Introduction
Odontoblasts are neural crest-derived, highly
differentiated cells aligned in a single layer at the periphery of
the dental pulp. The main function of these cells is to form
dentin, the largest part of the hard tissue in teeth. Following
differentiation from dental papilla mesenchymal cells, functional
odontoblasts synthesize and secrete collagenous and non-collagenous
matrix proteins that are essential for mineralized dentin formation
(1).
The differentiation of odontoblasts is a complex
process regulated by reciprocal epithelium-mesenchyme interactions.
A number of signaling factors are reported to be involved in this
process, including Notch, bone morphogenetic protein (BMP), Wnt and
transforming growth factor-β (TGF-β). Notch signaling is an
evolutionarily conserved pathway that is responsible for the
control of cell fate through local cell-cell interactions (2). It has been well documented that
during tooth development, Notch receptors and ligands are expressed
in dental epithelium, dental papilla mesenchyme, ameloblast or
odontoblast at different stages of tooth germ development (3,4).
Additionally, in the pulp of injured teeth, the expression of Notch
receptors and the Delta-1 ligand is significantly upregulated
(5,6). These results suggest that Notch
signaling is involved in primary and reparative dentinogenesis.
Further evidence has demonstrated that Notch signaling has a
critical role in dental pulp stem cell (DPSC) differentiation into
odontoblasts in vitro (7,8).
BMP signaling is also a potent regulator of odontoblast
differentiation. As one of the strongest signals stimulating
biomineralization, BMP-2 has been identified to be required for
odontoblast differentiation in vivo and in vitro
(9–11). Additionally, in vitro
studies have demonstrated that BMP-2 gene transfection enhances the
odontogenic differentiation of DPSC and stem cells from apical
papilla (12,13).
Hairy/enhancer-of-split related with YRPW motif 1
(Hey1), also known as CHF2, HRT1, Herp2 or Hesr1, is a member of
the basic helix-loop-helix family. Hey1 was first characterized as
a downstream effector of canonical Notch signaling (14), and further investigations
indicated that Hey1 was also induced by TGF-β/BMP signaling
independently of Notch (15,16). Numerous studies have demonstrated
that Hey1 is responsible for the development of various tissues,
including bone, nerve, heart, muscle and vascular tissues (17–21). Our preliminary study demonstrated
that Hey1 was expressed in dental pulp tissues and may affect
dentin sialophosphoprotein (DSPP) expression during odontogenesis
(22). In addition, substantial
evidence has demonstrated the regulatory roles of Hey1 in
mineralization (23,24). However, it remains unclear whether
Hey1 regulates odontoblastic differentiation.
In this study, the effects of Hey1 on the
differentiation of odontoblasts were investigated in an
odontoblast-lineage cell line (OLC) (25,26). The expression of Hey1 in OLCs was
first observed during odontogenic differentiation. Subsequently, a
plasmid encoding the full-length sequence of Hey1 or Hey1-silencing
short hairpin RNA (shRNA) were transfected into OLCs to compare the
differentiation and mineralization capabilities of cells expressing
different levels of Hey1. The findings suggested that Hey1 has an
important role in odontoblastic differentiation.
Materials and methods
Cell culture and differentiation
induction
OLC cell line was provided by Professor S. Arany
(Department of Biochemistry, Akita University School of Medicine,
Akita, Japan). It is a murine spontaneously immortalized cell line
which was developed by Arany et al (25). OLCs were cultured in α-minimum
essential medium (α-MEM) supplemented with 10% fetal bovine serum
(FBS; both from Hyclone; GE Healthcare Life Sciences, Logan, UT,
USA) at 37°C in a humidified atmosphere of 5% CO2 and
95% air. For differentiation induction, cells were plated at a
density of 2×104 cells/cm2 in 6-well plates
and were cultured until they reached 80% confluence. Cells were in
serum-free α-MEM for 24 h to be synchronized, then in
osteoblastic/odontoblastic differentiation medium [α-MEM
supplemented with 10% FBS, 50 μg/ml ascorbic acid (AA), 10
mM β-glycerol phosphate (β-GP) and 10−8 M dexamethasone
(DEX)] (27,28).
Establishment of stable
Hey1-overexpressing cell lines
The plasmid encoding the full-length sequence of
mouse Hey1 with a C-terminal His-tag, obtained from Dr Nobuyuki
Kawashima (Department of Endodontics and Dental Pulp Biology, Tokyo
Medical and Dental University, Tokyo, Japan), was constructed using
eukaryotic expression vector pEF-Dest51 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and was named pEF-Hey1.
Following confirmation by DNA sequencing, pEF-Hey1 was transfected
into OLCs using Lipofectamine® 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. Following 24 h of transfection, the cells were
subcultured at 1:12 for another 24 h and selected in growth medium
containing 4 μg/ml blasticidin (Invitrogen; Thermo Fisher
Scientific, Inc.). Single cell isolation was performed using
96-well plates. These single cell clones were amplified in
blasticidin selection medium and then processed for selection by
reverse-transcription polymerase chain reaction (RT-PCR) and
western blot analyses. Empty pEF-Dest51 vector was transfected into
OLCs as a mock negative control.
Construction and transient transfection
of shRNA expression vectors targeting Hey1
A mouse Hey1-targeting sequence
(5′-TGAAGGACTCGATGCCTCCGA-3′) was designed using Invitrogen's
online RNAi designer and was verified using BLAST to avoid
off-target gene silencing. Two pairs of oligonucleotides coding
shRNA, one pair containing mouse Hey1-targeting sequence and the
other containing a scrambled sequence (5′-GTTCTCCGAACGTGTCACGT-3′)
with no significant similarity to any mouse gene sequences, were
synthe-sized. Pairs of oligonucleotides were annealed and inserted
into the shRNA expression vector pGPU6/GFP/Neo (Shanghai GenePharma
Co., Ltd., Shanghai, China). Transient transfections into OLCs were
performed using Lipofectamine® 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) to yield OLC/Hey1-knockdown (KD)
and OLC/pGP-Mock.
