Introduction
Intervertebral disc (IVD) degeneration (IDD) is
widely known as a main contributor to low back pain (LBP) that is
prevalent worldwide (1,2). The etiological factors of IDD
involve aging, smoking, infection, abnormal mechanical stress,
diabetes, trauma and genetic predisposition (3–8).
Degenerative discs are structurally characterized by disc space
collapse, nucleus pulposus (NP) dehydration, annulus fibrosus (AF)
fissures and cartilage endplate calcification. Consideration of
massive socio-economic burdens caused by IDD-associated LBP,
elucidating the causes and pathogenesis of IDD in detail benefits
developing new measures for the prevention and treatment of
LBP.
The pathogenesis of IDD is complicated. The
microenvironment of IVDs is hypoxic but not entirely anaerobic (1%
O2 in central NP) (9,10).
Resident disc cells survive well in this hypoxic microenvironment
and still have oxygen-consumption metabolic processes (11–13). With IDD progression,
neovascularization in discs increases oxygen tension in the
microenvironment of IVDs (14,15). High oxygen tension is expected to
enhance reactive oxygen species (ROS) production and subsequently
to cause oxidative stress in the microenvironment of IVDs, which is
strongly associated with the establishment and progression of IDD
(16–18). However, the role of high oxygen
tension in the pathogenesis of IDD remains to be a relatively
poorly explored area. Furthermore, IDD is a disc cell-mediated
pathological process (19,20).
The initiation and progression of IDD depend on the viability and
function of disc cells. Although high oxygen tension has been shown
to reinforce the matrix catabolism and autophagy of NP cells
(21,22), the effect of high oxygen tension
on the viability and function of disc cells and the molecular
mechanism underlying the effect should be investigated further in
depth to elucidate the involvement of high oxygen tension in the
pathogenesis of IDD.
Alternative splicing (AS) is a regulatory process by
which the exons of pre-mRNAs are spliced in different ways to
synthesize structurally and functionally diverse mRNA and proteins.
Approximately >90% of multiexonic genes undergo AS regulation.
It is crucial to increasing the macromolecular and cellular
complexity of eukaryotic organisms without extending genome size
(23–25). The types of AS that are
responsible for RNA isoform generation includes cassette
alternative exon (exon skip or inclusion), intron retention,
alternative 5′/3′ splice sites, mutually exclusive alternative
exons, alternative promoter and first exon as well as alternative
poly-A site and terminal exon. Among them, cassette alternative
exon is the most common AS event and has been investigated
extensively (25). AS has been
reported to be involved in various diseases, including autoimmune
disease, neurodegenerative disease and carcinoma (26–28). With respect to IDD, the AS of
fibronectin, thrombospondins and versican has been determined to be
associated with matrix remodeling of degenerative discs (29–31). However, the role of AS in the
pathogenesis of IDD remains unclear. Besides, AS probably is an
essential mechanism mediating the effect of high oxygen tension on
disc cells.
In the present study, in order to investigate the
transcriptome and AS of disc cells in response to high oxygen
tension, rat NP cells were cultured at 1% O2, and then
were treated with 20% O2 (high oxygen tension). After
that, total RNA of NP cells was extracted to perform microarray
assays using the Affymetrix Rat Transcriptome Array 1.0. A
comparative analysis between NP cells at 1% O2 and NP
cells treated with high oxygen tension was performed to determine
differentially expressed genes (DEGs) and alternative splicing
genes (ASGs). The function of DSGs and ASGs was annotated through
Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway analysis. Moreover, increased ROS production
induced by high oxygen tension was observed in NP cells. We also
analyzed the growth, cell cycle distribution and matrix metabolism
of NP cells to support the results of microarray further. Our study
revealed the functional role of high oxygen tension in NP cells on
genome-wide scale. Also, we investigated the regulatory effect of
high oxygen tension on AS of NP cells, suggesting the involvement
of AS in the pathogenesis of IDD. This study contributes to
understanding the regulatory effect of high oxygen tension on NP
cells, which gives a novel insight into the establishment and
progression of IDD.
Materials and methods
Ethics statement
This study was approved by the Ethics Committee of
Xinqiao Hospital, Third Military Medical University. All
experimental procedures described in this study were in accordance
with the standards set forth in the eighth edition of Guide for the
Care and Use of Laboratory Animals published by the National
Academy of Sciences (The National Academies Press, Washington, DC,
USA).
NP cell culture at 1% O2 and
high oxygen tension (20% O2) treatment
NP tissues were harvested from caudal discs (C1-C10)
of adult (3-month-old) male Sprague-Dawley rats (Laboratory Animal
Research Center of Daping Hospital, Chongqing, China) after
sacrificed using excessive pentobarbital sodium. The isolated
tissues were incubated with Dulbecco's modified Eagle's medium
(DMEM)/F-12 (Invitrogen, Carlsbad, CA, USA) containing 0.2% type II
collagenase (Sigma, St. Louis, MO, USA) at 37°C and 1%
O2 for 2 h. A 70 µm cell mesh was used to remove
tissue debris. After centrifuging at 1,000 rpm for 5 min, the
supernatant of cell suspension was removed followed by resuspension
with DMEM/F12 containing 10% fetal bovine serum and 1%
penicillin/streptomycin (Invitrogen). NP cells grew as monolayer in
25 cm2 culture flasks (Corning, Inc., Corning, NY, USA)
at 37°C and 1% O2. Culture medium was changed twice a
week. When confluent, NP cells were sub-cultured. The second or
third culture passages were used in this study. For high oxygen
tension treatment, NP cells were transferred into an incubator with
20% O2 at 37°C for 7 days.
RTA 1.0 and bioinformatics analysis
NP cells without high oxygen tension treatment
served as the control. The high oxygen tension-treated samples
(n=3) and the control samples (n=3) lysed by TRIzol (Takara Bio,
Shiga, Japan) were sent to Bioassay Laboratory of CapitalBio Corp.
(Beijing, China). The global gene expression profile and
alternative splicing exons (ASEs) of NP cells were analyzed using
RTA 1.0 (Affymetrix Corp., Santa Clara, CA, USA). The
hybridization, scanning and data extraction of microarray were
performed in Bioassay Laboratory of CapitalBio Corp. according to
the protocol provided by Affymetrix Corp. Concisely, the
fluorescence signals of microarray scanned as DAT files were
transformed into CEL files via the AGCC software (Affymetrix
Corp.). Then, Affymetrix Expression Console software pretreated CEL
files through robust multichip analysis algorithm to obtain chp
files (32). The chp files were
analyzed by Affymetrix Transcriptome Analysis Console software to
determine DEGs and ASGs. DEGs were identified according to fold
changes (fold change ≥2 or ≤-2) and p-value (p<0.05). ASGs were
identified according to splicing index (SI, SI ≥2 or ≤-2) and
p-value (p<0.05). SI is the ratio of the exon signal intensities
normalized to the gene signal intensities between the high oxygen
tension-treated group and the control group. It generally
represents the exon exclusion/inclusion level (33,34). Herein, SI ≥2 is regarded as
general exon inclusion while SI ≤-2 is regarded as general exon
exclusion. GO analysis (http://www.geneontology.org) and KEGG pathway analysis
(http://www.genome.jp/kegg) were
performed to identify the cellular components, molecular functions,
biological processes and signaling pathways enriched by DEGs or
ASGs.
Reversed transcription-quantitative PCR
(RT-qPCR) and semi-quantitative RT-PCR
Total RNA of NP cells were extracted using TRIzol
reagent. The concentration of RNA was measured using a NanoDrop
ND-2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA,
USA). One microgram of RNA was reversely transcribed into cDNA
using a PrimeScript RT reagent kit (Takara Bio) according to the
protocol provided by the manufacturer. For semi-quantitative
RT-PCR, 1 µl cDNA was used for each reaction. The primers of
target exons were designed based on constitutively expressed exons
flanking the target exons (Table
I). Glyceraldehyde 3-phosphate dehydrogenase GAPDH served as
the internal reference gene. The representative ASEs for
semi-quantitative RT-PCR validation were selected according to
three criteria: i) |SI| >3, ii) cassette alternative exon and
iii) for the feasibility of primer design, not the first and the
last alternative exons. On the other hand, a ViiA™ 7 Real-Time PCR
system (Applied Biosystems, Thermo Fisher Scientific) was used to
perform real-time quantitative PCR in triplicate. Twenty
microliters SYBR® Premix Ex Taq™ II (Takara Bio)
reaction volume was composed of 10 µl SYBR, 6 µl
H2O, 0.4 µl ROX, 0.8 µl forward primer,
0.8 µl reverse primer and 2 µl cDNA. The cycle
parameters were 95°C for 30 sec, followed by 40 cycles of 95°C for
5 sec and 60°C for 30 sec. The relative fold changes of target
genes were analyzed according to the 2−ΔΔCt algorithm
(35). The internal reference
gene was GAPDH. The primers of target genes used in this study are
listed in Table II.
