Introduction
Idiopathic pulmonary fibrosis (IPF), also called
cryptogenic fibrosing alveolitis, is a devastating, age-associated
lung disease with a generally fatal outcome of undefined cause
(1,2). As an uncommon disease, IPF has an
incidence of 13–42 cases per 100,000 individuals, and it mainly
affects aged individuals (>50 years) (3). This type of disease was once
considered a chronic inflammatory condition, but a recent study
revealed that the fibrotic response is caused by abnormally
activated alveolar epithelial cells (4). The typical symptoms of IPF include
disabling fatigue and exertional breathlessness, and it is
continually accompanied by a non-productive cough (5). Numerous types of common pathologic
features have been reported for IPF patients, including
inflammation with lymphocytes, neutrophils and macrophages as well
as damage to endothelial and epithelial cells, which is followed by
proliferation of collagen deposition and fibroblasts (6). A previous study demonstrated that
IPF has a complex pathogenesis involving cytokines, mediators and
growth factors, and multiple cell types within the lung contribute
to IPF (7). No therapy has been
proven effective, and the median survival time for patients who
succumbed to IPF is ~3 years from the time of diagnosis (8,9).
As a result, the diagnosis and treatment of IPF have received
considerable attention.
Nucleotide-binding domain leucine-rich repeat
proteins (NLRPs) are involved in immunity and disease via their
ability to regulate inflammatory reactions to pathogen-derived and
endogenous damage signaling (10). Activation of NLRPs may result in
the formation of multiprotein inflammasome complexes, which may be
considered stages for the activation of inflammatory caspases by
their cleavage and recruitment (11). Previous evidence has revealed that
NLRP1 regulates the innate immune response, and its expression may
be detected in numerous immunocompetent cell types (12). In addition, NLRP1 is correlated
with certain autoimmune diseases, including type 1 diabetes,
generalized vitiligo, rheumatoid arthritis and Addison disease
(13). The NLRP1 inflammasome
consists of NLRP1, and it was the first to be discovered as an
apoptosis-associated speck-like protein, which contains one
caspase-activating recruitment domain and caspase-1 (14). Among the NLRP family members, the
NLRP3 inflammasome has been involved in distinguishing
non-microbial origin damage-associated molecular patterns (DAMPs),
including extracellular ATP, urate crystals, asbestos and silica.
β-amyloid NLRP3 is known to be present in several tissue and cell
types (15). As demonstrated by a
previous study, the NLRP3 inflammasome has a crucial role in the
pathogenesis of fibrotic respiratory diseases (16). The mechanistic involvement of the
NLRP1 and NLRP3 inflammasome pathways in pulmonary fibrosis has
remained to be demonstrated; therefore, the present study
investigated the effects of the NLRP1 and NLRP3 inflammasome
pathways on murine cytomegalovirus (MCMV)-induced pulmonary
fibrosis in mice.
Materials and methods
Animal experiment
A total of 77 BALB/c mice (age, 8 weeks; weight,
18.0±2.0 g), purchased from the Shanghai Experimental Animal Center
of the Chinese Academy of Sciences (Shanghai, China), were used in
the present study. All specific-pathogen-free (SPF) mice were kept
in an isolated cage (individual ventilated cages, Suzhou Science
& Education Equipment Co., Ltd, Suzhou, China) to prevent
infection from which 55 mice were randomly selected and divided
into the following 5 groups containing 11 mice in each group:
Control, bleomycin (BLM), MCMV, MCMV+BLM (BLM was added after MCMV
infection) and MCMV+BLM+CD4+ T-cell group (BLM was added
after MCMV infection and then CD4+ T-cells were
injected). All mice were fed with standard feed, free access to
food and water, the feeding temperature was 18–22°C with relative
humidity of 60±10%, under a 12 h light/dark. The details concerning
grouping and treatment are displayed in Table I. The MCMV Smith strain used in
the present study was purchased from the American Type Culture
Collection (Manassas, VA, USA). Virus multiplication was performed
in the salivary glands of an additional 20 BALB/c mice, house under
the aforementioned conditions; the strain was collected for the
subsequent experiment. The experimental method was approved by the
animal ethics committee of the First Affiliated Hospital of Anhui
Medical University (Hefei, China). The animal experiment was
performed in strict accordance with the Guidelines for the Care and
Use of Laboratory Animals (17).
 | Table ITreatment regimens. |
Table I
Treatment regimens.
| Group | Treatment
regimen |
|---|
| Control | Hank's solution
(200 µl) and PBS (80 µl) |
| BLM | Hank's solution
(200 µl) and 3 mg/kg BLM [vehicle: PBS (80 µl)] |
| MCMV | 105 PFU
MCMV [vehicle: Hank's solution (200 µl)] and PBS (80
µl) |
| MCMV + BLM | 105 PFU
MCMV and 3 mg/kg BLM |
| MCMV + BLM +
CD4+ T | 105 PFU
MCMV, 3 mg/kg BLM (80 µl) and 1×107
CD4+ T |
Isolation and culture of mouse spleen T
lymphocytes
Following an intraperitoneal injection of 1%
pentobarbital sodium (30 mg/kg), two mice were sacrificed by
cervical dislocation and their spleens were collected under sterile
conditions. These spleens were added into the phosphate buffer to
create a suspension. The suspension was added on the top of a
lymphocyte separation solution (1:2) for centrifugation at 1,200 ×
g for 20 min to collect the splenic lymphocytes (1.5×108
cells). Mouse lymphocytes were then isolated with 37°C pre-heated
PBS containing 5% fetal calf serum (Hyclone; GE Healthcare Life
Sciences, Logan, UT, USA) and diluted into 2 ml, which was followed
by placement on a nylon wool column (4 cm in height, Polysciences
Inc., Warrington, PA, USA) for incubation at 37°C for 60 min.
Subsequently, the column was washed with 5 ml nylon buffer solution
pre-heated at 37°C with a flow rate of 0.5 ml/min. The eluates were
collected and considered T lymphocytes. Another 5 ml of nylon
buffer solution was used to wash the remaining T lymphocytes. In
addition, another nylon wool column (4 com in height, Polysciences
Inc.) was used to separate and purify the cells, which was followed
by the collation of T lymphocytes. The T lymphocytes were added to
0.5 µg/ml soluble anti-CD3 antibody (1:25, AB_2228819,
eBiosciences; Thermo Fisher Scientific Inc., Waltham, MA, USA) to
collect activated CD4+ T-cells.