RNA preparation and RT-quantitative PCR
(RT-qPCR)
Total RNA was extracted from OLCs using TRIzol
reagent (Thermo Fisher Scientific, Inc.) and was quantified by
spectrophotometry using a NanoDrop 2000c spectrophotometer (Thermo
Fisher Scientific, Inc., Wilmington, DE, USA). Total RNA (1
μg) was reverse transcribed into cDNA using a First Strand
cDNA Synthesis kit (Thermo Fisher Scientific, Inc.) according to
the manufacturer's guidelines. qPCR was performed on 1 μl of
cDNA in a 20 μl reaction with SYBR Premix Ex Taq (Takara
Biotechnology Co., Ltd., Dalian, China) using the Bio-Rad CFX96
real-time PCR detection system (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The sense and anti-sense primers were as
follows: 5′-CGACGAGACCGAATCAATAAC-3′ and
5′-CAAACTCCGATAGTCCATAGCC-3′ for Hey1; 5′-AGCATCAAGAATAGCACCAACC-3′
and 5′-CCCATCAGTATCATCCAAACCT-3′ for DSPP;
5′-GACCCCTTCATTGACCTCA-3′ and 5′-GCTCCTGGAAGATGGTGA-3′ for GAPDH.
The protocol for the qPCR reactions consisted of an initial
denaturation step (95°C for 3 min), followed by 45 cycles of
denaturation (95°C for 10 sec), annealing (55°C for 10 sec), and
extension (72°C for 20 sec). GAPDH was used as the housekeeping
gene for template normalization. The relative expression level of
mRNA was calculated using 2−ΔΔCq analysis (29). All RT-qPCR reactions were
performed in triplicate.
Western blot analysis
The cells were washed with cold phosphate-buffered
saline and lysed on ice using radioimmunoprecipitation assay lysis
buffer (Thermo Fisher Scientific, Inc.) containing 1 mM
phenylmethylsulfonyl fluoride. The cell lysates were centrifuged at
4°C for 20 min at 13,500 × g, and the total protein content of the
supernatant was collected. Protein concentration was determined
using a bicinchoninic acid protein assay kit (Thermo Fisher
Scientific, Inc.). The protein samples were mixed with 5X loading
buffer and then were boiled for denaturation. Protein extracts (30
μg) from each sample were subjected to 8% SDS-PAGE and were
transferred to polyvinylidene difluoride membranes (EMD Millipore,
Billerica, MA, USA). The membranes were blocked in 5% BSA for 2 h
at 37°C and were incubated with rabbit anti-His-probe (cat. no.
sc-803; 1:500), anti-dentin sialoprotein (DSP; cat. no. sc-33587;
1:200) both from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA)
or anti-β-actin (cat. no. ab8227; 1:2,000; Abcam, Cambridge, UK)
primary antibodies overnight at 4°C. The membranes were then rinsed
in TBS-Tween and incubated with an HRP-conjugated goat anti-rabbit
secondary antibody (cat. no. AP307P, 1:5,000; EMD Millipore) for 1
h, and then detected using an enhanced chemiluminescence system (GE
ImageQuant 350; GE Healthcare, Piscataway, NJ, USA).
Semi-quantitative analyses of the bands were performed using
Image-Pro Plus 6.0 software (Media Cybernetics, Inc., Rockville,
MD, USA).
Immunofluorescence staining
OLCs in 35 mm glass bottom dishes were fixed with 4%
paraformaldehyde for 30 min at room temperature, permeabilized in
0.1% Triton X-100 for 5 min, and blocked with 2% goat serum (cat.
no. C0265; Beyotime Biotechnology, Shanghai, China) at 37°C for 1
h. Cells were then incubated with rabbit anti-Hey1 (cat. no.
ab22614; 1:50; Abcam) or anti-DSP (cat. no. sc-33587; 1:50; Santa
Cruz Biotechnology, Inc.) primary antibody at 4°C overnight.
Finally, cells were incubated with Alexa Fluor 594-conjugated goat
anti-rabbit IgG secondary antibody (cat. no. A11037; 1:400;
Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 1 h, and
nuclei were stained with 2 μg/ml DAPI (cat. no. C1002;
Beyotime Biotechnology) for 5 min at room temperature. Fluorescence
was examined using a FV1000 confocal laser scanning microscope
(Olympus Corporation, Tokyo, Japan). Fluorescence intensity was
determined with Image-Pro Plus 6.0 software (Media Cybernetics,
Inc.).
Mineralization assay
Mock OLCs and stable Hey1-overexpressing cells were
plated in 6-well plates and cultured by osteoblastic/odontoblastic
differentiation medium, respectively. Media were collected every 4
days for determination of alkaline phosphatase (ALP) activity using
an Alkaline Phosphatase assay kit (Jiancheng Bioengineering
Institute, Nanjing, China) as described in a previous study
(30). In brief, 20 μl of
cell culture medium mixed with 1 ml of reaction solution containing
18 mM 4-nitrophenyl phosphate and 0.5 M 2-amino-2-methyl-1-propanol
was incubated in the dark for 15 min at 37°C. ALP activity was
quantified by measuring the absorbance values of the reaction
solution at 405 nm using an absorbance microplate reader (BioTek
Instruments, Inc., Winooski, VT, USA). After culture for 32 days,
cells were rinsed with distilled water and fixed with 4%
paraformaldehyde for 30 min at room temperature. Mineralized
deposits were then stained with 40 mM alizarin red S (cat. no.
A5533; Sigma-Aldrich, St. Louis, MO, USA) for 30 min at room
temperature. The excess dye was removed by washing 3 times with
distilled water. Red stain of mineralized deposits was observed by
a light microscope (Olympus Corporation).