 | Table IPrimer sequences used in
semi-quantitative RT-PCR analysis. |
Table I
Primer sequences used in
semi-quantitative RT-PCR analysis.
| ASG (Exon) | Forward primer | Reverse primer |
|---|
| ADAMTS9 (Exon
31) |
CTACAGGCAAAGGCTGGTCTCC |
CAGGACAGTCCCTCAGGTAGCA |
| FNDC1 (Exon
11) |
GCAATCGGTCTTCGCTGAGGAG |
GCATCGGTGTGGTGGAGGAGTA |
| GDF15 (Exon 2) |
GCTGCTGTCACCTGGAGACTGT |
GCCGCTGTCTGTCCTGTGCATA |
| KRCC1 (Exon 3) |
GAGTTTGATTCCAGGCCCAGGT |
CTGCCTTTCTCTGCCCTCCTCT |
| NPTN (Exon 2) |
GGGTTTGTCAAGTCGCCCATGT |
TCAGCACACTCACGCCGTTTG |
| OGN (Exon 8) |
TGGTTGTAGCAGACCGCCATCT |
GGGAAGGTCACTGGGAGCACTT |
| PREB (Exon 4) |
TGGTTACTGTGGGCTGGGACTT |
CCTGGTAGCGGTACGGTGTGTT |
| SEC31A (Exon
23) |
ACCAGCCAGCCCAGCAGTATT |
TTCCAGGAGGCAGTGCATAAGC |
| TJP1 (Exon 27) |
GCAGAAGCCTCATCTCCAGTCC |
GCGACGGCAATGACACTCCTT |
| GAPDH |
GTCCATGCCATCACTGCCACTC |
GATGACCTTGCCCACAGCCTTG |
 | Table IIPrimer sequences used in the RT-qPCR
analysis. |
Table II
Primer sequences used in the RT-qPCR
analysis.
| Target gene | Forward primer | Reverse primer |
|---|
| MMP13 |
TACGAGCATCCATCCCGAGACC |
TGAACCGCAGCACTGAGCCT |
| BMP3 |
AGCCTTCAGACTCAGCCTCCTG |
TCGCCTCGCCTTCTTCAGTGT |
| BMP4 |
GACTTCGAGGCGACACTTCTGC |
GGTTCCCTGGCTCTGCTCTTCT |
| Neuropilin 1 |
TCGGTGGGATTGCTGTGGATGA |
TGCCTGGCTTCCTGGAGATGTT |
| ADAMTS5 |
GCTCCTCTTGGTGGCTGACTCT |
GCGTTCTTGCTCACCTCCAGAC |
| p15 |
GGCTTCCTGGACACGCTAATGG |
ATATCACGGTGGCCCTGCTCTT |
| GAS1 |
TTTCTGCTGCTCCTGCTGCTTG |
GGGTCAGTGCTCCCGATCATCT |
| Nox4 |
CGCACAGTCCTGGCTTACCTTC |
GGCAGCTACATGCACACCTGAG |
| PCNA |
CGCAACTCCGCCACCATGTT |
TTCACGCCGCCCGAACTGAT |
| VEGFA |
TCACCACCACACCACCATCGT |
ATCCAGTTCCACGAGGGACCAC |
| Cyclin I |
AGCAGCCTTCCACCTCCATCTC |
AGCCGCTTGATCCCGTCATACA |
| GDF15 |
CCTGCTGTTCCTGCTGCTCTTG |
TAGCTCGTCCGGGTTGAGTTGG |
| GPX1 |
AGGCTCACCCGCTCTTTACCTT |
TGGAACACCGTCTGGACCTACC |
| Aggrecan |
ATCCGCTGCTCCAGAAGTGAGT |
ACGGTGGTGCTGACGGTAACA |
| Collagen type
II |
CATGAACGGCGGCTTCCACTT |
GCTTCGTCCAGGTAGGCAATGC |
| HIF-1 |
CGCAACTGCCACCACTGATGAA |
GGCTGTCCGACTGTGAGTACCA |
| GAPDH |
GTCCATGCCATCACTGCCACTC |
GATGACCTTGCCCACAGCCTTG |
ROS detection
2′,7′-Dichlorfluorescein-diacetate (DCFH-DA) is
oxidized by ROS to generate dichlorofluorescein (DCF) with high
fluorescence. Therefore, ROS production in NP cells was detected
using DCFH-DA (Sigma). NP cell suspension was incubated with
H2DCF-DA (25 µM) dissolved in phosphate-buffered saline
(PBS) at 37°C and 5% CO2 for 30 min followed by washing
with serum-free DMEM/F12 three times. The mean fluorescence
intensity (MFI) of DCF in NP cells was detected using a flow
cytometer (Beckman Coulter, Inc., Brea, CA, USA).
Western blot analysis
NP cell proteins were extracted using the extraction
reagent (Thermo Fisher Scientific). Proteins were quantified using
a BCA kit (Beyotime, Shanghai, China) and were dissolved in loading
buffer (Invitrogen). After electrophoresis on 10% (w/v) sodium
dodecyl sulfate-polyacrylamide (SDS) gels, proteins were
transferred to polyvinylidenefluoride (PVDF) membranes (Millipore,
Billerica, MA, USA) followed by incubation with 5% milk proteins in
Tris-buffered saline containing 0.1% Triton X-100 at 37°C for 1 h.
Next, the membrane was incubated with primary antibodies against
GAPDH (1:1,000 dilution), matrix metalloproteinase 13 (MMP13, 1:500
dilution) and a disintegrin and metalloproteinase with
thrombospondin motifs 5 (ADAMTS5, 1:500 dilution) at 4°C overnight.
The mouse monoclonal anti-rat GAPDH (sc-47724) antibody, the rabbit
polyclonal anti-rat MMP13 (sc-30073) and ADAMTS5 (sc-134952)
antibodies were purchased from Santa Cruz Biotechnology, Inc.
(Santa Cruz, CA, USA). The goat polyclonal anti-mouse IgG (H+L)
horseradish peroxidase (HRP)-conjugated secondary antibody and the
goat anti-rabbit IgG (H+L) HRP-conjugated secondary antibody
(ZSGB-Bio, Beijing, China) were used. Immunolabeling was visualized
using electrochemiluminescence (ECL) reagent (Thermo Fisher
Scientific).
Cell growth assay
NP cells seeded in 96-well culture plates were
incubated with 10 µl of cell counting kit-8 reagent
(Dojindo, Tokyo, Japan) at 37°C for 2 h. The absorbance was
detected at 450 nm using a spectrophotometer (Varioskan Flash,
Thermo Fisher Scientific) daily for 7 days. Then, absorbance data
were used to calculate the number of NP cells based on the standard
curve. Population doubling (PD) was calculated in following ways:
PD = [log10(NH)-log10(NI)]/log10
(2). NH is the calculated number
of NP cells every day. NI is the initial number of NP cells seeded
in the 96-well plate. PD values were used to depict the growth
curve of NP cells.
Cell cycle analysis
NP cells were trypsinized and washed with PBS three
times. To analyze the cell cycle distribution of NP cells, NP cells
were resuspended in 1 ml propidium iodide working solution of Cell
Cycle Analysis kit (Beyotime). Then, the percentage of NP cells in
different cell cycle phases including G1, G2 and S was measured by
a flow cytometer (Beckman Coulter).
Statistical analysis
Independent experiments were performed at least
three times. All results were presented as mean or mean ± standard
error of the mean. Two-tailed Student's t-test was used for the
comparison between two independent groups. RT-qPCR results were
analyzed using Kruskal-Wallis nonparametric analysis and
Mann-Whitney U post-hoc tests. GraphPad Prism 6 (GraphPad Software
Inc., La Jolla, CA, USA) and SPSS version 22.0 (International
Business Machines Corp., Amonk, NY, USA) were used in this study.
P<0.05 was considered statistically significant.
Results
Analysis and functional annotation of
DEGs induced by high oxygen tension
DEGs were identified according to the criteria
mentioned above. There are 2499 DEGs in NP cells treated with high
oxygen tension compared with the control. The percentage of
upregulated genes (1243/2499=49.7%) was nearly the same as the
percentage of downregulated genes (1256/2499=50.3%). To validate
the results of the microarray, 13 genes were selected as the
representative DEGs for RT-qPCR analysis. Eleven of the 13
representative genes showed the expression pattern that was
consistent with the results of microarray (Fig. 1). The expression of MMP13,
ADAMTS5, nicotinamide adenine dinucleotide phosphate (NADPH)
oxidase 4 (Nox4), cyclin-dependent kinase inhibitor 2B (p15), bone
morphogenetic protein 3 (BMP3), BMP4, growth arrest specific 1
(GAS1) and neuropilin 1 was significantly upregulated together with
the markedly downregulation of vascular endothelial growth factor A
(VEGFA), cyclin I and glutathione peroxidase 1 (GPX1).