Model establishment and detection of
viral load
The MCMV passaged in salivary glands of BALB/c mice
was suspended in Hank's solution (Thermo Fisher Scientific Inc.)
containing 200 µl 3% fetal bovine serum (FBS, Hyclone; GE
Healthcare Life Sciences). A total of 33 mice were randomly
selected, and 105 PFU MCMV was intraperitoneally
injected for infection (18). The
remaining 22 mice were intraperitoneally injected with Hank's
solution containing 200 µl 3% FBS and were used as the
controls. After treatment with MCMV for 3, 14 or 28 days, infected
mice (n=3) and uninfected mice (n=3) were randomly selected to
assess the viral loads in the salivary glands, kidneys, liver and
lungs via semi-quantitative reverse-transcription polymerase chain
reaction (RT-PCR). The mouse specimens from salivary glands,
kidney, liver and lungs were homogenized, respectively. PBS was
added to the homogenate and then centrifuged at 1,500 × g for 10
min; the supernatant was then collected. Total RNA was extracted
from the supernatant using an RNeasy Mini kit (cat. no. 74106,
Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's
protocols and subsequently reverse-transcribed into cDNA using a
RT2 First Strand kit (330404, Qiagen, Inc.) according to the
manufacturer's protocols. The cDNA was subjected to amplification
by RT-PCR. The reaction system (25 µl) was as follows: 2.5
µl 10X buffer (containing 15 mmol/l Mg2+), 1.0
µl deoxynucleoside triphosphate (2.5 mmol/l), 0.35 µl
forward primer (25 µmol/l), 0.35 µl reverse primer
(25 µmol/l), 0.3 µl Taq DNA polymerase (5
U/µl), 19.5 µl double-distilled H2O and 1
µl template. The forward primer was
5′-ATCTGGTGCTCCTCAGATCAGCTAA-3′ and the reverse primer was
5′-ATTGTTCATTGCCTGGGGAGTTT-3′. GAPDH served as the internal
reference: Forward, 5′-CCACAGTCCATGCCATCACT-3′ and reverse,
5′-TCCACCACCCTGTTGCTGTAG-3′. The PCR reaction conditions were as
follows: 40 cycles of 95°C for 10 min, 94°C for 1 min, 63°C for 30
sec and 72°C for 1 min with a final elongation at 70°C for 7 min
and 4°C at 5 min to stop the reaction. The experiment was performed
three times; the PCR product was subjected to 2% agarose gel
electrophoresis with ethidium bromide staining and observed under
an ultraviolet lamp, and the gray value was detected by ImageJ
software (v1.44, National Institutes of Health, Bethesda, MD, USA).
Following treatment with MCMV for 4 weeks, mice infected with MCMV
were divided into the MCMV, MCMV+BLM and MCMV+BLM+CD4+ T
groups, and mice without MCMV infection were assigned to the
control and BLM groups with 11 mice in each group. Mice in the
MCMV, MCMV+BLM and MCMV+BLM+CD4+ T groups were treated
by sevoflurane inhalation anesthesia (Abbott Laboratories, Abbott
Park, IL, USA), and the supine position was used for treatment with
80 µl BLM (0.75 U/ml) via tracheal intubation. At the same
time, mice in the MCMV+BLM+CD4+ T group were
intravenously injected with 1×107/ml (1 ml)
CD4+ T (19). Mice in
the MCMV and control groups were injected with an isodose of PBS,
and animals were sacrificed after 4 weeks.
Detection of the weight, lung coefficient
and hydroxyproline (HYP)
The mice were weighed prior to model establishment
and at the end of the experiment. After the model was established
on the 28th day, all mice were sacrificed and the trachea was
intubated, which was followed by lavage of lung tissues using 0.5
ml PBS. Subsequently, the lung tissues were weighed and the lung
coefficient was calculated using the following formula: Lung
coefficient=wet weight of both lungs (mg)/weight (g). The HYP
content in mouse lung tissues was detected by alkaline hydrolysis
and spectrophotography according to the instructions of the HYP kit
(A030-2, Nanjing Jiancheng Bioengineering Institute, Nanjing,
China).
Histopathological observation
Following an intraperitoneal injection of 1%
pentobarbital sodium (30 mg/kg), the mice were killed by 3–5-min
exsanguination via the abdominal aorta. The anocelia was opened
immediately, and the right main bronchus was ligated, which was
followed by injection of 4% paraformaldehyde (Wuhan Boster
Biological Technology Ltd., Wuhan, China) via the trachea to fix
the right lung tissues. Subsequently, the lung tissues were
immediately removed, fixed in 4% paraformaldehyde at 4°C for 24 h,
dehydrated by gradient ethanol (75, 85, 95 and 100%, for 3 min
each), embedded in paraffin and sectioned into 5-µm serial
tissue sections. The sections were dewaxed in dimethylbenzene and
hydrated in gradient ethanol (100, 95, 85 and, 75% for 3 min each).
At room temperature, hematoxylin and eosin (HE) staining was
performed to evaluate histopathological changes of lung tissues and
Masson staining was performed to evaluate collagen deposition.
Alveolitis and the degree of pulmonary fibrosis were scored by HE
and Masson staining as in the study by Szapiel et al
(20): 1 point-no; 2 points-mild;
3 points-moderate; 4 points-severe.
RT-quantitative PCR analysis
Total RNA of lung tissues was extracted according to
the instructions of RNeasy Mini kit. The RNA concentration was
determined via optical density (OD) measurement at 260/280 nm using
an ultraviolet spectrophotometer, and the sample was preserved at
−80°C. RT was performed to synthetize complementary DNA according
to the RT2 First Strand kit's instructions (cat. no. 330404,
Qiagen, Inc.). Primers were designed based on the gene sequence
published in the GenBank database. The sequences of the primers are
presented in Table II and were
synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). The PCR
system (20 µl) had the following components: 10 µl
SYBR PremixExTaq (2X), 0.8 µl forward primer (10 µM),
0.8 µl reverse primer (10 µM), 0.4 µl ROX
Reference Dye II (50X), 2 µl DNA template and 6.0 µl
double-distilled H2O. The reaction conditions were as
follows: Pre-denaturation at 95°C for 30 sec, denaturation at 95°C
for 5 sec, annealing at 60°C for 30 sec and elongation at 72°C for
30 sec for a total of 40 cycles. GAPDH was used as an internal
reference. The reliability of the PCR results was evaluated by a
dissolution curve. The Cq value (the inflection point on the
amplification power curve) was determined, and the relative
expression of the target gene was calculated as 2−∆∆Cq
(21).
 | Table IIPrimer sequences for polymerase chain
reaction. |
Table II
Primer sequences for polymerase chain
reaction.
