR-130b
inhibitor exhibited a significant reduction in the number of cells
that traversed the membrane compared with that of the NC
(P<0.05; Fig. 5A and B). The
wound healing assay also demonstrated that cell migration was
inhibited in the U87 cells transfected with the miR-130b inhibitor
compared with the NC (Fig. 5C and
D).
Inhibition of the AKT signaling pathway
by an miR-130b inhibitor via PTEN upregulation
The miR-130b inhibitor was transiently transfected
into U87 cells to reduce miR-130b expression, and then total
cellular RNA and protein were extracted following 48 h of
transfection. PTEN expression at the RNA and protein levels was
measured. The results demonstrated that compared with cells
transfected with NC, when miR-130b expression was downregulated via
transfection with the miR-130b inhibitor, PTEN was clearly
upregulated at the protein level (Fig. 6A), but no significant difference
in the level of mRNA expression was observed (Fig. 6B). The phosphorylation of AKT was
significantly inhibited, and the tumor suppressor gene p27 was
upregulated by the miR-130b inhibitor. In addition, cyclin D1,
MMP-2 and MMP-9 expression was decreased.
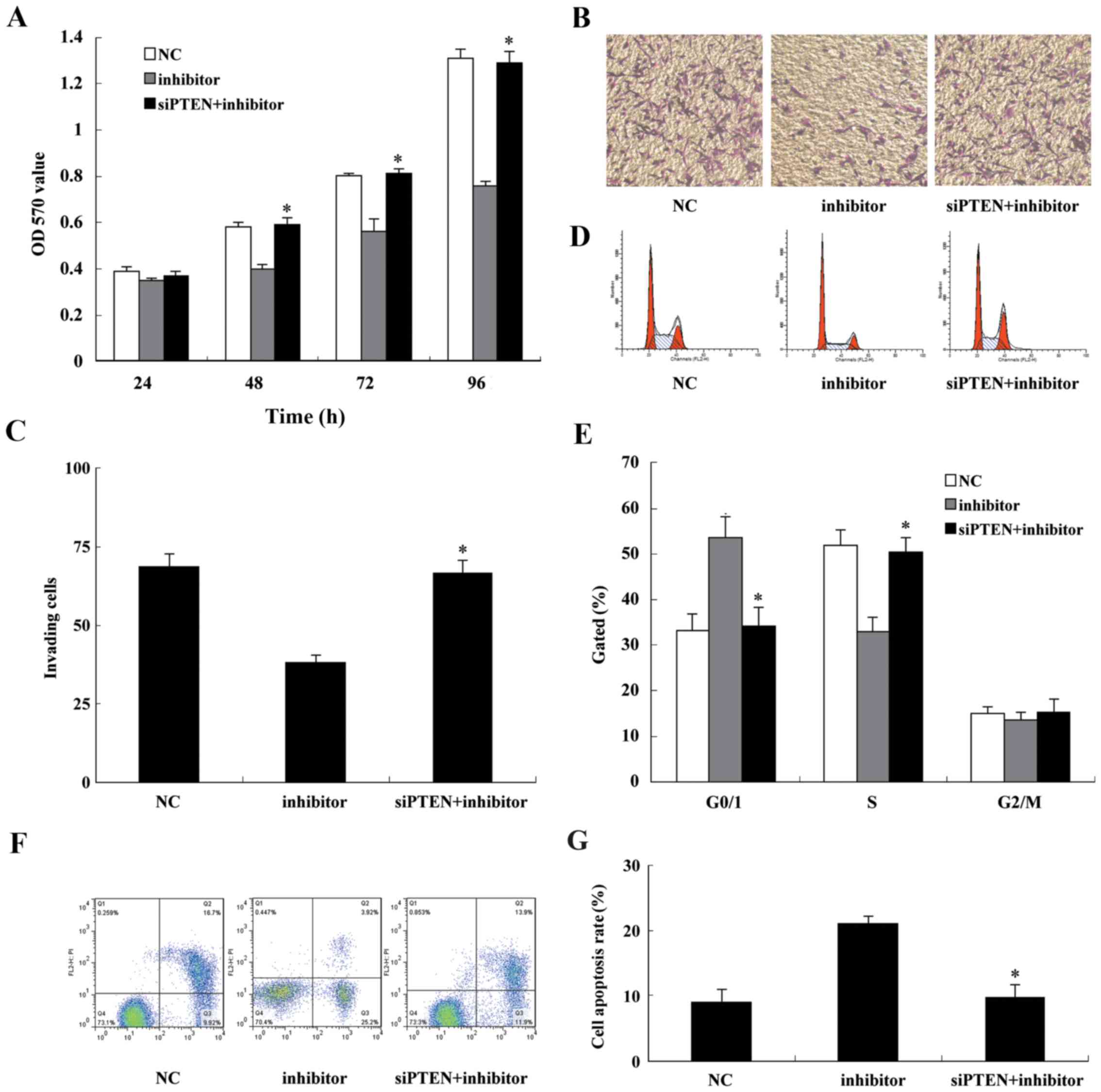
Knockdown of endogenous PTEN reverses the
effect of the miR-130b inhibitor
To confirm that miR-130b and PTEN serve a role in
glioma cell proliferation and invasion, U87 cells were
co-transfected with the miR-130b inhibitor and siPTEN. At 48 h
after transfection, total proteins were extracted and western
blotting demonstrated that the protein expression level of PTEN was
markedly reduced (Fig. 7). The
protein of PTEN was markedly reduced when U87 cells were
co-transfected with the miR-130b inhibitor and siPTEN, the
phosphorylation of AKT was significantly activated, and the tumor
suppressor gene p27 was downregulated. In addition, cyclin D1,
MMP-2 and MMP-9 expression was increased. CCK-8 and Transwell assay
results indicated that following co-transfection with the miR-130b
inhibitor and siPTEN U87, cell proliferation and cell invasion were