Statistical analysis
All values presented are expressed as the mean ±
standard deviation. One-way analysis of variance was used to
analyze the differences between the groups. The differences between
groups were detected with post hoc Student-Newman-Keuls tests.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Odontoblastic differentiation medium
induces an increase in Hey1 expression
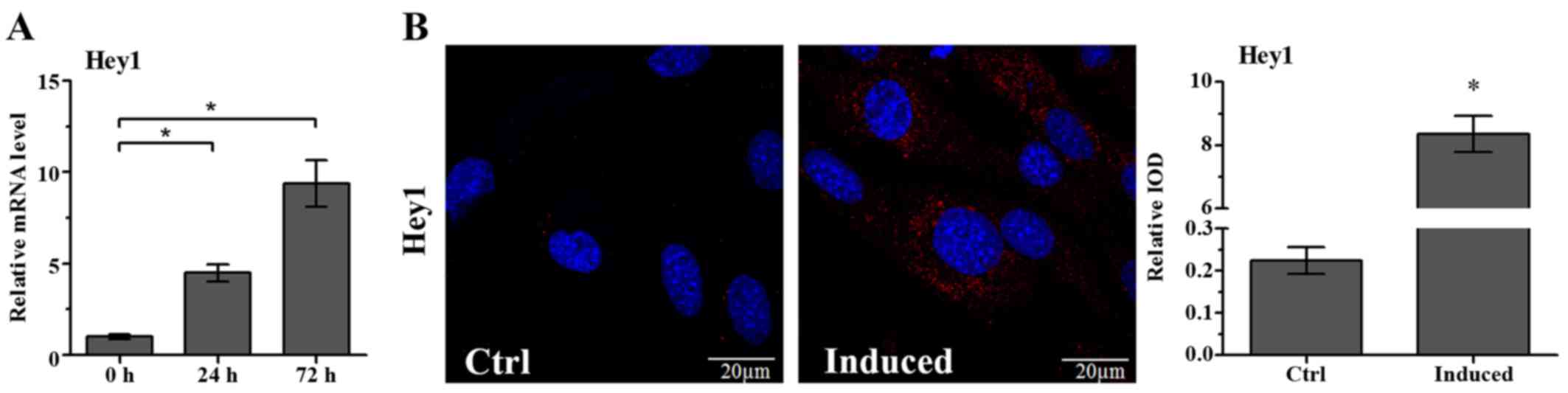
Hey1 mRNA levels in the OLCs were significantly
increased on the day 1 and 3 of culture in
osteoblastic/odontoblastic differentiation medium containing AA,
β-GP and DEX (Fig. 1A).
Additionally, immunofluorescence staining was performed to evaluate
the protein levels of Hey1 during differentiation induction. There
was almost no positive staining in the untreated OLCs. However, the
expression of Hey1 protein was observed in OLCs following
stimulation with differentiation medium for 5 days (Fig. 1B).
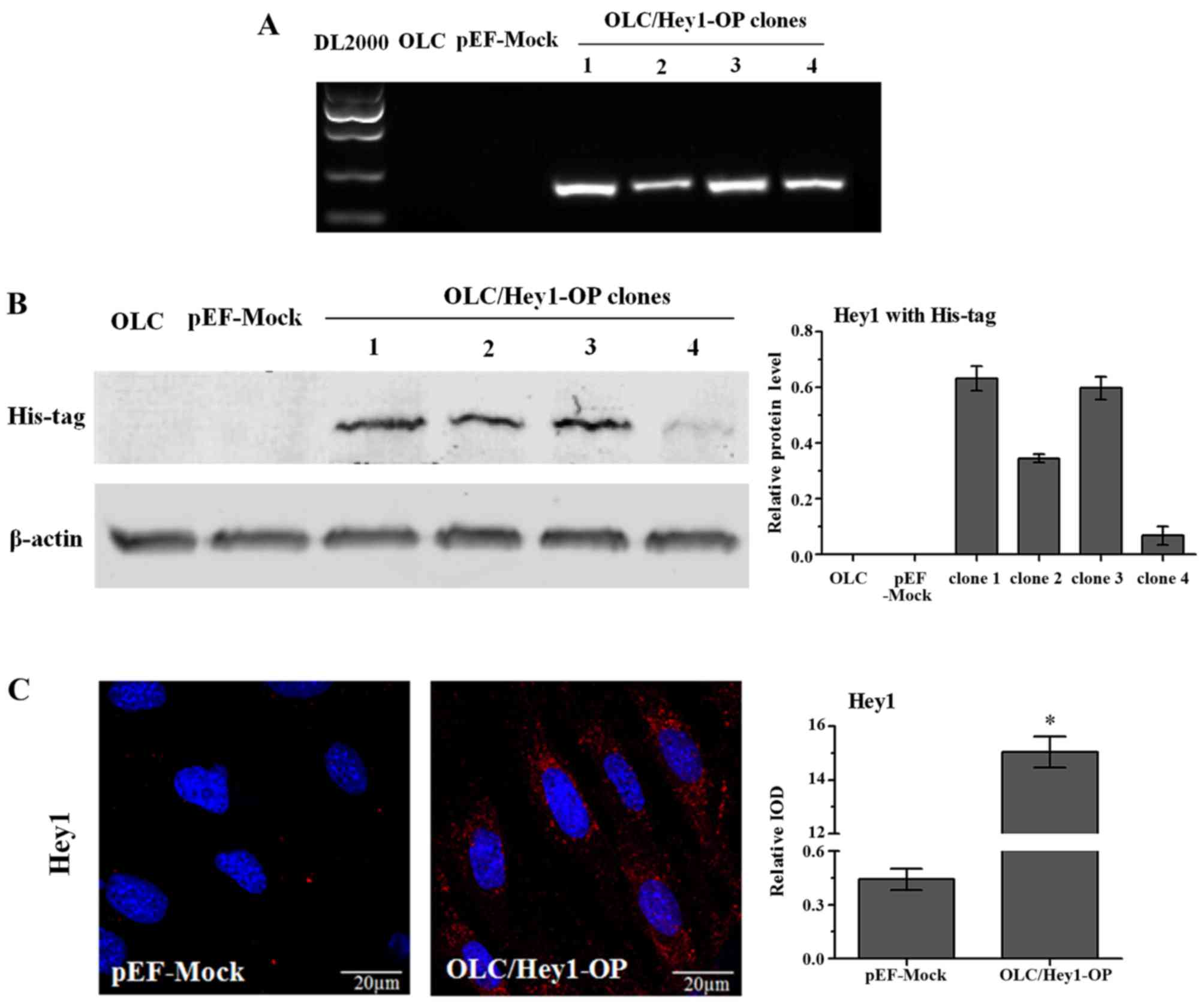
Stable Hey1-overexpressing cells line was
established
To investigate the effects of Hey1 on the
differentiation of odontoblasts, pEF-Hey1 and the control vector
pEF-Dest51 were transfected into OLCs. Following pEF-Hey1
transfection, four single cell clones that were resistant to
blasticidin were amplified. RT-PCR demonstrated that Hey1 mRNA
expression was barely detected in normal OLCs (untransfected) or
mock-transfected cells (transfected with empty pEF-Dest51), whereas
the four single cell clones transfected with pEF-Hey1 expressed
much higher levels of Hey1 mRNA (Fig.