| mRNA | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| IL-1β |
TCTTTGAAGTTGACGGACCC |
TGAGTGATACTGCCTGCCTG |
| IL-18 | ACATCCGAAGCAACAAGC
C |
GAAGTGAGAAGGCAACA |
| Caspase-1 |
TGGAAGGTAGGCAAGACT |
ATAGTGGGCATCTGGGTC |
| GAPDH |
ACCACAGTCCATGCCATCAC |
TCCACCACCCTGTTGCTGTA |
Western blot analysis
The lung tissues (20 mg) were ground in liquid
nitrogen using a mortar and pestle, and dissociated in 250
µl radioimmunoprecipitation assay buffer (P0013C, Beyotime
Institute of Biotechnology, Jiangsu, China) for the extraction of
total protein. The concentration of the total protein was detected
based on the instructions of the bicinchoninic acid kit (Wuhan
Boster Biological Technology, Ltd.). After the addition of sample
buffer solution, the extracted protein was heated at 95°C for 10
min, and separated and purified using 10% SDS-PAGE (Wuhan Boster
Biological Technology, Ltd.) with 30 µg total protein loaded
per well. The protein was transferred to a polyvinylidene fluoride
membrane (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) using the
semi-dry film method, followed by blocking with 5% bovine serum
albumin (ST023, Beyotime Institute of Biotechnology) at room
temperature for 1 h. Subsequently, primary antibodies to mature
caspase-1 (1:1,000; ab62698), pro-caspase-1 (1:1,000; ab179515),
interleukin (IL)-1β (1:1,000, ab106035) and IL-18 (1:1,000,
ab207323) and GAPDH (1:2,500, ab9485) (all from Abcam) were added,
followed by incubation at 4°C overnight. The membrane was washed 3
times with Tris-buffered saline containing Tween-20 for 5 min each
time, followed by addition of relevant secondary antibody, goat
anti-rabbit immunoglobulin G (1:2,000, ab205718, Abcam) for
incubation at room temperature for 1 h. The membrane was washed 3
times for 5 min each and developed using a chemiluminescence
reagent (36222ES60, Yeasen Biotechnology, Co., Ltd., Shanghai,
China). GAPDH was used as an internal reference. The Bio-Rad Gel
Dol EZ Imager (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was
used for development. Gray value analysis of protein bands was
performed using ImageJ software (v1.44, National Institutes of
Health).
ELISA
At the end of the experiment, a total of 3–5 ml
blood was collected from the abdominal aorta and preserved at 4°C
in a refrigerator, which was followed by centrifugation at 2,400 ×
g for 10 min at 4°C. Subsequently, the serum phase was collected
and placed in numbered tubes with 200 µl in each tube, which
was followed by preservation at −80°C. The analysis of the serum
levels of caspase-1 (AG-45B-0002), TNF-α (ADI-900-047), IL-1β
(ADI-900-132A) and IL-18 (bsk00297) was performed in strict
accordance with the ELISA kit instructions (Shanghai Meilian
Biotechnology Co., Ltd., Shanghai, China).
Statistical analysis
SPSS 18.0 software (SPSS Inc, Chicago, IL, USA) was
applied for statistical analysis. Values are expressed as the mean
± standard deviation. Comparison of measurement data that followed
a normal distribution between two groups was evaluated by the
Student's two-tailed t-test, and comparison among multiple groups
was performed by one-way analysis of variance. Post hoc multiple
comparisons were achieved by means of least significance difference
method if homogeneity of variance was appropriate otherwise the
non-parametric Kruskal-Wallis test was employed. P<0.05 was
considered to indicate a statistically significant difference.
Results
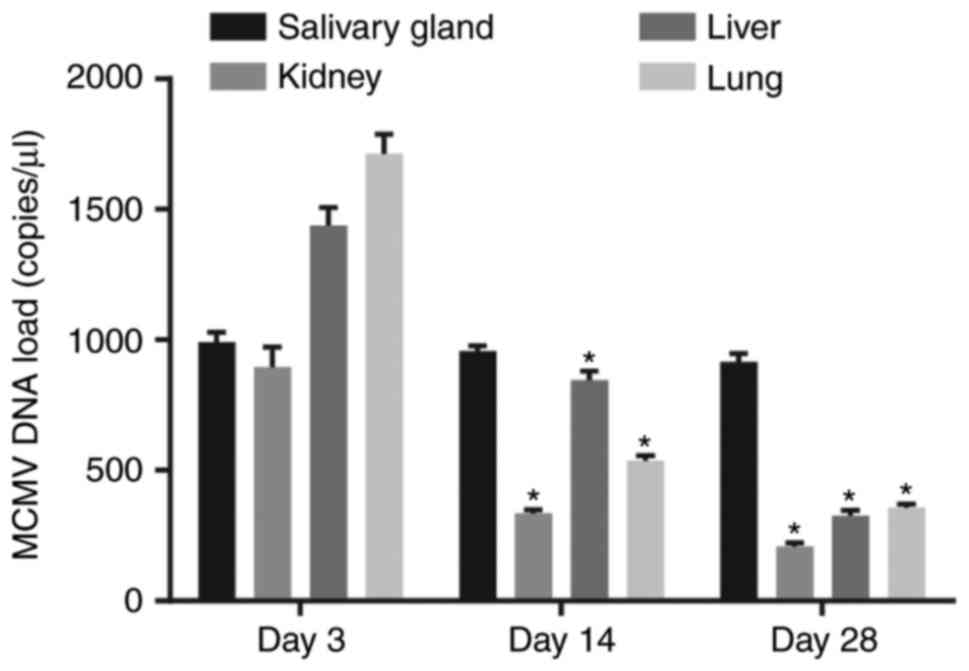
MCMV DNA load is stable in the salivary
glands of mice
The viral load in various tissue types of the
MCMV-infected mice at 3, 14, and 28 days following treatment is
presented in Fig. 1. MCMV DNA was
not detected in mice without MCMV treatment, while it was
detectable in the salivary glands, kidneys, liver and lungs of mice
treated with MCMV. The MCMV DNA content in the kidney, liver and
lungs of mice treated with MCMV was highest at 3 days after
inoculation. In addition to the salivary glands, the viral load was
gradually reduced and maintained at a low level in other tissues,
which was similar to the observations in humans with latent
infection.