markedly increased compared with that of the cells transfected with
miR-130b inhibitor alone (Fig.
8A–C). Cell cycle analysis demonstrated that U87 cells
co-transfected with the miR-130b inhibitor and siPTEN presented a
lower percentage of cells in the G1 phase and a higher percentage
of cells in the S phase (Fig.
8D). The analysis of apoptosis by flow cytometry revealed that
the knockdown of endogenous PTEN attenuated the apoptosis-inducing
effect of the miR-130b inhibitor (Fig. 8E and F). These findings confirm
that miR-130b inhibits glioblastoma cell proliferation and invasion
and induces their apoptosis via the downregulation of PTEN
expression.
Discussion
Glioma is the most common malignancy and the most
lethal adult brain tumor (3). Due
to the abnormal cellular and molecular heterogeneity of tumor
cells, the treatment of glioma using surgery combined with
radiotherapy and comprehensive chemotherapy with temozolomide has
remained unsatisfactory, often resulting in short survival times
and poor prognosis. In the past decade, the median survival time of
patients with glioblastoma was only 15 months (14,15). Research on the diagnosis and
treatment of glioma is now particularly focused at the molecular
mechanism underlying glioma tumorigenesis and development,
particularly in the evolution of a variety of genes. Therefore, the
discovery of the fundamental disease genes associated with glioma
may provide an effective treatment for the targeted prevention and
control of the disease at the genetic level. A number of studies
have shown that miRNAs serve an important role in the initiation
and progression of human cancer, acting as either oncogenes or
tumor suppressor genes (16,17). It has been confirmed that a single
miRNA molecule may act on multiple or even hundreds of target
genes, and several different miRNAs may be able to regulate the
same gene, so that miRNAs and their targeting proteins form a
complex regulatory network that is important in regulating growth,
development and tumorigenesis (18).