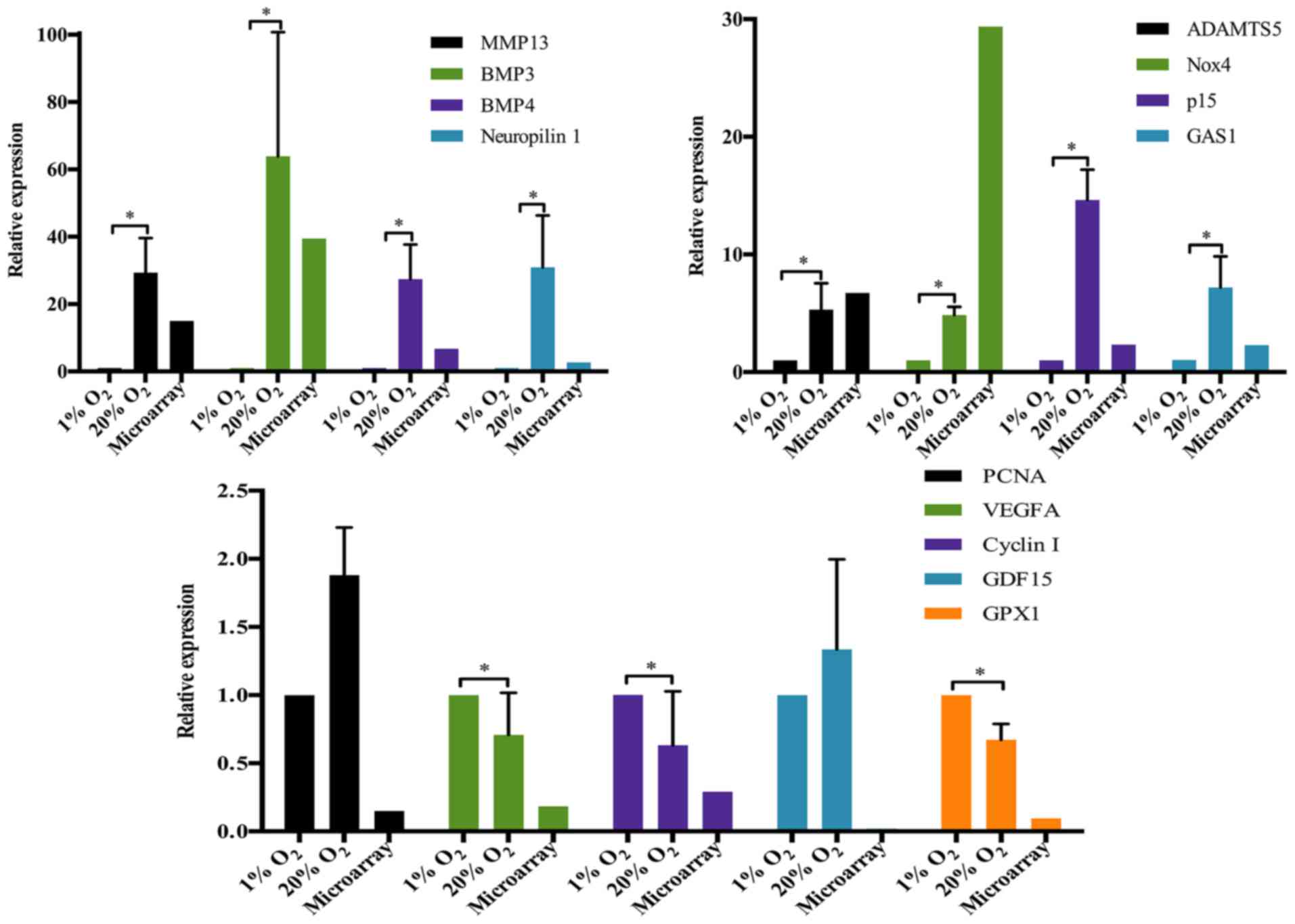 | Figure 1Validation of the representative
differentially expressed genes by RT-qPCR. *p-value
<0.05, error bars represent standard error. MMP13, matrix
metalloproteinase 13; ADAMTS5, a disintegrin and metalloproteinase
with thrombospondin motifs 5; BMP, bone morphogenetic protein;
Nox4, nicotinamide adenine dinucleotide phosphate (NADPH) oxidase
4; GAS1, growth arrest specific 1; PCNA, proliferating cell nuclear
antigen; VEGFA, vascular endothelial growth factor A; GDF15, growth
and differentiation factor 15; GPX1, glutathione peroxidase 1. |
GO analysis was performed to annotate the functions
of DEGs. Many GO terms were related to the carbohydrate metabolism
of NP cells, such as carbohydrate biosynthetic process, glycolytic
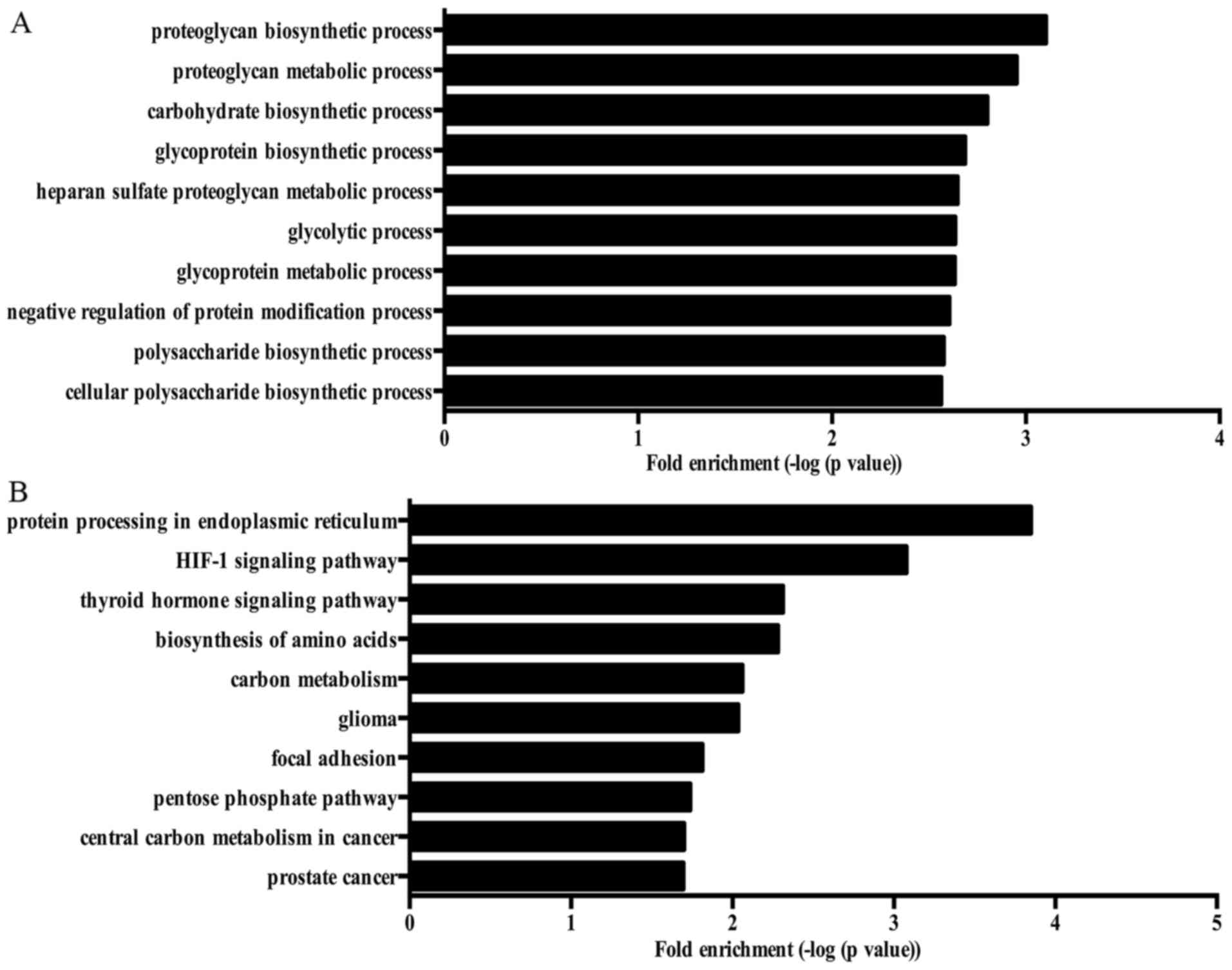
process, gluconeogenesis and pyruvate metabolic process. Fig. 2A showed the top 10 biological
responses enriched by DEGs in NP cells treated with high oxygen
tension.
KEGG pathway analysis showed that various pathways
were involved in the response of NP cells to high oxygen tension,
including protein processing in endoplasmic reticulum, carbon
metabolism, mTOR signaling pathway and PI3K-Akt signaling pathway.
The top 10 KEGG pathways enriched by DEGs are shown in Fig. 2B.