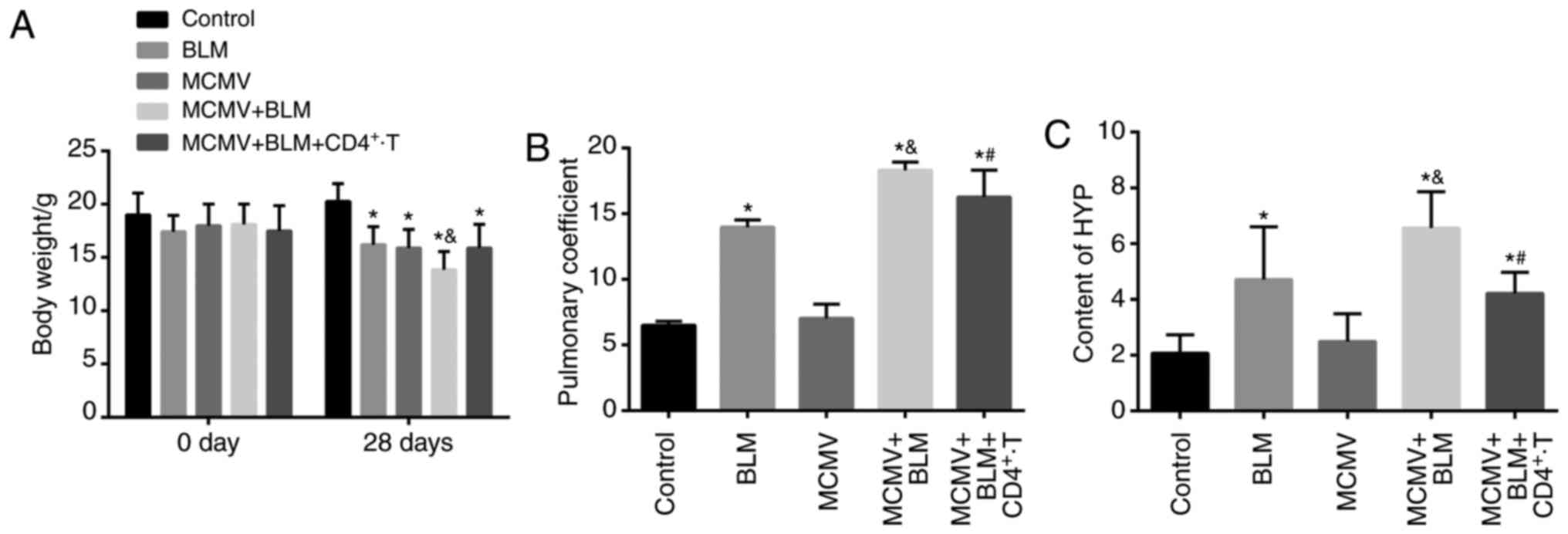
BLM exacerbates but CD4+
T-cell injection ameliorates lung tissue damage associated with
latent respiratory virus infection
The weight, lung coefficient and HYP content of mice
in each group are displayed in Fig.
2. Prior to modeling, no significant difference was identified
in the murine weight (P>0.05). Compared with the control group,
the weight of mice in the BLM, MCMV, BLM+MCMV and
MCMV+BLM+CD4+ T groups was significantly decreased at
the end of the experiment, and the weight in the MCMV+BLM group was
significantly lower than that in the BLM, MCMV and
MCMV+BLM+CD4+ T groups (P<0.05). No significant
difference in body weight was identified among the mice in the BLM,
MCMV and MCMV+BLM+CD4+ T groups (P>0.05; Fig. 2A). The lung coefficient and HYP
content were significantly higher in the BLM, BLM+MCMV and
MCMV+BLM+CD4+ T groups compared with those in the
control group (P<0.05). No such significant difference was
identified between the HYP content and lung coefficient between the
MCMV and control groups (P>0.05). Compared with the BLM group,
mice in the MCMV+BLM group had an elevated lung coefficient and HYP
content P<0.05. Of note, compared with the MCMV+BLM group, mice
in the MCMV+BLM+CD4+ T group had a decreased lung
coefficient and HYP content (P<0.05; Fig. 2B and C).
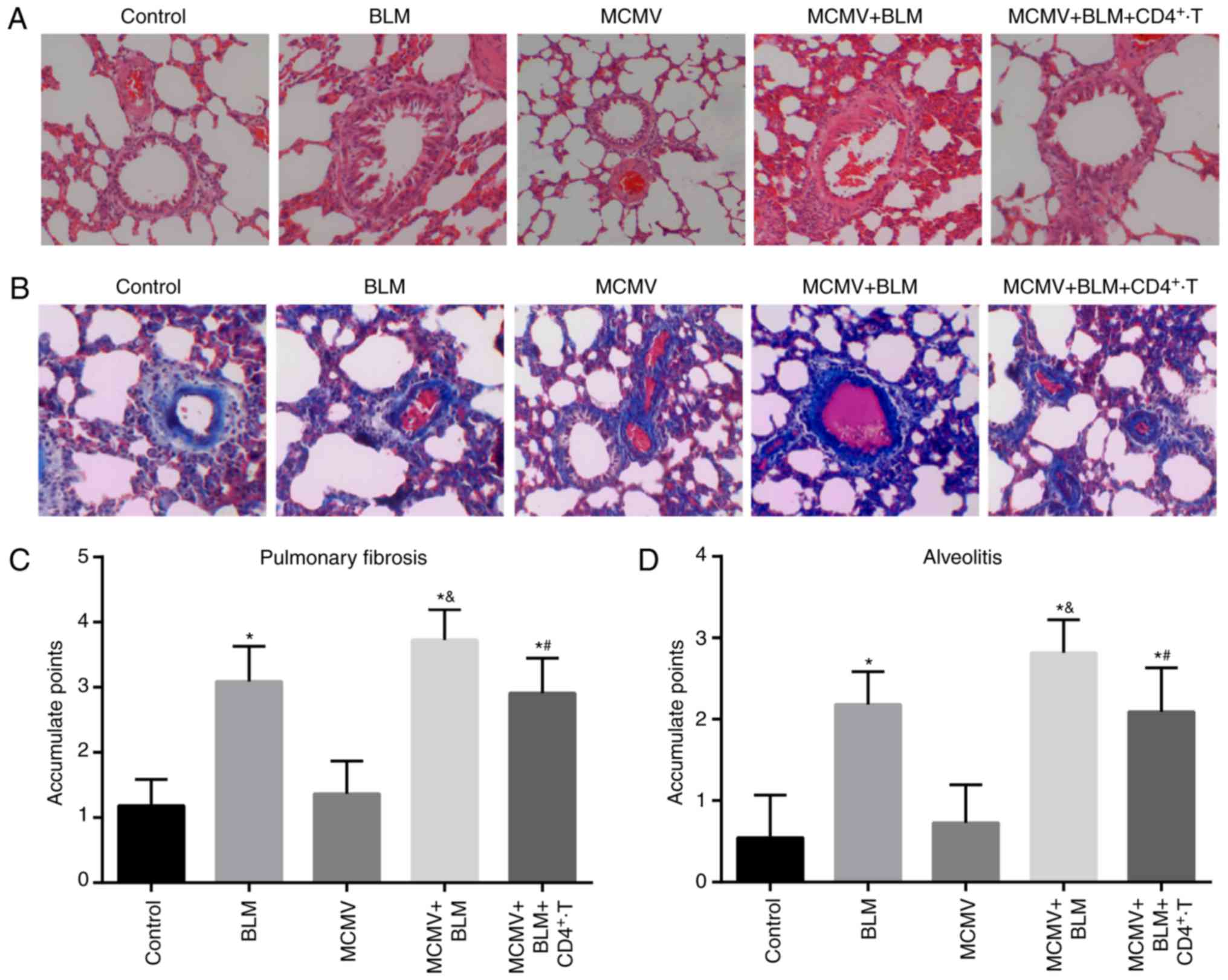
BLM exacerbates but CD4+
T-cell injection ameliorates pulmonary fibrosis associated with
latent respiratory virus infection
HE staining and Masson staining of lung tissues in
the control group revealed normal structures of the alveolar wall
without inflammatory cell infiltration around the alveoli, and only
few blue collagen fibers were present in the alveolar space
(Fig. 3A and B). The histology
results for the MCMV group were similar to those for the control
group. In the BLM group, moderate damage to structures of the
alveolar wall, infiltration of inflammatory cells in the alveoli
and alveolar cavities, hyperemia and edema in the lung
interstitium, fibroblast proliferation and an increased number of
blue collagen fibers were observed. In the MCMV+BLM group, severe
damage to the alveolar structure with massive inflammatory cell
infiltration in the alveoli and alveolar cavities, severe hyperemia
and edema in the lung interstitium, a high level of fibroblast
proliferation and a large amount of blue collagen fibers were
observed. Compared with the MCMV+BLM group, the damage in the
MCMV+BLM+CD4+ T group was ameliorated. Quantitative
scoring revealed that compared with the control group, mice in the
BLM, MCMV+BLM and MCMV+BLM+CD4+ T groups had obviously
increased alveolitis and a higher degree of pulmonary fibrosis. In
the MCMV+BLM group, these features were also more pronounced than
in the BLM group (all P<0.05). No significant difference was
identified between the MCMV and control groups. Compared with the
MCMV+BLM group, the MCMV+BLM+CD4+ T group exhibited
obviously reduced alveolitis and pulmonary fibrosis (Fig. 3C and D).