2A). Western blot analysis with an anti-His-tag antibody was
performed to verify the effectiveness of the plasmid to induce Hey1
overexpression. Among the four clones, clone one synthesized the
highest levels of Hey1 protein (Fig.
2B) and was designated as OLC/Hey1-OP for further
investigations. Immunofluorescence staining with an anti-Hey1
antibody further confirmed that the OLC/Hey1-OP cell line expressed
a higher level of Hey1 protein compared with mock transfection
cells (Fig. 2C).
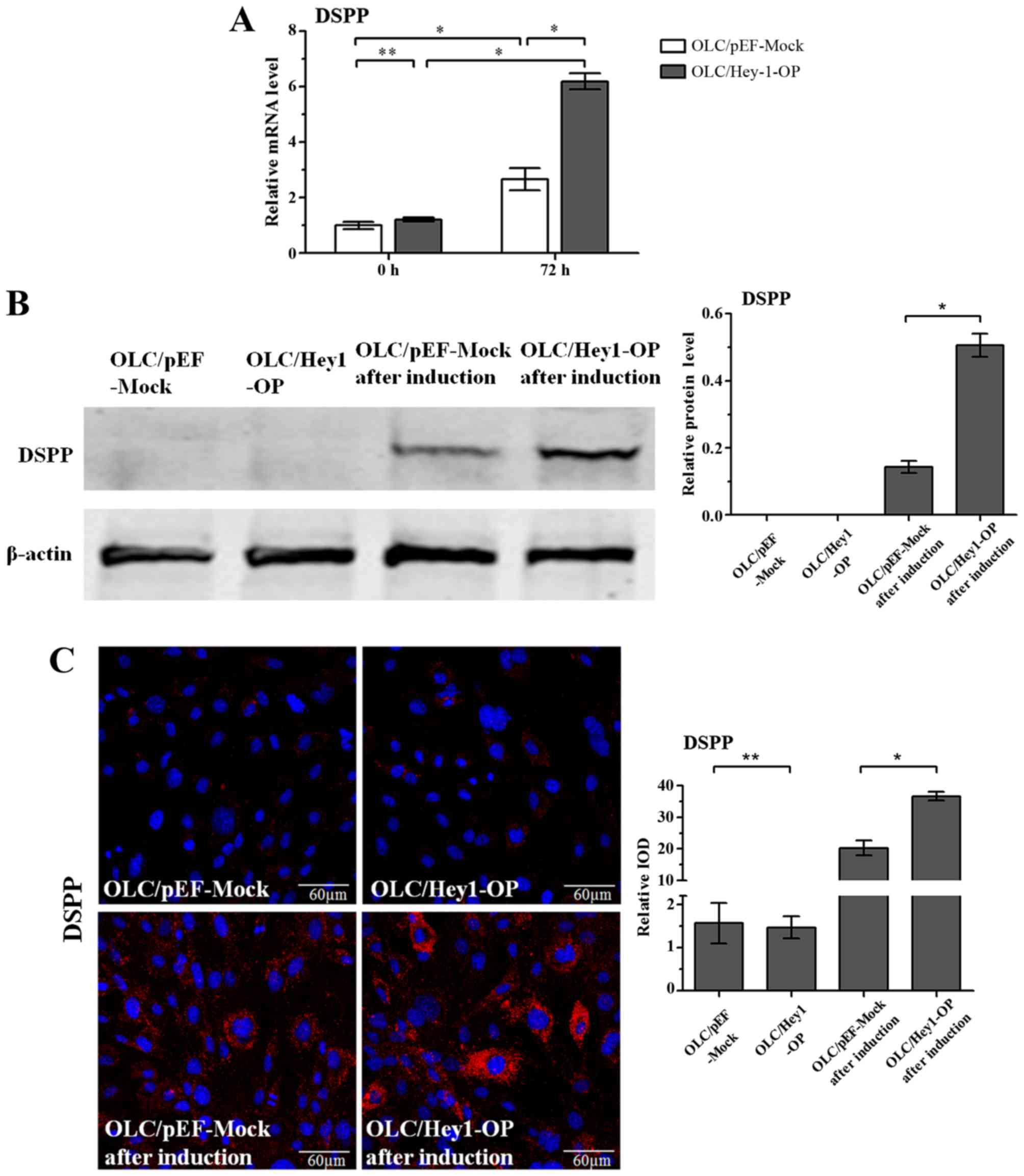
Stable Hey1-overexpressing cells line
exhibit increased differentiation capabilities
The differentiation capabilities between the stable
Hey1-overexpressing and mock cell lines were compared. The results
of RT-qPCR demonstrated that overexpressing Hey1 alone did not
affect the mRNA levels of DSPP, the odontoblastic differentiation
marker, in OLCs (P>0.1). However, following culture in
differentiation medium for 3 days, OLC/Hey1-OP expressed a
significantly higher mRNA level of DSPP than the mock cells
(Fig. 3A). Since DSPP is a large
protein that can be specifically cleaved into two fractions, DSP
and dentin phosphoprotein, an antibody against the DSP portion of
DSPP was used to detect protein expression of the full-length DSPP.