Analysis and functional annotation of
ASGs induced by high oxygen tension
According to the filter criteria, 27004 ASEs derived
from 8451 ASGs were identified. Among them, 10434 ASEs underwent
general exon inclusion (SI ≥2). 16570 ASEs underwent general exon
exclusion (SI ≤-2). 87.5% (23619/27004) of ASEs were derived from
54.6% (4616/8451) of ASGs, suggesting that a gene contains multiple
ASEs in NP cells. For example, both aggrecan and ADAMTS5 had 7
ASEs, respectively. Furthermore, 1986 of the 8451 ASGs were
differentially expressed. Semi-quantitative RT-PCR results of 10
representative genes are shown in Fig
3. Nine of the 10 genes had cassette alternative exons.
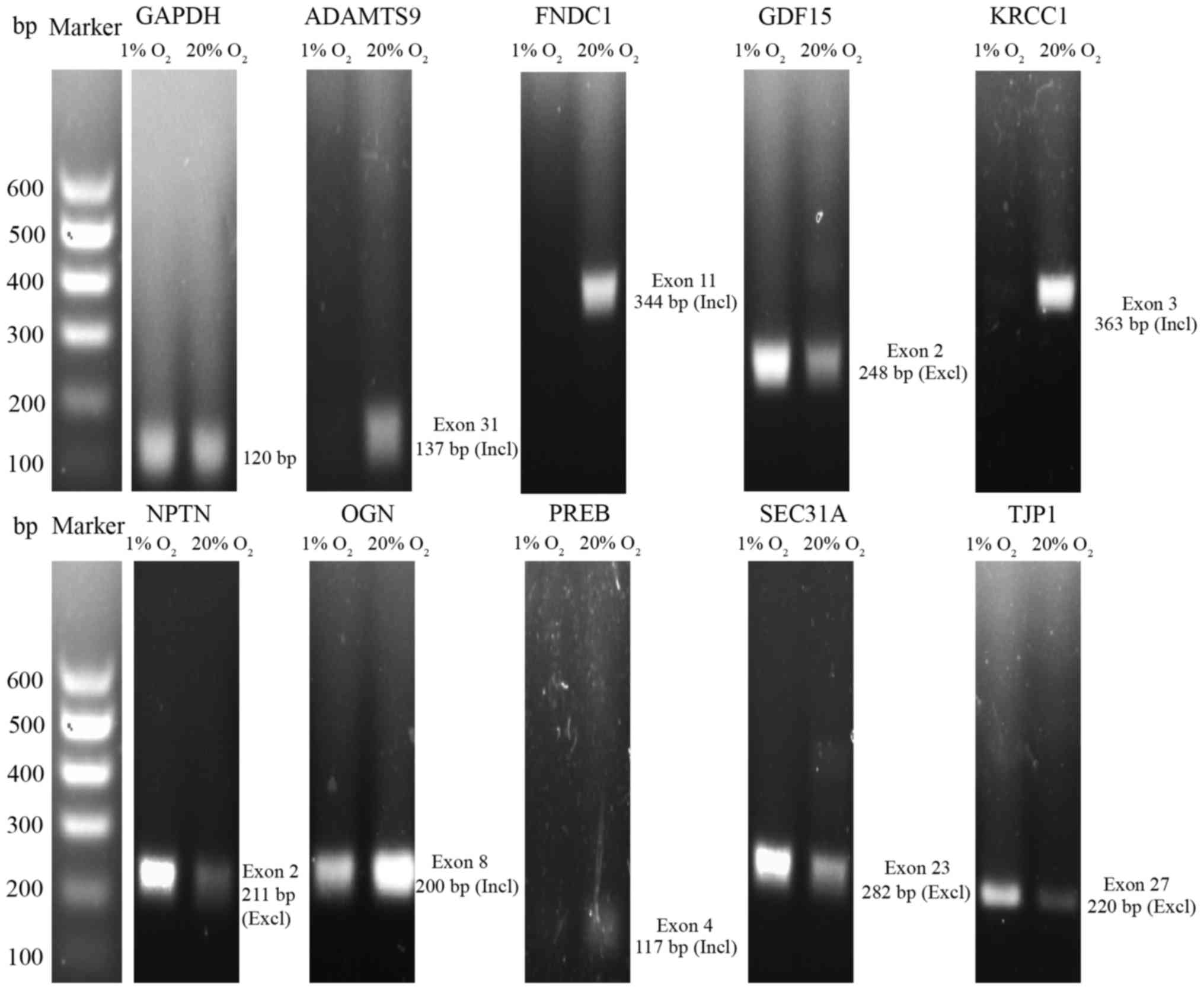 | Figure 3Validation of the representative
alternative splicing genes by semi-quantitative RT-PCR. ADAMTS9, a
disintegrin and metalloproteinase with thrombospondin motifs 9;
FNDC1, fibronectin type III domain containing 1; GDF15, growth and
differentiation factor 15; KRCC1, lysine-rich coiled-coil 1; NPTN,
neuroplastin; OGN, osteoglycin; PREB, prolactin regulatory element
binding; SEC31A, SEC31 homolog A, COPII coat complex component;
TJP1, tight junction protein 1; Incl, inclusion; Excl,
exclusion. |
GO analysis showed that various biological processes
were enriched by ASGs, including cellular macromolecule metabolic
process, macromolecule modification, cell cycle, cellular response
to oxidative stress, stress-activated MAPK cascade, angiogenesis,
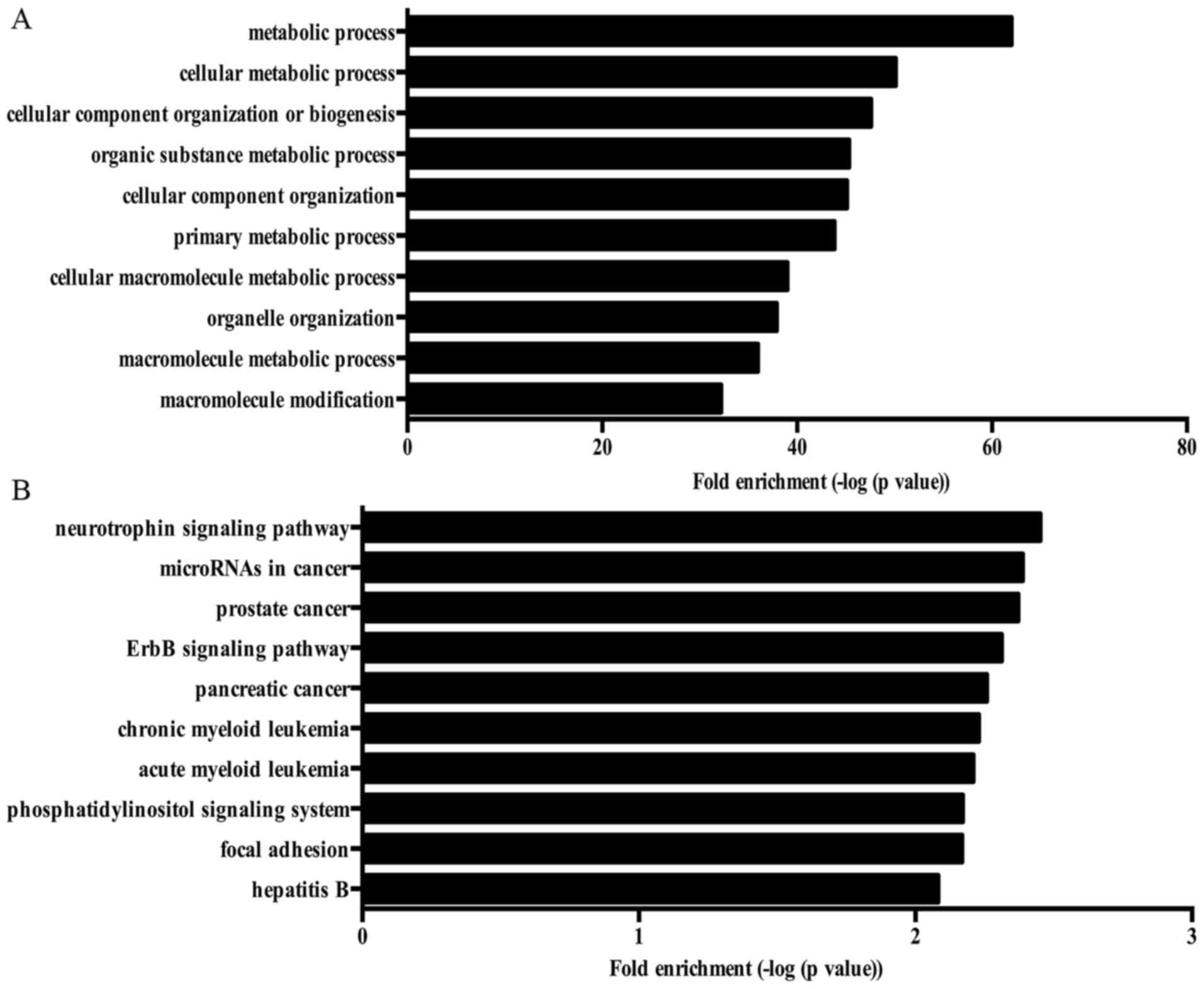
cell aging and regulation of autophagy. Fig. 4A showed the top 10 biological
processes enriched by ASGs in NP cells treated with high oxygen
tension. Moreover, several KEGG pathways associated with
pathogenesis of IDD were enriched by ASGs, such as neurotrophin
signaling pathway and mTOR signaling pathway. Fig. 4B shows the top 10 KEGG pathway
enriched by ASGs.