The miR-130 family includes miR-130a and b. The
human miRNA-130b gene consists of 22 nucleotides (5′-CAGUGCA
AUGAUGAAAGGGCAU-3′). miR-130a and miR-130b are co-localized on
chromosome 22, located 10 kb apart, and can be expressed in cells
in tandem, suggesting that they may have similar biological
functions (19). The present
study detected abnormal miR-130b expression in a variety of
gliomas, which was consistent with the increased expression of
miR-130b in melanoma (20),
gastric cancer (21), bladder
cancer (22), colorectal cancer
(23), metastatic renal carcinoma
(24) and glioma (10), whereas miR-130b has been found to
be downregulated in papillary thyroid carcinomas (25), endometrial cancer (9), pituitary adenoma (26) and pancreatic cancer (27). In addition, miR-130b has an
association with neurodegenerative diseases such as Alzheimer's and
Parkinson's diseases (28), and
inhibits lipogenesis (20). Tu
et al (29) detected the
increased expression of miR-130b in human hepatocellular carcinoma
(HCC) and liver cancer cell lines, which was associated with a poor
prognosis in liver cancer patients. They also demonstrated that the
inhibition of miR-130b expression upregulated its target genes
through PPAR-γ, which resulted in inhibition of the invasion and
migration of HCC. Furthermore, a study conducted by Zhao et
al (27) demonstrated that
the expression of miR-130b was reduced in pancreatic cancer, and
identified an association of the downregulation of miR-130b with
poor prognosis in pancreatic cancer patients. The study also found
that the over-expression of miR-130b inhibited the proliferation
and invasion of pancreatic cancer cells by downregulating the
target gene signal transducer and activator of transcription 3.
However, the expression pattern of miR-130b and the specific
mechanisms underlying its effects in the pathogenesis of glioma
have not been fully elucidated. The purpose of the present study
was to investigate the mechanism by which miR-130b acts in glioma
cell proliferation and invasion, and to determine whether it
regulates the target gene, PTEN. In the present study RT-qPCR
detected significantly higher miR-130b expression in human glioma
tissues than in normal brain tissues, as well as significantly
higher expression in the LN229, U87 and U251 cell lines compared
with the NHA cell line. The downregulation of miR-130b in U87
glioma cells inhibited cell proliferation, invasion and migration
and induced apoptosis. These findings indicate that miR-130b
functions as an oncogene in glioma cells.
The PTEN gene is a tumor suppressor gene that is
located on chromosome 10q23.3 and has dual specificity (30). PTEN, as a lipid phosphatase,
dephosphorylates the second messenger phosphatidylinositol
3,4,5-triphosphate, blocks the phosphoinositide 3-kinase (PI3K)/Akt
signaling pathway, upregulates the tumor suppressor gene p27 and
inhibits cyclin D1 accumulation to regulate the cell cycle, which
in turn causes cell G0/G1 arrest that prevent cells from entering
the S phase. p27 is the key mediator in the repression of the G1
phase by PTEN, as well as a downstream target for PTEN-dependent
cell cycle arrest (31,32). PTEN also reduces AKT
phosphorylation and inhibits MMP secretion, thereby inhibiting
tumor invasion and migration (33). Human malignant glioma (astrocytoma
and glioblastoma) has a high level of PTEN gene deletions and
mutations, which leads to abnormal activation of the PI3K/Akt
pathway, particularly an increase in Akt phosphorylation and
activity that promotes tumor cell growth, proliferation, invasion
and metastasis (34). The present
study demonstrated that the transfection of U87 cells with an
miR-130b inhi bitor promoted the expression of the target gene PTEN
and reduced the phosphorylation of AKT. Glioma cells transfected
with the miR-130b inhibitor also exhibited increased expression of
the tumor suppressor gene p27, suppressed cyclin D1 expression and
reduced cell proliferation with G0/G1 arrest, which prevented cells
from entering the S phase. Finally, the miR-130b inhibitor also
reduced Akt phosphorylation levels to inhibit MMP-2 and -9
expression, thereby suppressing U87 invasion and migration. The
siRNA silencing of PTEN reversed the inhibition of cell
proliferation and invasion and promoted the apoptosis-inducing
effect of the miR-130b inhibitor. Thus, the present study
demonstrated that miR-130b functions in the tumorigenesis and
development of glioma via the PTEN/AKT signaling pathway.