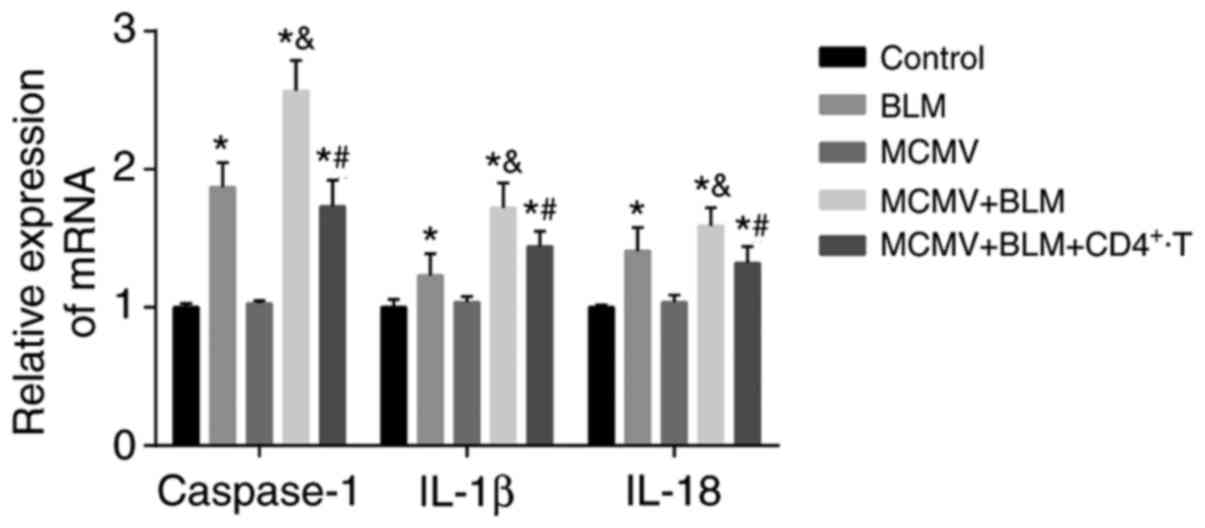
Latent respiratory virus infection
activates NLRP1/NLRP3 inflammasome pathway, evaluated by
RT-PCR
Compared with the control group, the BLM, MCMV+BLM
and MCMV+BLM+CD4+ T groups had increased mRNA expression
levels of caspase-1, IL-1β and IL-18 in lung tissues (all
P<0.05), while no significant difference was identified between
the MCMV and control groups (P>0.05). The mRNA expression levels
of caspase-1, IL-1β and IL-18 in the lung tissues of the MCMV+BLM
group were significantly higher than those in the BLM group (all
P<0.05). Compared with the MCMV+BLM group, the
MCMV+BLM+CD4+ T group had decreased mRNA expression
levels of caspase-1, IL-1β and IL-18 (all P<0.05; Fig. 4).
Latent respiratory virus infection
activates NLRP1/NLRP3 inflammasome pathway, evaluated by western
blot analysis
No significant difference was identified in the
protein expression levels of pro-caspase-1, pro-IL-1β and pro-IL-18
among all groups (all P<0.05). The BLM, MCMV+BLM and
MCMV+BLM+CD4+ T groups had increased protein expression
levels of caspase-1 (p20), mature IL-1β and mature IL-18 in lung
tissues compared with those in the control group (all P<0.05).
No such significant difference was identified between the MCMV and
control groups (P>0.05). The protein expression levels of
caspase-1 (p20), mature IL-1β and mature IL-18 in lung tissues of
the MCMV+BLM group were significantly higher than those in the BLM
group (all P<0.05). Compared with the MCMV+BLM group, the
MCMV+BLM+CD4+ T group had decreased protein expression
levels of caspase-1 (p20), mature IL-1β and mature IL-18 (all
P<0.05; Fig. 5).
BLM increases but CD4+ T-cell
injection reduces inflammatory response
Fig. 6 displays
the caspase-1, TNF-α, IL-1β and IL-18 levels in the serum of mice
in each group. Compared with those in the control group, the mice
in the BLM, MCMV+BLM and MCMV+BLM+CD4+ T groups had
increased serum levels of caspase-1, TNF-α, IL-1β and IL-18 (all
P<0.05). No such significant difference was identified between
the MCMV and control groups (P>0.05). The caspase-1, TNF-α,
IL-1β and IL-18 levels in the serum of mice in the MCMV+BLM group
were significantly higher than those in the BLM group (all
P<0.05). Compared with the MCMV+BLM group, the
MCMV+BLM+CD4+ T group had decreased caspase-1, TNF-α,
IL-1β and IL-18 levels (all P<0.05).