The results of western blot analysis and immunofluorescence
staining further revealed that Hey1 overexpression increased DSPP
protein expression in OLCs when the cells were induced by
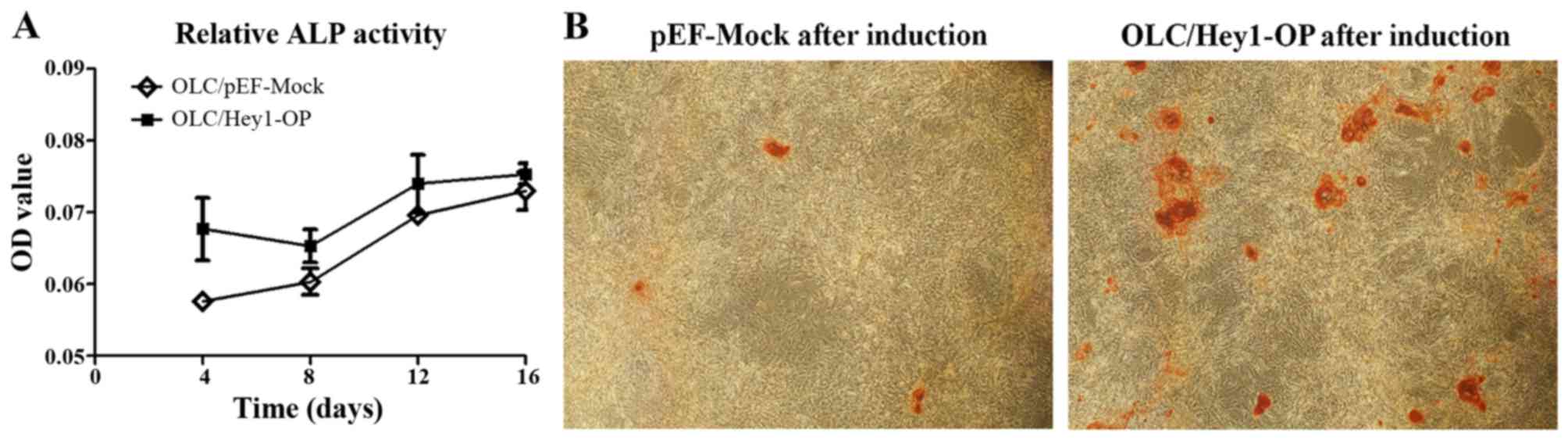
differentiation medium for 7 days (Fig. 3B and C). Furthermore, when
cultured in differentiation medium, the ALP activities of the
OLC/Hey1-OP cells were much higher than that of the mock cells
(Fig. 4A). Alizarin red S
staining revealed that the OLC/Hey1-OP cells formed more and larger
mineralized nodules than the mock cells (Fig. 4B).
Transient Hey1 knockdown inhibited OLC
differentiation
To further determine whether Hey1 is critical for
odontoblastic differentiation, a Hey1-targeting shRNA expression
vector was constructed, and transient transfection into OLCs was
performed. To evaluate the efficiency of the RNA interference in
silencing Hey1 expression, the mRNA and protein expression levels
of Hey1 in transfected OLCs and mock cells were measured using
RT-qPCR and immunofluorescence staining, respectively, following
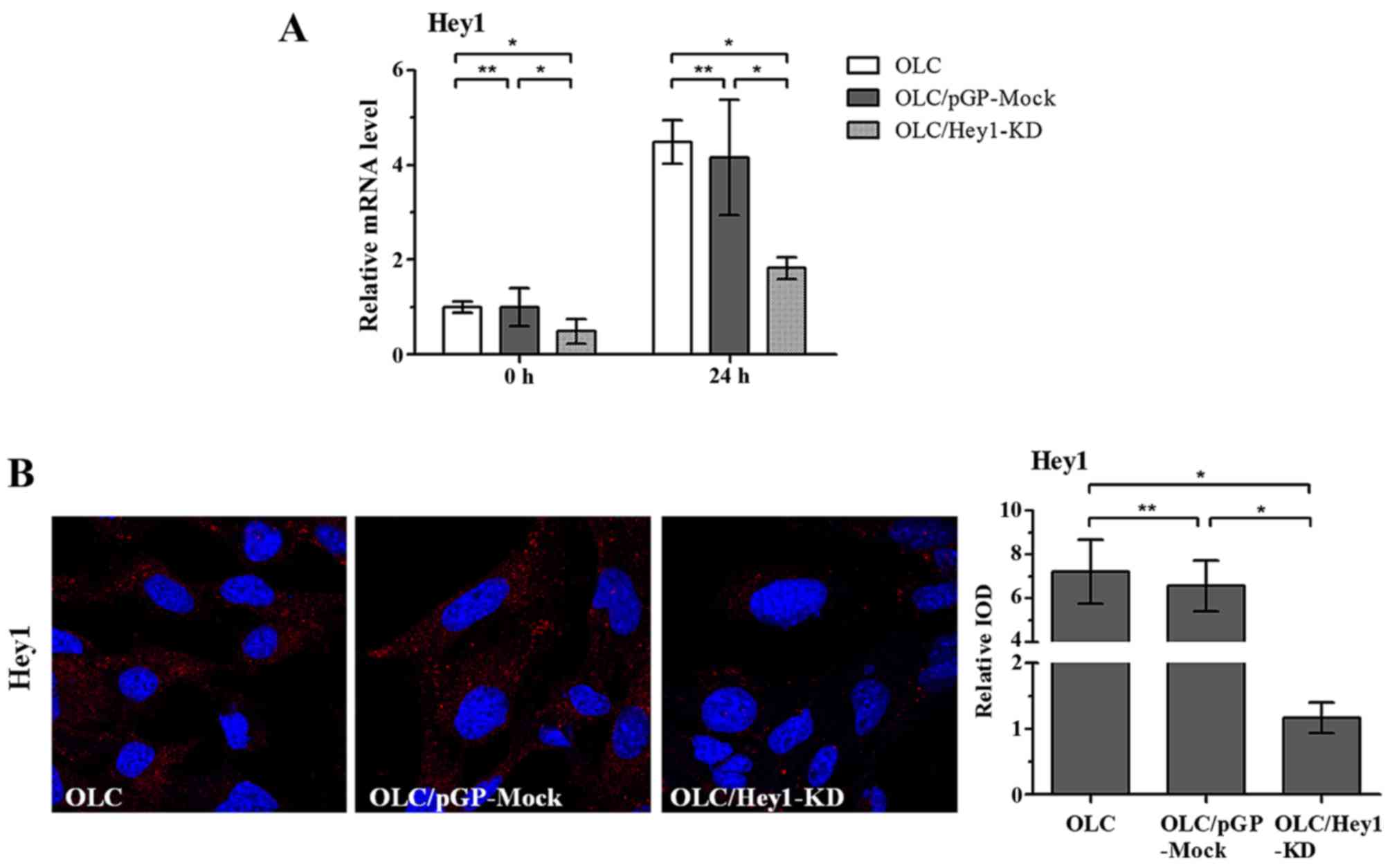
stimulation in differentiation medium. OLC/Hey1-KD cells expressed
lower levels of Hey1 mRNA and protein than OLC/pGP-Mock cells
(P<0.05; Fig. 5). Further
investigations revealed that DSPP expression was decreased in
OLC/Hey1-KD cells compared with the mock cells following
differentiation induction (Fig.
6). Following culturing in differentiation medium for 3 days,
the mRNA level of DSPP in OLC/Hey1-KD cells was significantly lower
than OLC/pGP-Mock cells (Fig.
6A). Furthermore, after culture in differentiation medium for 7
days, DSPP protein expression in OLC/Hey1-KD cells was
significantly lower than in OLC/pGP-Mock cells (Fig. 6B and C).
Discussion
Differentiation of the progenitor cells derived from
dental pulp tissue into odontoblasts has a vital role in the
dentinal regeneration process. Elucidating the underlying
mechanisms will facilitate the development of therapeutic
approaches for injured dentin-pulp complex. Numerous methods have
been used to induce odontoblast differentiation in vitro.
The application of osteoblastic/odontoblastic differentiation
medium (containing AA, β-GP and DEX) has been demonstrated to be an
effective method and has been adopted in numerous studies (7,8,27,28,31–33). Previous in vitro studies
have demonstrated the involvement of Notch and BMP signaling in
odontoblast differentiation induced by AA + β-GP + DEX (7,8,32,33). However, the underlying mechanisms
remain unclear. The present study revealed that Hey1 expression in