In conclusion, the present study has elucidated the
oncogenic functions of miR-130b in glioma, which involved the
negatively regulation of the expression of PTEN. These results
indicate that miR-130b is important in the proliferation, apoptosis
and invasion of glioma, and suggest that the inhibition of miR-130b
expression may be an effective treatment approach for human
glioma.
References
|
1
|
Ohgaki H and Kleihues P: Epidemiology and
etiology of gliomas. Acta Neuropathol. 109:93–108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhou X, Ren Y, Moore L, Mei M, You Y, Xu
P, Wang B, Wang G, Jia Z, Pu P, et al: Downregulation of miR-21
inhibits EGFR pathway and suppresses the growth of human
glioblastoma cells independent of PTEN status. Lab Invest.
90:144–155. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu Y, Yan W, Zhang W, Chen L, You G, Bao
Z, Wang Y, Wang H, Kang C and Jiang T: miR-218 reverses high
invasiveness of glioblastoma cells by targeting the oncogenic
transcription factor LEF1. Oncol Rep. 28:1013–1021. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gu JJ, Gao GZ and Zhang SM: miR-218
inhibits the migration and invasion of glioma U87 cells through the
Slit2-Robo1 pathway. Oncol Lett. 9:1561–1566. 2015.PubMed/NCBI
|
|
7
|
Jiang L, Wang C, Lei F, Zhang L, Zhang X,
Liu A, Wu G, Zhu J and Song L: miR-93 promotes cell proliferation
in gliomas through activation of PI3K/Akt signaling pathway.
Oncotarget. 6:8286–8299. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Borgdorff V, Lleonart ME, Bishop CL,
Fessart D, Bergin AH, Overhoff MG and Beach DH: Multiple microRNAs
rescue from Ras-induced senescence by inhibiting p21(Waf1/Cip1).
Oncogene. 29:2262–2271. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dong P, Karaayvaz M, Jia N, Kaneuchi M,
Hamada J, Watari H, Sudo S, Ju J and Sakuragi N: Mutant p53
gain-of-function induces epithelial-mesenchymal transition through
modulation of the miR-130b-ZEB1 axis. Oncogene. 32:3286–3295. 2013.
View Article : Google Scholar
|
|
10
|
Malzkorn B, Wolter M, Liesenberg F,
Grzendowski M, Stühler K, Meyer HE and Reifenberger G:
Identification and functional characterization of microRNAs
involved in the malignant progression of gliomas. Brain Pathol.
20:539–550. 2010. View Article : Google Scholar
|
|
11
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
13
|
Yu T, Cao R, Li S, Fu M, Ren L, Chen W,
Zhu H, Zhan Q and Shi R: MiR-130b plays an oncogenic role by
repressing PTEN expression in esophageal squamous cell carcinoma
cells. BMC Cancer. 15:292015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Y and Jiang T: Understanding high
grade glioma: molecular mechanism, therapy and comprehensive
management. Cancer Lett. 331:139–146. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Carlsson SK, Brothers SP and Wahlestedt C:
Emerging treatment strategies for glioblastoma multiforme. EMBO Mol
Med. 6:1359–1370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li R and Li X, Ning S, Ye J, Han L, Kang C
and Li X: Identification of a core miRNA-pathway regulatory network
in glioma by therap eutically targeting miR-181d, miR-21, miR-23b,
β-catenin, CBP, and STAT3. PLoS One. 9:e1019032014. View Article : Google Scholar
|
|
17
|
Jun GJ, Zhong GG and Ming ZS: miR-218
inhibits the proliferation of glioma U87 cells through the
inactivation of the CDK6/cyclin D1/p21Cip1/Waf1 pathway.
Oncol Lett. 9:2743–2749. 2015.PubMed/NCBI
|
|
18
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar :
|
|
19
|
Lee EK, Lee MJ, Abdelmohsen K, Kim W, Kim
MM, Srikantan S, Martindale JL, Hutchison ER, Kim HH, Marasa BS, et
al: miR-130 suppresses adipogenesis by inhibiting peroxisome
proliferator-activated receptor gamma expression. Mol Cell Biol.