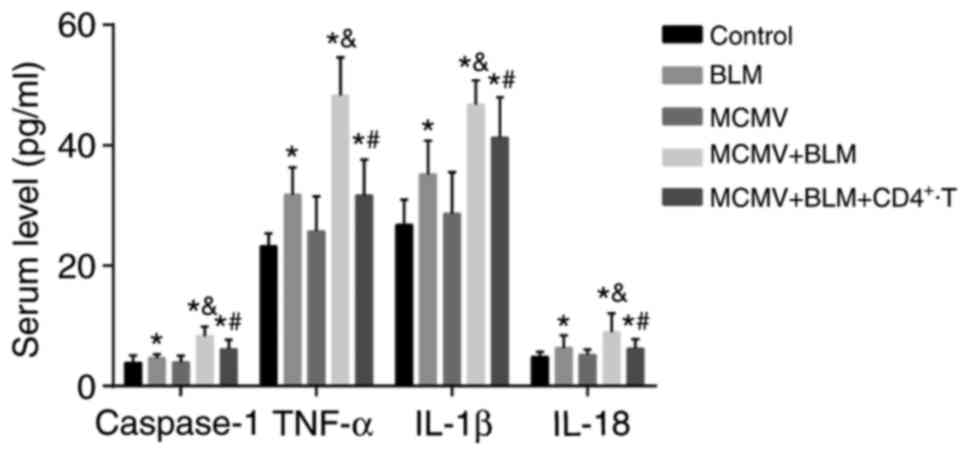 | Figure 6Comparison of the caspase-1, TNF-α,
IL-1β and IL-18 levels in serum among the three groups.
*P<0.05, compared with the control group;
&P<0.05, compared with the BLM group; and
#P<0.05, compared with the MCMV+BLM group. BLM,
bleomycin; T, T-tells; MCMV, murine cytomegalovirus; IL,
interleukin; TNF, tumor necrosis factor. |
Discussion
Previous studies have suggested a crucial role of
inflammasomes in processing IL-1β, affecting the pathogenesis of
autoimmune disease, as well as central nervous system infection and
injury (22–25). Therefore, the present in
vivo study was performed to investigate the role of the
NLRP1/NLRP3 inflammasome pathways in MCMV-induced pulmonary
fibrosis in mice. The results demonstrated that the NLRP1 and NLRP3
inflammasome pathways are upregulated in MCMV-induced pulmonary
fibrosis in mice, which is ameliorated by treatment with
CD4+ T cells.
The results of the present study indicated that BLM
exacerbates but CD4+ T-cell injection ameliorates
pulmonary fibrosis associated with latent respiratory virus
infection. Cytomegalovirus (CMV), a major human pathogen, is
generally controlled by cellular immune responses (26). CMV, a herpes virus, may contribute
to life-threatening pulmonary infections in immunocompromised
patients (27). The BLM-induced
fibrosis model in mice and rodents has been widely employed by
experimental studies for >20 years due to its important
characteristics of closely mimicking IPF (28,29). Consistent with the present study,
Gwinn et al (30) also
demonstrated that mice treated with BLM had a reduced weight. HYP
is a common index for collagen quantification (31). A previous study verified that
BLM-induced fibrosis was associated with an increased lung
coefficient and HYP content (32). CD4+ T (helper) cells
have an important role in the immune system, which not only help
macrophages respond to intracellular antigens but also help B cells
form germinal centers (33). In
addition, previous evidence also demonstrated that T cells
inhibited the innate immune response via suppressing NLRP3 and
NLRP1 inflammasomes (34). In
accordance with this, the MCMV+BLM+CD4+ T group of the
present study had a decreased lung coefficient and HYP content
compared with the MCMV+BLM group.
In addition, the present study revealed that NLRP1
and NLRP3 inflammasome pathways were activated when latent
respiratory virus infection occurred. Inflammasomes have a crucial
role in mediating caspase-1, which promotes the release of IL-1β
and IL-18 (35). Previous studies
have demonstrated that BLM treatment induced the production of
reactive oxygen species, which also activated the NLRP3
inflammasome in numerous cases (36,37). In addition, the production of
IL-1β and lung inflammation induced by BLM relies on
apoptosis-associated speck-like protein containing a caspase
recruitment domain (ASC) (38).
Inflammasomes, which are multiprotein complexes, stimulate
caspase-1, and inflammasome assembly occurs in response to
metabolic reprogramming, which is similar to that triggered by
cellular transformation and infection with viruses (including CMV)
(39). Furthermore, NLRs only
function as sensors of endogenous or exogenous damage-associated
molecules, but they also activate caspase-1 and promote subsequent
cleavage of pro-IL-1β and pro-IL-18 cytokines into their mature
forms (40). In addition,
post-translational cleavage of pro-IL-1β to mature IL-1β is
required for its functional activity, which is achieved by the
inflammasome via caspase-1 activation (41). Furthermore, activation of T-cells
and antigenic stimulation a considered a necessary precondition for
blocking NLRP inflammasomes, which agrees with the prior migration
of activated T cells to inflammatory sites 42).
Furthermore, the present study revealed that BLM
increases but CD4+ T-cell injection reduces inflammatory
response. Cytokines are mainly involved in the pathophysiological
and homeostatic regulation of connective tissue, and a complex
network consisting of several cytokines has an important role in
local injury and the inflammatory response in the lung as well as
subsequent tissue repair and fibrosis (43). Previous evidence has demonstrated
that TNF-α promoted the development of lung fibrosis (44). The activation and assembly of the
inflammasome (consisting of an NLR family member, ASC and
pro-caspase-1) was reported to induce the production of caspase-1
(45). Another noteworthy study
reported that pharmacologically induced NLRP3 activation may result
in interstitial lung disease (46). IL-18, a member of the highly
inflammatory cytokines of the IL-1 family, has a central role in
regulating lung inflammation (47). Furthermore, suppression of
caspase-1 in lung fibroblasts obviously reduced the expression
levels of IL-1β and IL-18 (48).
As reflected in one study performed by Huang et al (49), the levels of TNF-α, IL-1β and
IL-18 were evidently increased in mice after BLM treatment.
Previous studies have therefore revealed that BLM stimulates
inflammasomes. Finally, the production of active IL-1β activates
transforming growth factor-β, which participates in the development
of lung fibrosis (50,51). In addition, CMV infection of
immune cells induces the production of a variety of cytokines,
including TNF-α, IL-1β and IL-6 (52,53). Furthermore, CD41 effector and
memory T cells were demonstrated to selectively block the NLRP1 and
NLRP3 inflammasomes in an antigen-dependent manner (34). Collectively, NLRP1 and NLRP3
inflammasomes may have increased the inflammatory response in mice
with pulmonary fibrosis caused by latent MCMV infection.
In conclusion, employing a mouse model of pulmonary
fibrosis, the present study provided evidence that the activation
of the NLRP1 and NLRP3 inflammasome pathways may contribute to
pulmonary fibrosis caused by latent MCMV infection in mice.
Signaling pathways may be affected by various factors and further
study should be performed to confirm these conclusions.
Acknowledgments
Not applicable.