High oxygen tension increases ROS
generation in NP cells
Several biological responses enriched by DEGs or
ASGs were correlated with oxygen tension, such as cellular response
to oxygen levels and cellular response to hypoxia. Also, biological
responses related to redox homeostasis, including positive
regulation of ROS metabolic process, glutathione metabolic process
and cellular response to ROS, were identified. In fact, ROS
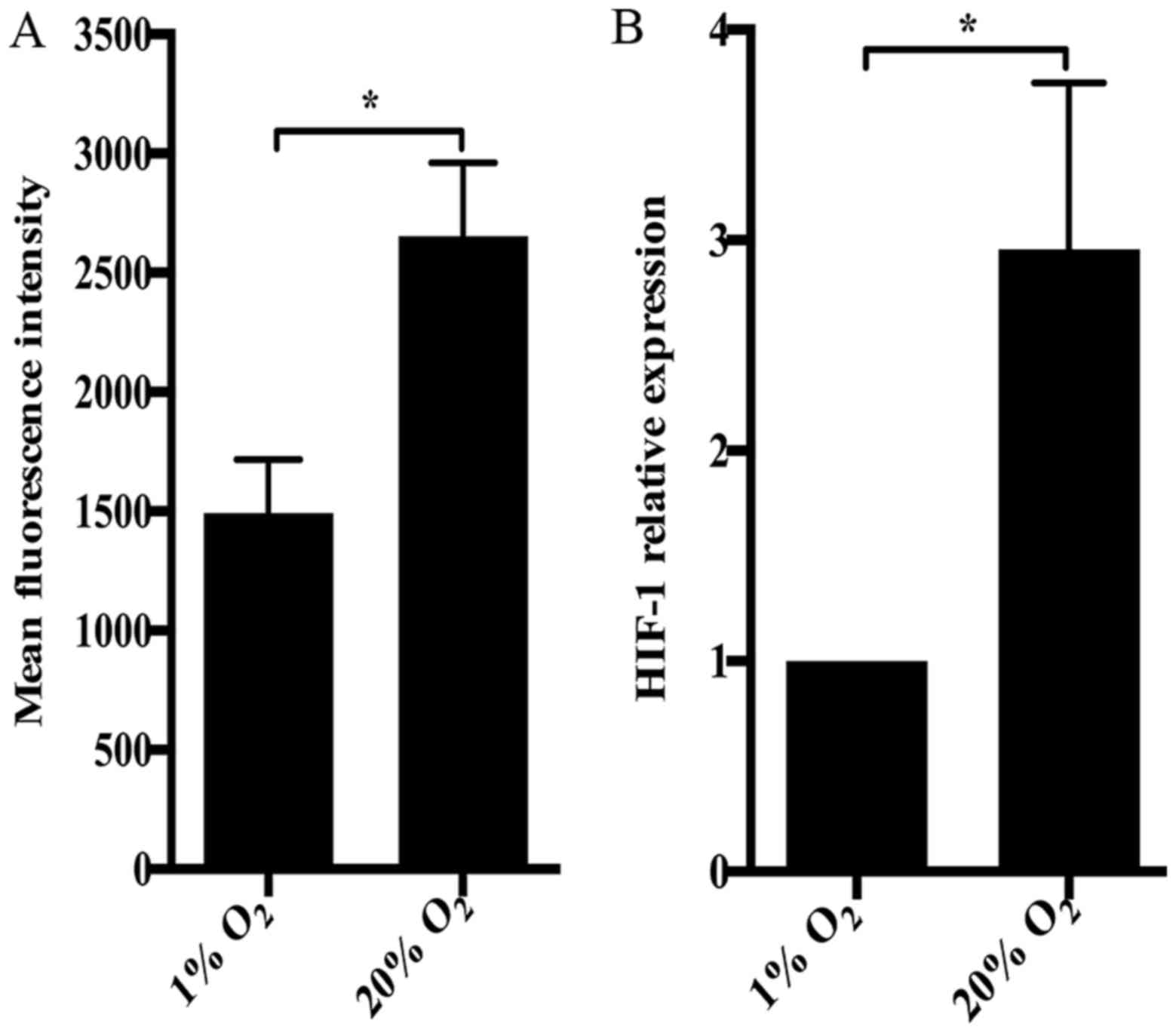
production was dramatically increased by high oxygen tension in NP
cells (Fig. 5A). It was
consistent with the upregulation of Nox4 and the downregulation of
GPX1 in NP cells treated with high oxygen tension (Fig. 1). Notably, hypoxia inducible
factor-1 (HIF-1) signaling pathway was enriched by DEGs and ASGs.
Therefore, we investigated the expression of HIF-1 in NP cells
using RT-qPCR. Paradoxically, the expression of HIF-1 in NP cells
subjected to 20% O2 was significantly higher than that
in NP cells under 1% O2 (Fig. 5B).
Effect of high oxygen tension on matrix
metabolism of NP cells
GO terms and KEGG pathways associated with matrix
metabolism of disc cells have been shown to be enriched by DEGs,
including proteoglycan metabolic process, tissue remodeling and
glycosaminoglycan biosynthesis. Thus, we investigated the
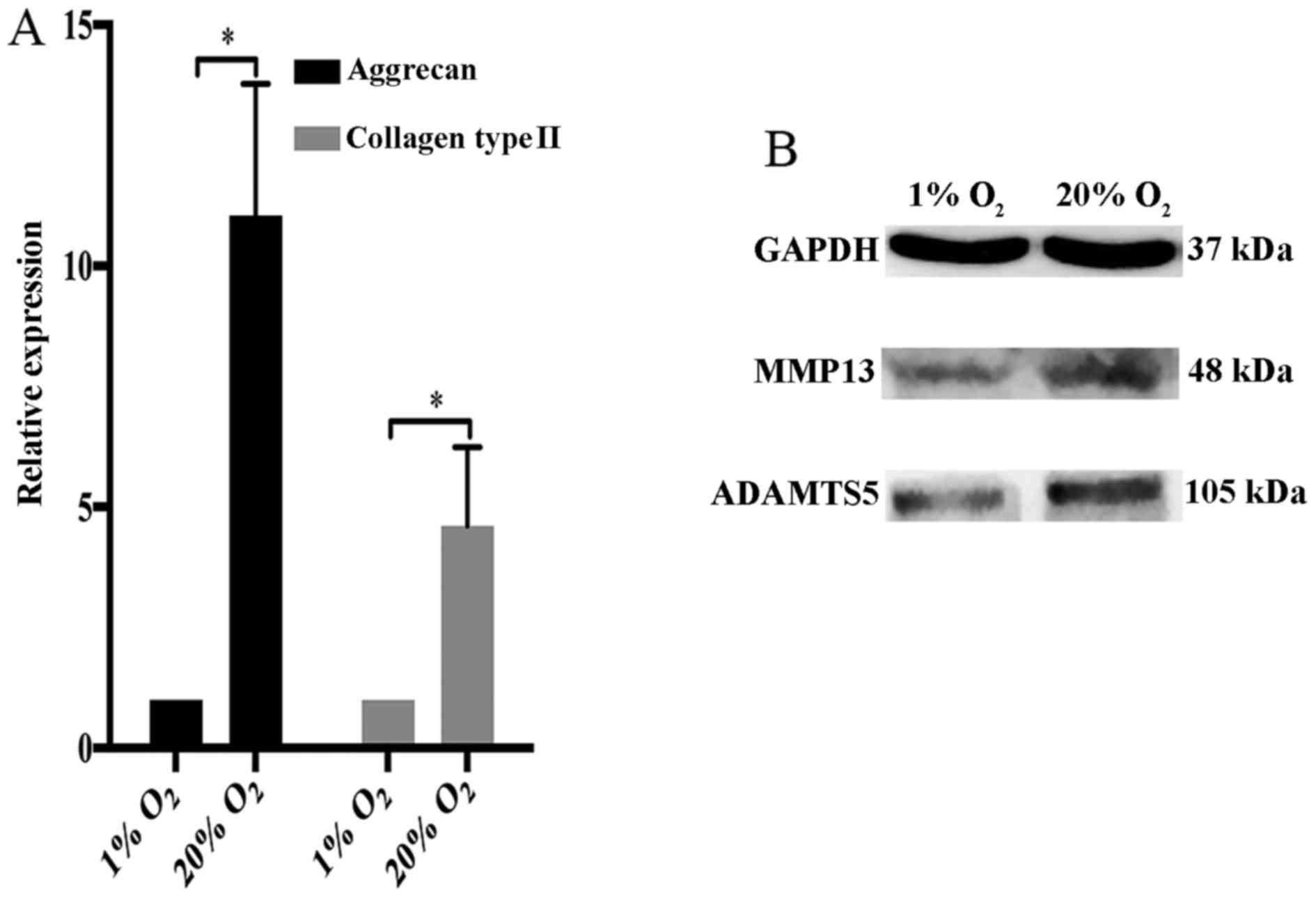
expression of collagen type II and aggrecan in NP cells. We found
that collagen type II and aggrecan were markedly upregulated by
high oxygen tension (Fig. 6A).