31:626–638. 2011. View Article : Google Scholar
|
|
20
|
Sand M, Skrygan M, Sand D, Georgas D,
Gambichler T, Hahn SA, Altmeyer P and Bechara FG: Comparative
microarray analysis of microRNA expression profiles in primary
cutaneous malignant melanoma, cutaneous malignant melanoma
metastases, and benign melanocytic nevi. Cell Tissue Res.
351:85–98. 2013. View Article : Google Scholar
|
|
21
|
Lai KW, Koh KX, Loh M, Tada K, Subramaniam
MM, Lim XY, Vaithilingam A, Salto-Tellez M, Iacopetta B, Ito Y, et
al: Singapore Gastric Cancer Consortium: MicroRNA-130b regulates
the tumour suppressor RUNX3 in gastric cancer. Eur J Cancer.
46:1456–1463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Scheffer AR, Holdenrieder S, Kristiansen
G, von Ruecker A, Müller SC and Ellinger J: Circulating microRNAs
in serum: novel biomarkers for patients with bladder cancer. World
J Urol. 32:353–358. 2014. View Article : Google Scholar
|
|
23
|
Colangelo T, Fucci A, Votino C, Sabatino
L, Pancione M, Laudanna C, Binaschi M, Bigioni M, Maggi CA, Parente
D, et al: MicroRNA-130b promotes tumor development and is
associated with poor prognosis in colorectal cancer. Neoplasia.
15:1218–1231. 2013. View Article : Google Scholar
|
|
24
|
Wu X, Weng L, Li X, Guo C, Pal SK, Jin JM,
Li Y, Nelson RA, Mu B, Onami SH, et al: Identification of a
4-microRNA signature for clear cell renal cell carcinoma metastasis
and prognosis. PLoS One. 7:e356612012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yip L, Kelly L, Shuai Y, Armstrong MJ,
Nikiforov YE, Carty SE and Nikiforova MN: MicroRNA signature
distinguishes the degree of aggressiveness of papillary thyroid
carcinoma. Ann Surg Oncol. 18:2035–2041. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Leone V, Langella C, D'Angelo D, Mussnich
P, Wierinckx A, Terracciano L, Raverot G, Lachuer J, Rotondi S,
Jaffrain-Rea ML, et al: miR-23b and miR-130b expression is
downregulated in pituitary adenomas. Mol Cell Endocrinol. 390:1–7.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao G, Zhang JG, Shi Y, Qin Q, Liu Y,
Wang B, Tian K, Deng SC, Li X, Zhu S, et al: miR-130b is a
prognostic marker and inhibits cell proliferation and invasion in
pancreatic cancer through targeting STAT3. PLoS One. 8:e738032013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shacka JJ, Roth KA and Zhang J: The
autophagy-lysosomal degradation pathway: role in neurodegenerative
disease and therapy. Front Biosci. 13:718–736. 2008. View Article : Google Scholar
|
|
29
|
Tu K, Zheng X, Dou C, Li C, Yang W, Yao Y
and Liu Q: MicroRNA-130b promotes cell aggressiveness by inhibiting
peroxisome proliferator-activated receptor gamma in human
hepatocellular carcinoma. Int J Mol Sci. 15:20486–20499. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen Z, Trotman LC, Shaffer D, Lin HK,
Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al:
Crucial role of p53-dependent cellular senescence in suppression of
Pten-deficient tumorigenesis. Nature. 436:725–730. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sansal I and Sellers WR: The biology and
clinical relevance of the PTEN tumor suppressor pathway. J Clin
Oncol. 22:2954–2963. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chung JH and Eng C: Nuclear-cytoplasmic
partitioning of phosphatase and tensin homologue deleted on
chromosome 10 (PTEN) differentially regulates the cell cycle and
apoptosis. Cancer Res. 65:8096–8100. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yamada KM and Araki M: Tumor suppressor
PTEN: modulator of cell signaling, growth, migration and apoptosis.
J Cell Sci. 114:2375–2382. 2001.PubMed/NCBI
|
|
34
|
Vivanco I and Sawyers CL: The
phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat Rev
Cancer. 2:489–501. 2002. View
Article : Google Scholar : PubMed/NCBI
|