odontoblast-lineage cells was significantly upregulated by AA +
β-GP + DEX stimulation, suggesting that Hey1 may be involved in
their differentiation. Because Hey1 has been reported to regulate
osteoblast differentiation and matrix mineralization (23,24,30), it is assumed that Hey1 may have an
important role in odontoblast differentiation. To verify this
hypothesis, Hey1 overexpression and knockdown models were
established in vitro to investigate the effects of Hey1 on
odontoblast differentiation.
DSPP, a non-collagenous protein that is
predominantly expressed in odontoblasts or dentin, was demonstrated
to be critical for dentin mineralization. DSPP is synthesized and
secreted by differentiated odontoblasts and is regarded as a marker
of odontogenic differentiation (34,35). In the present study, OLCs cultured
in differentiation medium exhibited increased expression of DSPP, a
result is consistent with previous in vitro studies.
Furthermore, the results revealed that overexpression of Hey1 did
not directly upregulate DSPP expression in OLCs but enhanced the
upregulatory effect of AA + β-GP + DEX stimulation on DSPP
expression. Previous studies have demonstrated that Hey1 not only
regulated downstream targets as a transcriptional repressor, but
also functioned through interaction with other transcription
factors (36,37). Similarly, it has been shown that
Hey2 overexpression alone is not sufficient to induce strong
changes in downstream gene expression, but needs additional
cofactors (38). Therefore,
according to the results of the present study, Hey1 may regulate
DSPP expression indirectly by interacting with other cofactors
activated in the process of odontoblastic differentiation; this
requires further investigations.
OLCs in which Hey1 was exogenously overexpressed
exhibited upregulation of ALP activity and increased nodule
formation. These results indicate that Hey1 may be a positive
regulator of odontoblast cell mineralization. These findings are
consistent with previous studies showing that Hey1 enhanced
osteogenic differentiation and mineralization of mesenchymal stem
cells induced by BMP-9 or BMP-7 (23,39). However, Zamurovic et al
(24) reported that Hey1
inhibited mineralization of the MC3T3 cell line. This discrepancy
may result from differences in the source of cells. Certain
transcription factors, such as runt related transcription factor 2
and nuclear factor erythroid 2-related factor 1, have demonstrated
different effects on the differentiation of odontoblasts and
osteoblasts (40,41). The function of Hey1 may also be
dependent on cell type.