However, two crucial matrix degradation proteases in IVDs, MMP13
and ADAMTS5 also were upregulated by high oxygen tension in NP
cells (Fig. 6B).
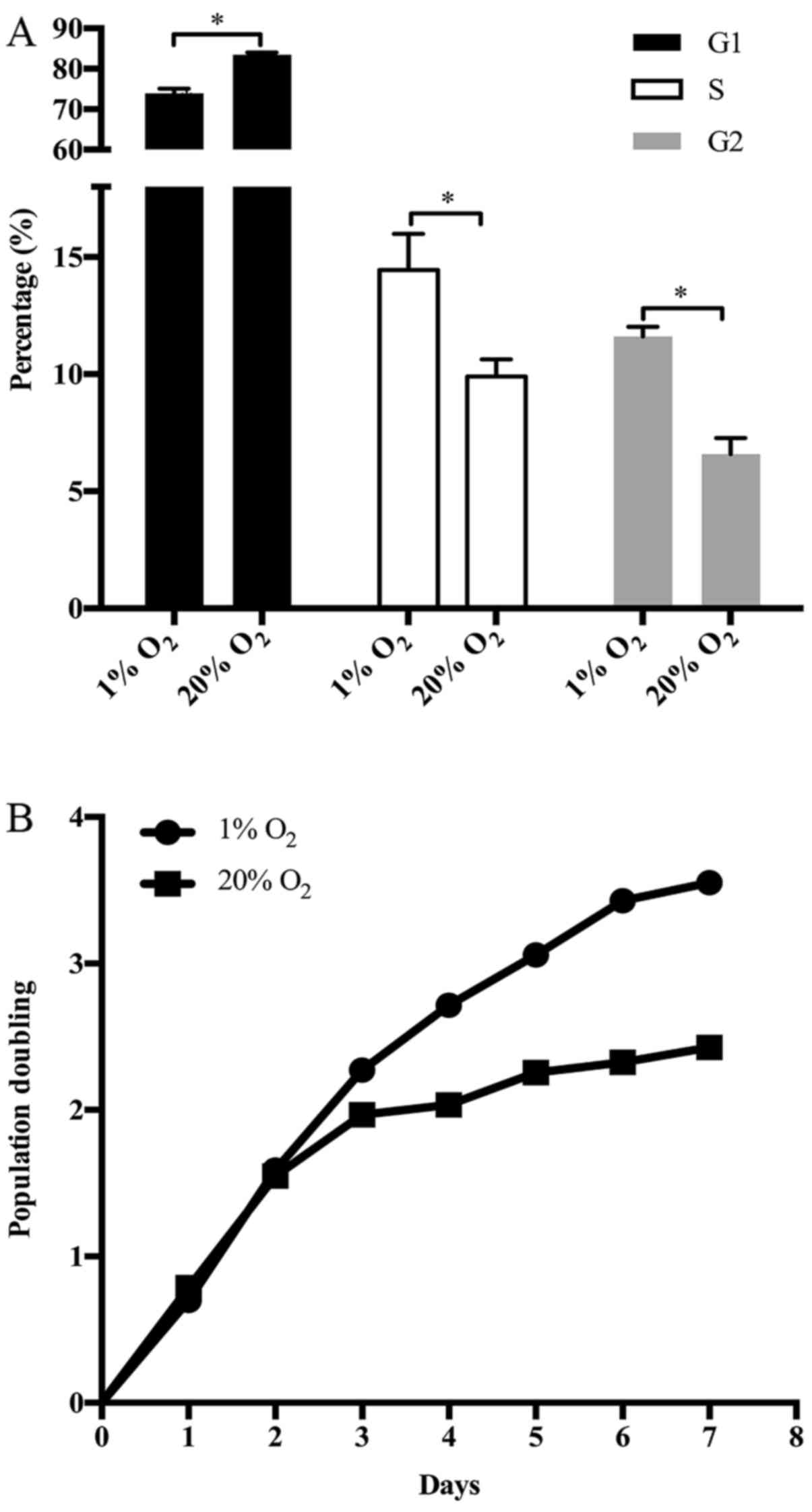
High oxygen tension arrested the G1/S
transition of NP cell cycle
The proliferation of disc cells is crucial to the
structural and functional maintenance of IVDs. The proliferation of
NP cells depends on cell cycle progression. Interestingly, various
GO terms and KEGG pathways enriched by DEGs or ASGs were associated
with the cell cycle of NP cells, including cell cycle arrest, G1/S
phase transition of cell cycle, regulation of cell growth, mitotic
G1/S transition checkpoint and regulation of cyclin-dependent
protein serine/threonine kinase activity involved in G1/S
transition of mitotic cell cycle. Actually, the upregulation of p15
and GAS1 together with the downregulation of cyclin I in NP cells
treated with high oxygen tension have been mentioned (Fig. 1). At the same time, the G1/S phase
transition of NP cell cycle was retarded by high oxygen tension
(Fig. 7A). As a result, the
growth of NP cells was suppressed by high oxygen tension (Fig. 7B). On the other hand, several GO
terms, such as DNA damage response, G1 DNA damage checkpoint,
signal transduction by p53 class mediator resulting in
transcription of p21 class mediator and p53 signaling pathway,
indicate that the cell cycle arrest of NP cells induced by high
oxygen tension is potently associated with DNA damage checkpoint in
G1/S transition.
Discussion
The viability and function of disc cells are
essential to the homeostasis maintenance system of IVDs (36). Investigating the effect of high
oxygen tension on disc cells contributes to elucidating the role of
high oxygen tension in the pathogenesis of IDD. Exposure of NP
cells to high oxygen tension has been demonstrated as an effective
method to increase ROS production and consequently cause oxidative
stress in NP cells. Excessive ROS generation induced by high oxygen
tension has been shown to reinforce the matrix catabolism and
autophagy of NP cells (21,22). However, to our knowledge, there
were no studies that analyze the gene expression profile of disc
cells in response to high oxygen tension on a genome-wide scale.
Moreover, the involvement of AS in regulating the response of disc
cells to high oxygen tension remains unknown. Therefore, this
study, for the first time, performed a genome-wide analysis to
determine the global gene expression profile and AS in NP cells
subjected to high oxygen tension. More than two thousand genes were
significantly differentially expressed in NP cells subjected to
high oxygen tension. The results of bioinformatics analysis
revealed a wide role of high oxygen tension in regulating the
viability and function of disc cells.
The involvement of AS in various diseases has
aroused the interest of many studies. However, few studies
investigated the role of AS in the pathogenesis of IDD. Our study
provides a genome-wide view to analyze ASGs in NP cells treated
with high oxygen tension, and 27004 ASEs derived from 8451 ASGs
were determined. Nine of the 10 representative ASGs were confirmed
by semi-quantitative RT-PCR. The results suggest the effect of high
oxygen tension on the AS of NP cells. Among the 10 representative
ASGs, ADAMTS9 is an aggrecanase, and growth and differentiation
factor 15 is a member of TGF superfamily. Matrix proteases and
growth factors have been demonstrated to be strongly associated
with IDD (37,38). Furthermore, numerous GO terms and
KEGG pathways enriched by ASGs were correlated with the
pathogenesis of IDD, including regulation of autophagy, mTOR
signaling pathway and neurotrophin signaling pathway. Disc cell
autophagy has been demonstrated to be an essential event in the
process of IDD. It is a double-edged sword. The appropriate
autophagy promotes the survival of disc cells through
self-digestion that protects disc cells from various stresses.
However, excessive autophagy causes disc cell death to decrease the
number of viable and functional cells in discs (20,22,39). With regard to the mechanism
underling disc cell autophagy, mTOR pathway is a crucial regulatory
signaling pathway (40,41). Besides, neurotrophin is
responsible for neuronal ingrowth in degenerative discs that
contributes to pain sensation in LBP (42,43). In short, AS induced by high oxygen
tension is widely involved in regulating the biological process and
signaling pathways of disc cells and consequently affects the
initiation and progression of IDD.