Notes
[1]
Funding
No funding was received.
[2] Availability
of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
[3] Authors'
contributions
YHL, XW, SJ, SYG and SMZ contributed to the
conception of the work and conduction of the study. SYG and SMZ
revised the draft. All authors approved the final version of the
manuscript, and agreed for all aspects of the work.
[4] Ethics
approval and consent to participate
The experimental method was approved by the animal
ethics committee of the First Affiliated Hospital of Anhui Medical
University (Hefei, China). The animal experiment was performed in
strict accordance with the Guidelines for the Care and Use of
Laboratory Animals (17).
[5] Consent for
publication
Not applicable.
[6] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Huang H, Peng X and Zhong C: Idiopathic
pulmonary fibrosis: The current status of its epidemiology,
diagnosis, and treatment in China. Intractable Rare Dis Res.
2:88–93. 2013.PubMed/NCBI
|
|
2
|
Loveman E, Copley VR, Scott DA, Colquitt
JL, Clegg AJ and O'Reilly KM: Comparing new treatments for
idiopathic pulmonary fibrosis-a network meta-analysis. BMC Pulm
Med. 15:372015. View Article : Google Scholar
|
|
3
|
Lynch JP III, Huynh RH, Fishbein MC,
Saggar R, Belperio JA and Weigt SS: Idiopathic Pulmonary fibrosis:
Epidemiology, clinical features, prognosis, and management. Semin
Respir Crit Care Med. 37:331–357. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
King TE Jr, Pardo A and Selman M:
Idiopathic pulmonary fibrosis. Lancet. 378:1949–1961. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Strookappe B, Elfferich M, Swigris J,
Verschoof A, Veschakelen J, Knevel T and Drent M: Benefits of
physical training in patients with idiopathic or end-stage
sarcoidosis-related pulmonary fibrosis: A pilot study. Sarcoidosis
Vasc Diffuse Lung Dis. 32:43–52. 2015.PubMed/NCBI
|
|
6
|
Dempsey OJ, Kerr KM, Gomersall L, Remmen H
and Currie GP: Idiopathic pulmonary fibrosis: An update. QJM.
99:643–654. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Spagnolo P, Rossi G and Cavazza A:
Pathogenesis of idiopathic pulmonary fibrosis and its clinical
implications. Expert Rev Clin Immunol. 10:1005–1017. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Collard HR, Ryerson CJ, Corte TJ, Jenkins
G, Kondoh Y, Lederer DJ, Lee JS, Maher TM, Wells AU, Antoniou KM,
et al: Acute exacerbation of idiopathic pulmonary fibrosis. An
International Working Group Report. Am J Respir Crit Care Med.
194:265–275. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fernández Pérez ER, Daniels CE, Schroeder
DR, St Sauver J , Hartman TE, Bartholmai BJ, Yi ES and Ryu JH:
Incidence, prevalence, and clinical course of idiopathic pulmonary
fibrosis: A population-based study. Chest. 137:129–137. 2010.
View Article : Google Scholar
|
|
10
|
Finger JN, Lich JD, Dare LC, Cook MN,
Brown KK, Duraiswami C, Bertin J and Gough PJ: Autolytic
proteolysis within the function to find domain (FIIND) is required
for NLRP1 inflammasome activity. J Biol Chem. 287:25030–25037.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Saresella M, La Rosa F, Piancone F, Zoppis
M, Marventano I, Calabrese E, Rainone V, Nemni R, Mancuso R and
Clerici M: The NLRP3 and NLRP1 inflammasomes are activated in
Alzheimer's disease. Mol Neurodegener. 11:232016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
D'Osualdo A and Reed JC: NLRP1, a
regulator of innate immunity associated with vitiligo. Pigment Cell
Melanoma Res. 25:5–8. 2012. View Article : Google Scholar
|
|
13
|
Levandowski CB, Mailloux CM, Ferrara TM,
Gowan K, Ben S, Jin Y, McFann KK, Holland PJ, Fain PR, Dinarello CA
and Spritz RA: NLRP1 haplotypes associated with vitiligo and
autoimmunity increase interleukin-1β processing via the NLRP1
inflammasome. Proc Natl Acad Sci USA. 110:2952–2956. 2013.
View Article : Google Scholar
|
|
14
|
Wang YC, Li WZ, Wu Y, Yin YY, Dong LY,
Chen ZW and Wu WN: Acid-sensing ion channel 1a contributes to the
effect of extracellular acidosis on NLRP1 inflammasome activation
in cortical neurons. J Neuroinflammation. 12:2462015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vandanmagsar B, Youm YH, Ravussin A,
Galgani JE, Stadler K, Mynatt RL, Ravussin E, Stephens JM and Dixit
VD: The NLRP3 inflammasome instigates obesity-induced inflammation
and insulin resistance. Nat Med. 17:179–188. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rastrick J and Birrell M: The role of the
inflammasome in fibrotic respiratory diseases. Minerva Med.
105:9–23. 2014.PubMed/NCBI
|
|
17
|
National Research Council (US) Committee
for the Update of the Guide for the Care and Use of Laboratory
Animals: Guide for the care and use of laboratory animals. Eighth
Edition. Guide for the Care and Use of Laboratory Animals, National
Academies Press (US); Washington: 103. pp. 1072–1073. 2010
|
|
18
|
Li Y, Gao J, Wang G and Fei G: Latent
cytomegalovirus infection exacerbates experimental pulmonary
fibrosis by activating TGF-β1. Mol Med Rep. 14:1297–1301. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Banczyk D, Kalies K, Nachbar L, Bergmann
L, Schmidt P, Bode U, Teegen B, Steven P, Lange T, Textor J, et al:
Activated CD4+ T cells enter the splenic T-cell zone and
induce autoantibody-producing germinal centers through bystander
activation. Eur J Immunol. 44:93–102. 2014. View Article : Google Scholar
|
|
20
|
Szapiel SV, Elson NA, Fulmer JD,
Hunninghake GW and Crystal RG: Bleomycin-induced interstitial
pulmonary disease in the nude, athymic mouse. Am Rev Respir Dis.
120:893–899. 1979.PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
22
|
Labzin LI, Lauterbach MA and Latz E:
Interferons and inflammasomes: Cooperation and counterregulation in
disease. J Allergy Clin Immunol. 138:37–46. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Walsh JG, Muruve DA and Power C:
Inflammasomes in the CNS. Nat Rev Neurosci. 15:84–97. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Masood H: Inflammasomes in the
pathophysiology of kidney diseases. Kidney Dis (Basel). 1:187–193.
2015. View Article : Google Scholar
|
|
25
|
Yang CA and Chiang BL: Inflammasomes and
human autoimmunity: A comprehensive review. J Autoimmun. 61:1–8.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sierro S, Rothkopf R and Klenerman P:
Evolution of diverse antiviral CD8+ T cell populations
after murine cytomegalovirus infection. Eur J Immunol.