An in vivo study demonstrated that Hey1
single knockout mice exhibited no major developmental or obvious
functional impairments. However, the dental phenotypes of
Hey1-deficient mice have not been analyzed (42). The present in vitro study
revealed that RNA interference-mediated knockdown of Hey1
expression led to decreased DSPP expression compared with the
control mock cells following differentiation induction, suggesting
that Hey1 is critical for odontoblastic differentiation. Previous
in vitro studies demonstrated that knockdown of Hey1
promoted myogenesis and inhibited osteogenic differentiation
(23,43). Taken together, Hey1 may
participate in the regulation of cell fate.
In conclusion, the present study demonstrated that
Hey1 was involved in the differentiation of odontoblast-lineage
cells. Hey1 overexpression increased DSPP expression during
odontoblastic differentiation and further augmented mineralization
of OLCs. Additionally, knockdown of Hey1 diminished DSPP expression
induced by odontoblastic differentiation medium. The findings of
the current study indicate that Hey1 functions as a positive
regulator of odontogenic differentiation. This study broadens our
understanding of odontoblast differentiation and
biomineralization.
Acknowledgments
This study was supported by the National Nature
Science Foundation of China (grant no. 81070832) and the National
Nature Science Foundation of China (grant no. 81371139).
References
|
1
|
Ruch JV, Lesot H and Bègue-Kirn C:
Odontoblast differentiation. Int J Dev Biol. 39:51–68.
1995.PubMed/NCBI
|
|
2
|
Artavanis-Tsakonas S, Rand MD and Lake RJ:
Notch signaling: Cell fate control and signal integration in
development. Science. 284:770–776. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mitsiadis TA, Lardelli M, Lendahl U and
Thesleff I: Expression of Notch 1, 2 and 3 is regulated by
epithelial-mesenchymal interactions and retinoic acid in the
developing mouse tooth and associated with determination of
ameloblast cell fate. J Cell Biol. 130:407–418. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cai X, Gong P, Huang Y and Lin Y: Notch
signalling pathway in tooth development and adult dental cells.
Cell Prolif. 44:495–507. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Løvschall H, Tummers M, Thesleff I,
Füchtbauer EM and Poulsen K: Activation of the Notch signaling
pathway in response to pulp capping of rat molars. Eur J Oral Sci.
113:312–317. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mitsiadis TA, Fried K and Goridis C:
Reactivation of Delta-Notch signaling after injury: Complementary
expression patterns of ligand and receptor in dental pulp. Exp Cell
Res. 246:312–318. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang X, He F, Tan Y, Tian W and Qiu S:
Inhibition of Delta1 promotes differentiation of odontoblasts and
inhibits proliferation of human dental pulp stem cell in vitro.
Arch Oral Biol. 56:837–845. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang C, Chang J, Sonoyama W, Shi S and
Wang CY: Inhibition of human dental pulp stem cell differentiation
by Notch signaling. J Dent Res. 87:250–255. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang W, Harris MA, Cui Y, Mishina Y,
Harris SE and Gluhak-Heinrich J: Bmp2 is required for odontoblast
differentiation and pulp vasculogenesis. J Dent Res. 91:58–64.
2012. View Article : Google Scholar :
|
|
10
|
Cho YD, Yoon WJ, Woo KM, Baek JH, Park JC
and Ryoo HM: The canonical BMP signaling pathway plays a crucial
part in stimulation of dentin sialophosphoprotein expression by
BMP-2. J Biol Chem. 285:36369–36376. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Washio A, Kitamura C, Morotomi T,
Terashita M and Nishihara T: Possible involvement of Smad signaling
pathways ininduction of odontoblastic properties in KN-3 cells by
bone morphogenetic protein-2: A growth factor to induce dentin
regeneration. Int J Dent. 2012:2584692012. View Article : Google Scholar
|
|
12
|
Zhang W, Zhang X, Ling J, Liu W, Zhang X,
Ma J and Zheng J: Proliferation and odontogenic differentiation of
BMP2 gene transfected stem cells from human tooth apical papilla:
An in vitro study. Int J Mol Med. 34:1004–1012. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang X, van der Kraan PM, van den Dolder
J, Walboomers XF, Bian Z, Fan M and Jansen JA: STRO-1 selected rat
dental pulp stem cells transfected with adenoviral-mediated human
bone morphogenetic protein 2 gene show enhanced odontogenic
differentiation. Tissue Eng. 13:2803–2812. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Iso T, Kedes L and Hamamori Y: HES and
HERP families: Multiple effectors of the Notch signaling pathway. J
Cell Physiol. 194:237–255. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zavadil J, Cermak L, Soto-Nieves N and
Böttinger EP: Integration of TGF-beta/Smad and Jagged1/Notch
signalling in epithelial-to-mesenchymal transition. EMBO J.
23:1155–1165. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wöltje K, Jabs M and Fischer A: Serum
induces transcription of Hey1 and Hey2 genes by Alk1 but not Notch
signaling in endothelial cells. PLoS One. 10:e01205472015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Salie R, Kneissel M, Vukevic M, Zamurovic
N, Kramer I, Evans G, Gerwin N, Mueller M, Kinzel B and Susa M:
Ubiquitous overexpression of Hey1 transcription factor leads to
osteopenia and chondrocyte hypertrophy in bone. Bone. 46:680–694.
2010. View Article : Google Scholar
|
|
18
|
Sakamoto M, Hirata H, Ohtsuka T, Bessho Y
and Kageyama R: The basic helix-loop-helix genes Hesr1/Hey1 and
Hesr2/Hey2 regulate maintenance of neural precursor cells in the
brain. J Biol Chem. 278:44808–44815. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kokubo H, Tomita-Miyagawa S, Hamada Y and
Saga Y: Hesr1 and Hesr2 regulate atrioventricular boundary
formation in the developing heart through the repression of Tbx2.