BMP4 has been reported to be associated with IDD
(44). The possible involvement
of BMP4 and its receptor in the chondrogenic progress of
spondylosis also has been mentioned (45). However, the precious role of BMP4
in the pathogenesis of IDD remains unclear. Another member of the
BMP family, BMP3, has not been investigated in IVDs so far. Herein,
both BMP3 and BMP4 were dramatically upregulated by high oxygen
tension in NP cells. The GO term, regulation of BMP signaling
pathway, was enriched by both DEGs and ASGs, suggesting that BMP3
and BMP4 are involved in regulating the responses of NP cells to
high oxygen tension. Revealing the relationship between BMP and
high oxygen tension extends our knowledge on the role of BMP in the
pathogenesis of IDD.
Enhanced blood vessel ingrowth into NP is a crucial
pathological characteristic of degenerative discs.
Neovascularization leads to the exposure of disc cells to higher
oxygen tension. VEGF, an angiogenic factor, has been demonstrated
to induce neovascularization in herniated discs (43,46). NP cells upregulate the expression
of VEGF in response to mechanical strain and pro-inflammatory
cytokines, which enhances angiogenesis in IVDs (47,48). On the contrary, herein, we found
that the expression of VEGF in NP cells was downregulated by high
oxygen tension, suggesting that high oxygen tension suppresses
angiogenesis in discs, probably forming a negative feedback.
Therefore, the effect of high oxygen tension on neovascularization
in degenerative discs should be assessed in further studies.
Consistent with previous studies (21,22), 20% O2 significantly
increased ROS levels in NP cells. The GO terms enriched by DEGs or
ASGs, positive regulation of ROS metabolic process, cellular
response to ROS and glutathione metabolic process, suggest that
oxygen tension regulates the balance between antioxidant generation
and ROS production in NP cells. The differential expression of Nox4
and GPX1 supports this idea further. Nox4 is a professional
ROS-generating enzyme and a major source of intracellular ROS
(49). The upregulation of Nox4
induced by high oxygen tension can explain increased ROS production
in NP cells subjected to high oxygen tension. On the other hand,
GPX1, a major antioxidant gene, was markedly downregulated by high
oxygen tension, indicating a decline in antioxidants of NP cells.
In conclusion, high oxygen tension disturbs the redox homeostasis
to arouse oxidative stress in NP cells. Besides, the expression of
HIF-1 was paradoxically upregulated in NP cells exposed to high
oxygen tension. This issue should be investigated further in
depth.
The structural and functional maintenance of IVDs
depends on the balance between the ECM anabolism and catabolism of
discs. Noticeably, high oxygen tension not only upregulated the
expression of aggrecan and collagen type II, but also upregulated
the expression of MMP13 and ADAMTS5 in NP cells. High oxygen
tension showed both anabolic and catabolic effects on the matrix
metabolism of NP cells. Thus, we hypothesize that high oxygen
tension accelerates matrix turnover of IVDs. Nevertheless, we still
need more in vivo evidence to elucidate the comprehensive
effect of high oxygen tension on matrix homeostasis of IVDs.
The proliferation of disc cells is crucial to
maintaining the number of functional cells in IVDs. It relies on
cell cycle progression of disc cells. In this study, as the GO
terms, cell cycle arrest and G1/S phase transition of cell cycle,
suggested, high oxygen tension regulated the expression of p15,
GAS1 and cyclin I, and subsequently retarded the G1/S phase
transition of NP cell cycle. As a consequence, the growth of NP
cells was arrested. The detrimental effect of high oxygen tension
on NP cell proliferation indicates that high oxygen tension may
cause a decrease in the number of functional and viable cells in
discs. Mechanistically, DNA damage is a widely known intrinsic
trigger of cell cycle arrest. It activates the G1/S cell cycle
checkpoint to retard cell cycle progression. Furthermore, ROS have
been demonstrated as potent genotoxic agents (17,50). Therefore, it can be speculated
that oxidative stress induced by high oxygen tension enhances DNA
damage to arrest the cell cycle progression of NP cells from G1 to
S phase.
There are several limitations in the current study.
One limitation is that we investigated the effect of high oxygen
tension on the global gene expression profile and AS of rat NP
cells. Further studies based on human disc cells should be
performed. On the other hand, this study elucidated the involvement
of high oxygen tension in the establishment and progression of IDD
sketchily. The precise roles of high oxygen tension in regulating
the viability and function of disc cells and the pathogenesis of
IDD are required to be discussed in detail.
In conclusion, high oxygen tension is widely
involved in regulating various biological processes of NP cells
through transcription regulation and AS regulation. Several
processes are potently associated with the process of IDD.
Specifically, it disturbs the redox homeostasis of NP cells and
promotes matrix turnover in discs. Furthermore, high oxygen tension
retards NP cell cycle progression from G1 to S phase, which
consequently suppresses the growth of NP cells. High oxygen tension
is a crucial driver to the disc cell-mediated IDD process.
Acknowledgments
We thank Dr Yi Zha for his help in analyzing the
microarray data.
Abbreviations:
|
IVD
|
intervertebral disc
|
|
IDD
|
intervertebral disc degeneration
|
|
LBP
|
low back pain
|
|
NP
|
nucleus pulposus
|
|
AF
|
annulus fibrosus
|
|
AS
|
alternative splicing
|
|
ECM
|
extracellular matrix
|
|
Nox4
|
NADPH oxidase 4
|
|
DEG
|
differentially expressed gene
|
|
ASG
|
alternative splicing gene
|
|
SI
|
splicing index
|
|
ADAMTS
|
a disintegrin and metalloproteinase
with thrombospondin motifs
|
|
MMP
|
matrix metalloproteinase
|
|
BMP
|
bone morphogenetic protein
|
|
ROS
|
reactive oxygen species
|
|
GAS1
|
growth arrest specific 1
|
|
VEGFA
|
vascular endothelial growth factor
A
|
|
GPX1
|
glutathione peroxidase 1
|
|
HIF-1
|
hypoxia inducible factor-1
|
|
PBS
|
phosphate-buffered saline
|
|
ASE
|
alternative splicing exon
|
Notes
[1]
Funding
The design of the study and collection, analysis,
and interpretation of data and writing of the manuscript study were
supported by the National Natural Science Foundation of China
(grant nos. 81672215, 81572186, 81271982, 81472076 and
81401801).
[2] Authors'
contributions
CF contributed to the conception and design,
acquisition of data, analysis and interpretation of data, and
manuscript writing. YaZ contributed to the acquisition of data and
provision of study material or patients. MY and ML contributed to
the acquisition of data and analysis and interpretation of data. BH
contributed to the conception and design and final approval of the
version to be published. HL gave the final approval of the version
to be published, and contributed to the conception and design,
financial and administrative support. YuZ gave the final approval
of the version to be published, and contributed to the conception
and design, financial and administrative support. All authors read
and approved the final manuscript.
[3] Availability
of data and material
All data generated or analyzed during this study are
included in this published article.
[4] Ethics
approval and consent to participate
This research was approved by the Ethics Committee
of Xinqiao Hospital. All procedures described in this study were in
accordance with the standards set forth in the eighth edition of
the Guide for the Care and Use of Laboratory Animals published by
the National Academy of Sciences (Washington, DC, USA).
[5] Consent for
publication
Not applicable.
[6] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Vos T, Flaxman AD, Naghavi M, Lozano R,
Michaud C, Ezzati M, Shibuya K, Salomon JA, Abdalla S, Aboyans V,
et al: Years lived with disability (YLDs) for 1160 sequelae of 289
diseases and injuries 1990-2010: A systematic analysis for the
Global Burden of Disease Study 2010. Lancet. 380:2163–2196. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hong J, Reed C, Novick D and Happich M:
Costs associated with treatment of chronic low back pain: An
analysis of the UK General Practice Research Database. Spine.
38:75–82. 2013. View Article : Google Scholar
|
|
3
|
Roberts S, Evans H, Trivedi J and Menage
J: Histology and pathology of the human intervertebral disc. J Bone
Joint Surg Am. 88(Suppl 2): 10–14. 2006.PubMed/NCBI
|
|
4
|
Battié MC, Videman T, Kaprio J, Gibbons
LE, Gill K, Manninen H, Saarela J and Peltonen L: The Twin Spine
Study: Contributions to a changing view of disc degeneration. Spine
J. 9:47–59. 2009. View Article : Google Scholar
|
|
5
|
Xing QJ, Liang QQ, Bian Q, Ding DF, Cui
XJ, Shi Q and Wang YJ: Leg amputation accelerates senescence of rat
lumbar intervertebral discs. Spine. 35:E1253–E1261. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang D, Nasto LA, Roughley P, Leme AS,
Houghton AM, Usas A, Sowa G, Lee J, Niedernhofer L, Shapiro S, et
al: Spine degeneration in a murine model of chronic human tobacco
smokers. Osteoarthritis Cartilage. 20:896–905. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stirling A, Worthington T, Rafiq M,
Lambert PA and Elliott TS: Association between sciatica and
Propionibacterium acnes. Lancet. 357:2024–2025. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Park EY and Park JB: Dose- and
time-dependent effect of high glucose concentration on viability of
notochordal cells and expression of matrix degrading and fibrotic
enzymes. Int Orthop. 37:1179–1186. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartels EM, Fairbank JC, Winlove CP and
Urban JP: Oxygen and lactate concentrations measured in vivo in the
intervertebral discs of patients with scoliosis and back pain.