35:1113–1123. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Restrepo-Gualteros SM, Jaramillo-Barberi
LE, Gonzalez-Santos M, Rodriguez-Martinez CE, Perez GF, Gutierrez
MJ and Nino G: Characterization of cytomegalovirus lung infection
in non-HIV infected children. Viruses. 6:2038–2051. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hoshino T, Nakamura H, Okamoto M, Kato S,
Araya S, Nomiyama K, Oizumi K, Young HA, Aizawa H and Yodoi J:
Redox-active protein thioredoxin prevents proinflammatory cytokine-
or bleomycin-induced lung injury. Am J Respir Crit Care Med.
168:1075–1083. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Boger DL and Cai H: Bleomycin: Synthetic
and mechanistic studies. Angewandte Chemie Int Edition. 38:448–476.
2010. View Article : Google Scholar
|
|
30
|
Gwinn WM, Kapita MC, Wang PM, Cesta MF and
Martin WJ II: Synthetic liposomes are protective from
bleomycin-induced lung toxicity. Am J Physiol Lung Cell Mol
Physiol. 301:L207–L217. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hofman K, Hall B, Cleaver H and Marshall
S: High-throughput quantification of hydroxyproline for
determination of collagen. Anal Biochem. 417:289–291. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhan H, Huang F, Ma W, Zhao Z, Zhang H and
Zhang C: Protective effect of ginsenoside Rg1 on bleomycin-induced
pulmonary fibrosis in rats: Involvement of caveolin-1 and TGF-β1
signal pathway. Biol Pharm Bull. 39:1284–1292. 2016. View Article : Google Scholar
|
|
33
|
Banczyk D, Kalies K, Nachbar L, Bergmann
L, Schmidt P, Bode U, Teegen B, Steven P, Lange T, Textor J, et al:
Activated CD4+ T cells enter the splenic T-cell zone and
induce autoantibody-producing germinal centers through bystander
activation. Eur J Immunol. 44:93–102. 2014. View Article : Google Scholar
|
|
34
|
Guarda G, Dostert C, Staehli F, Cabalzar
K, Castillo R, Tardivel A, Schneider P and Tschopp J: T cells
dampen innate immune responses through inhibition of NLRP1 and
NLRP3 inflammasomes. Nature. 460:269–273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Viganò E and Mortellaro A: Caspase-11: The
driving factor for noncanonical inflammasomes. Eur J Immunol.
43:2240–2245. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cheresh P, Kim SJ, Tulasiram S and Kamp
DW: Oxidative stress and pulmonary fibrosis. Biochim Biophys Acta.
1832:1028–1040. 2013. View Article : Google Scholar :
|
|
37
|
Saïd-Sadier N and Ojcius DM: Alarmins,
inflammasomes and immunity. Biomed J. 35:437–449. 2012. View Article : Google Scholar
|
|
38
|
Gasse P, Mary C, Guenon I, Noulin N,
Charron S, Schnyder-Candrian S, Schnyder B, Akira S, Quesniaux VF,
Lagente V, et al: IL-1R1/MyD88 signaling and the inflammasome are
essential in pulmonary inflammation and fibrosis in mice. J Clin
Invest. 117:3786–3799. 2007.PubMed/NCBI
|
|
39
|
Guerville F, Daburon S, Marlin R, Lartigue
L, Loizon S, Pitard V, Couzi L, Moreau JF, Déchanet-Merville J and
Faustin B: TCR-dependent sensitization of human gammadelta T cells
to non-myeloid IL-18 in cytomegalovirus and tumor stress
surveillance. Oncoimmunology. 4:e10030112015. View Article : Google Scholar
|
|
40
|
Schroder K and Tschopp J: The
inflammasomes. Cell. 140:821–832. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mankan AK, Kubarenko A and Hornung V:
Immunology in clinic review series; focus on autoinflammatory
diseases: Inflammasomes: Mechanisms of activation. Clin Exp
Immunol. 167:369–381. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bromley SK, Mempel TR and Luster AD:
Orchestrating the orchestrators: Chemokines in control of T cell
traffic. Nat Immunol. 9:970–980. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Luzina IG, Todd NW, Sundararajan S and
Atamas SP: The cytokines of pulmonary fibrosis: Much learned, much
more to learn. Cytokine. 74:88–100. 2015. View Article : Google Scholar
|
|
44
|
Zhou XM, Wen GY, Zhao Y, Liu YM and Li JX:
Inhibitory effects of alkaline extract of Citrus reticulata on
pulmonary fibrosis. J Ethnopharmacol. 146:372–378. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yu HB and Finlay BB: The caspase-1
inflammasome: A pilot of innate immune responses. Cell Host
Microbe. 4:198–208. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kong H, Wang Y, Zeng X, Zhu Q, Xie W and
Dai S: Involvement of NLRP3 inflammasome in rituximab-induced
interstitial lung disease: A case report. J Clin Pharm Ther.
39:691–694. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
De Nardo D, De Nardo CM and Latz E: New
insights into mechanisms controlling the NLRP3 inflammasome and its
role in lung disease. Am J Pathol. 184:42–54. 2014. View Article : Google Scholar :
|
|
48
|
Artlett CM, Sassi-Gaha S, Rieger JL,
Boesteanu AC, Feghali-Bostwick CA and Katsikis PD: The inflammasome
activating caspase 1 mediates fibrosis and myofibroblast
differentiation in systemic sclerosis. Arthritis Rheum.
63:3563–3574. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huang TT, Lai HC, Ko YF, Ojcius DM, Lan
YW, Martel J, Young JD and Chong KY: Hirsutella sinensis mycelium
attenuates bleomycin-induced pulmonary inflammation and fibrosis in
vivo. Sci Rep. 5:152822015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dostert C, Pétrilli V, Van Bruggen R,
Steele C, Mossman BT and Tschopp J: Innate immune activation
through Nalp3 inflammasome sensing of asbestos and silica. Science.
320:674–677. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xu JF, Washko GR, Nakahira K, Hatabu H,
Patel AS, Fernandez IE, Nishino M, Okajima Y, Yamashiro T, Ross JC,
et al: Statins and pulmonary fibrosis: The potential role of NLRP3
inflammasome activation. Am J Respir Crit Care Med. 185:547–556.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Tong CY, Bakran A, Williams H, Cuevas LE,
Peiris JS and Hart CA: Association of tumour necrosis factor alpha
and interleukin 6 levels with cytomegalovirus DNA detection and
disease after renal transplantation. J Med Virol. 64:29–34. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Contreras A, Botero JE and Slots J:
Biology and pathogenesis of cytomegalovirus in periodontal disease.
Periodontol 2000. 64:40–56. 2014. View Article : Google Scholar
|