Development. 134:747–755. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Buas MF, Kabak S and Kadesch T: The Notch
effector Hey1 associates with myogenic target genes to repress
myogenesis. J Biol Chem. 285:1249–1258. 2010. View Article : Google Scholar :
|
|
21
|
Itoh F, Itoh S, Goumans MJ,
Valdimarsdottir G, Iso T, Dotto GP, Hamamori Y, Kedes L, Kato M and
ten Dijke Pt P: Synergy and antagonism between Notch and BMP
receptor signaling pathways in endothelial cells. EMBO J.
23:541–551. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun H, Kawashima N, Xu J, Takahashi S and
Suda H: Expression of Notch signalling-related genes in normal and
differentiating rat dental pulp cells. Aust Endod J. 36:54–58.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sharff KA, Song WX, Luo X, Tang N, Luo J,
Chen J, Bi Y, He BC, Huang J, Li X, et al: Hey1 basic
helix-loop-helix protein plays an important role in mediating
BMP9-induced osteogenic differentiation of mesenchymal progenitor
cells. J Biol Chem. 284:649–659. 2009. View Article : Google Scholar :
|
|
24
|
Zamurovic N, Cappellen D, Rohner D and
Susa M: Coordinated activation of notch, Wnt, and transforming
growth factor-beta signaling pathways in bone morphogenic protein
2-induced osteogenesis. Notch target gene Hey1 inhibits
mineralization and Runx2 transcriptional activity. J Biol Chem.
279:37704–37715. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Arany S, Nakata A, Kameda T, Koyota S,
Ueno Y and Sugiyama T: Phenotype properties of a novel
spontaneously immortalized odontoblast-lineage cell line. Biochem
Biophys Res Commun. 342:718–724. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang H, Kawashima N, Iwata T, Xu J,
Takahashi S, Sugiyama T and Suda H: Differentiation of odontoblasts
is negatively regulated by MEPE via its C-terminal fragment.
Biochem Biophys Res Commun. 398:406–412. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Han N, Zheng Y, Li R, Li X, Zhou M, Niu Y
and Zhang Q: β-catenin enhances odontoblastic differentiation of
dental pulp cells through activation of Runx2. PLoS One.
9:e888902014. View Article : Google Scholar
|
|
28
|
Salmon B, Bardet C, Khaddam M, Naji J,
Coyac BR, Baroukh B, Letourneur F, Lesieur J, Decup F, Le Denmat D,
et al: MEPE-derived ASARM peptide inhibits odontogenic
differentiation of dental pulp stem cells and impairs
mineralization in tooth models of X-linked hypophosphatemia. PLoS
One. 8:e567492013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
30
|
Zeng Z, Yin X, Zhang X, Jing D and Feng X:
Cyclic stretch enhances bone morphogenetic protein-2-induced
osteoblastic differentiation through the inhibition of Hey1. Int J
Mol Med. 36:1273–1281. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu J, Yu B, Hong C and Wang CY: KDM6B
epigenetically regulates odontogenic differentiation of dental
mesenchymal stem cells. Int J Oral Sci. 5:200–205. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen Z, Li W, Wang H, Wan C, Luo D, Deng
S, Chen H and Chen S: Klf10 regulates odontoblast differentiation
and mineralization via promoting expression of dentin matrix
protein 1 and dentin sialophosphoprotein genes. Cell Tissue Res.
363:385–398. 2016. View Article : Google Scholar :
|
|
33
|
Wang X, He H, Wu X, Hu J and Tan Y:
Promotion of dentin regeneration via CCN3 modulation on Notch and
BMP signaling pathways. Biomaterials. 35:2720–2729. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Prasad M, Butler WT and Qin C: Dentin
sialophosphoprotein in biomineralization. Connect Tissue Res.
51:404–417. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yamakoshi Y: Dentinogenesis and dentin
sialophosphoprotein (DSPP). J Oral Biosci. 51:134–142. 2009.
View Article : Google Scholar
|
|
36
|
Weber D, Wiese C and Gessler M: Hey bHLH
transcription factors. Curr Top Dev Biol. 110:285–315. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Heisig J, Weber D, Englberger E, Winkler
A, Kneitz S, Sung WK, Wolf E, Eilers M, Wei CL and Gessler M:
Target gene analysis by microarrays and chromatin
immunoprecipitation identifies HEY proteins as highly redundant
bHLH repressors. PLoS Genet. 8:e10027282012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Aranguren XL, Agirre X, Beerens M,
Coppiello G, Uriz M, Vandersmissen I, Benkheil M, Panadero J,
Aguado N, Pascual-Montano A, et al: Unraveling a novel
transcription factor code determining the human arterial-specific
endothelial cell signature. Blood. 122:3982–3992. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lavery K, Hawley S, Swain P, Rooney R,
Falb D and Alaoui-Ismaili MH: New insights into BMP-7 mediated
osteoblastic differentiation of primary human mesenchymal stem
cells. Bone. 45:27–41. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jacob A, Zhang Y and George A:
Transcriptional regulation of dentin matrix protein 1 (DMP1) in
odontoblasts and osteoblasts. Connect Tissue Res. 55(Suppl 1):
107–112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li S, Kong H, Yao N, Yu Q, Wang P, Lin Y,
Wang J, Kuang R, Zhao X, Xu J, et al: The role of runt-related
transcription factor 2 (Runx2) in the late stage of odontoblast
differentiation and dentin formation. Biochem Biophys Res Commun.
410:698–704. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fuke S, Minami N, Kokubo H, Yoshikawa A,
Yasumatsu H, Sasagawa N, Saga Y, Tsukahara T and Ishiura S: Hesr1
knockout mice exhibit behavioral alterations through the
dopaminergic nervous system. J Neurosci Res. 84:1555–1563. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Belyea BC, Naini S, Bentley RC and
Linardic CM: Inhibition of the Notch-Hey1 axis blocks embryonal
rhabdomyosarcoma tumorigenesis. Clin Cancer Res. 17:7324–7336.
2011. View Article : Google Scholar : PubMed/NCBI
|