Spine. 23:1–7; discussion 8. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Urban JP, Smith S and Fairbank JC:
Nutrition of the intervertebral disc. Spine. 29:2700–2709. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee DC, Adams CS, Albert TJ, Shapiro IM,
Evans SM and Koch CJ: In situ oxygen utilization in the rat
intervertebral disc. J Anat. 210:294–303. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Risbud MV, Guttapalli A, Stokes DG,
Hawkins D, Danielson KG, Schaer TP, Albert TJ and Shapiro IM:
Nucleus pulposus cells express HIF-1 alpha under normoxic culture
conditions: A metabolic adaptation to the intervertebral disc
microenvironment. J Cell Biochem. 98:152–159. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Agrawal A, Guttapalli A, Narayan S, Albert
TJ, Shapiro IM and Risbud MV: Normoxic stabilization of HIF-1alpha
drives glycolytic metabolism and regulates aggrecan gene expression
in nucleus pulposus cells of the rat intervertebral disk. Am J
Physiol Cell Physiol. 293:C621–C631. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ali R, Le Maitre CL, Richardson SM,
Hoyland JA and Freemont AJ: Connective tissue growth factor
expression in human intervertebral disc: implications for
angiogenesis in intervertebral disc degeneration. Biotech
Histochem. 83:239–245. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Furusawa N, Baba H, Miyoshi N, Maezawa Y,
Uchida K, Kokubo Y and Fukuda M: Herniation of cervical
intervertebral disc: Immunohistochemical examination and
measurement of nitric oxide production. Spine. 26:1110–1116. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Suzuki S, Fujita N, Hosogane N, Watanabe
K, Ishii K, Toyama Y, Takubo K, Horiuchi K, Miyamoto T, Nakamura M,
et al: Excessive reactive oxygen species are therapeutic targets
for intervertebral disc degeneration. Arthritis Res Ther.
17:3162015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dimozi A, Mavrogonatou E, Sklirou A and
Kletsas D: Oxidative stress inhibits the proliferation, induces
premature senescence and promotes a catabolic phenotype in human
nucleus pulposus intervertebral disc cells. Eur Cell Mater.
30:89–103. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hou G, Lu H, Chen M, Yao H and Zhao H:
Oxidative stress participates in age-related changes in rat lumbar
intervertebral discs. Arch Gerontol Geriatr. 59:665–669. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Adams MA and Roughley PJ: What is
intervertebral disc degeneration, and what causes it? Spine.
31:2151–2161. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ding F, Shao ZW and Xiong LM: Cell death
in intervertebral disc degeneration. Apoptosis. 18:777–785. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nasto LA, Robinson AR, Ngo K, Clauson CL,
Dong Q, St Croix C, Sowa G, Pola E, Robbins PD, Kang J, et al:
Mitochondrial-derived reactive oxygen species (ROS) play a causal
role in aging-related intervertebral disc degeneration. J Orthop
Res. 31:1150–1157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen JW, Ni BB, Zheng XF, Li B, Jiang SD
and Jiang LS: Hypoxia facilitates the survival of nucleus pulposus
cells in serum deprivation by downregulating excessive autophagy
through restricting ROS generation. Int J Biochem Cell Biol.
59:1–10. 2015. View Article : Google Scholar
|
|
23
|
Pan Q, Shai O, Lee LJ, Frey BJ and
Blencowe BJ: Deep surveying of alternative splicing complexity in
the human transcriptome by high-throughput sequencing. Nat Genet.
40:1413–1415. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Black DL: Mechanisms of alternative
pre-messenger RNA splicing. Annu Rev Biochem. 72:291–336. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Blencowe BJ: Alternative splicing: New
insights from global analyses. Cell. 126:37–47. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lukong KE, Chang KW, Khandjian EW and
Richard S: RNA-binding proteins in human genetic disease. Trends
Genet. 24:416–425. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Martinez-Contreras R, Cloutier P, Shkreta
L, Fisette JF, Revil T and Chabot B: hnRNP proteins and splicing
control. Adv Exp Med Biol. 623:123–147. 2007. View Article : Google Scholar
|
|
28
|
Ng B, Yang F, Huston DP, Yan Y, Yang Y,
Xiong Z, Peterson LE, Wang H and Yang XF: Increased noncanonical
splicing of autoantigen transcripts provides the structural basis
for expression of untolerized epitopes. J Allergy Clin Immunol.
114:1463–1470. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Anderson DG, Markova D, Adams SL, Pacifici
M, An HS and Zhang Y: Fibronectin splicing variants in human
intervertebral disc and association with disc degeneration. Spine.
35:1581–1588. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hirose Y, Chiba K, Karasugi T, Nakajima M,
Kawaguchi Y, Mikami Y, Furuichi T, Mio F, Miyake A, Miyamoto T, et
al: A functional polymorphism in THBS2 that affects alternative
splicing and MMP binding is associated with lumbar-disc herniation.
Am J Hum Genet. 82:1122–1129. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sztrolovics R, Grover J, Cs-Szabo G, Shi
SL, Zhang Y, Mort JS and Roughley PJ: The characterization of
versican and its message in human articular cartilage and
intervertebral disc. J Orthop Res. 20:257–266. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Purdom E, Simpson KM, Robinson MD, Conboy
JG, Lapuk AV and Speed TP: FIRMA: A method for detection of
alternative splicing from exon array data. Bioinformatics.
24:1707–1714. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Srinivasan K, Shiue L, Hayes JD, Centers
R, Fitzwater S, Loewen R, Edmondson LR, Bryant J, Smith M and
Rommelfanger C: Detection and measurement of alternative splicing
using splicing-sensitive microarrays. Methods. 37:345–359. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Clark TA, Sugnet CW and Ares M Jr:
Genomewide analysis of mRNA processing in yeast using
splicing-specific microarrays. Science. 296:907–910. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
36
|
Sakai D and Andersson GB: Stem cell
therapy for intervertebral disc regeneration: Obstacles and
solutions. Nat Rev Rheumatol. 11:243–256. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vo NV, Hartman RA, Yurube T, Jacobs LJ,
Sowa GA and Kang JD: Expression and regulation of
metalloproteinases and their inhibitors in intervertebral disc
aging and degeneration. Spine J. 13:331–341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Feng C, Liu H, Yang Y, Huang B and Zhou Y:
Growth and differentiation factor-5 contributes to the structural
and functional maintenance of the intervertebral disc. Cell Physiol
Biochem. 35:1–16. 2015. View Article : Google Scholar
|
|
39
|
Shen C, Yan J, Jiang LS and Dai LY:
Autophagy in rat annulus fibrosus cells: Evidence and possible
implications. Arthritis Res Ther. 13:R1322011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen JW, Ni BB, Li B, Yang YH, Jiang SD
and Jiang LS: The responses of autophagy and apoptosis to oxidative
stress in nucleus pulposus cells: implications for disc
degeneration. Cell Physiol Biochem. 34:1175–1189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jiang L, Jin Y, Wang H, Jiang Y and Dong
J: Glucosamine protects nucleus pulposus cells and induces
autophagy via the mTOR-dependent pathway. J Orthop Res.
32:1532–1542. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Binch AL, Cole AA, Breakwell LM, Michael
AL, Chiverton N, Creemers LB, Cross AK and Le Maitre CL: Nerves are
more abundant than blood vessels in the degenerate human
intervertebral disc. Arthritis Res Ther. 17:3702015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Binch AL, Cole AA, Breakwell LM, Michael
AL, Chiverton N, Cross AK and Le Maitre CL: Expression and
regulation of neurotrophic and angiogenic factors during human
intervertebral disc degeneration. Arthritis Res Ther. 16:4162014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Takae R, Matsunaga S, Origuchi N, Yamamoto
T, Morimoto N, Suzuki S and Sakou T: Immunolocalization of bone
morphogenetic protein and its receptors in degeneration of
intervertebral disc. Spine. 24:1397–1401. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Nakase T, Ariga K, Miyamoto S, Okuda S,
Tomita T, Iwasaki M, Yonenobu K and Yoshikawa H: Distribution of
genes for bone morphogenetic protein-4, -6, growth differentiation
factor-5, and bone morphogenetic protein receptors in the process
of experimental spondylosis in mice. J Neurosurg. 94(Suppl 1):
68–75. 2001.PubMed/NCBI
|
|
46
|
Haro H, Kato T, Komori H, Osada M and
Shinomiya K: Vascular endothelial growth factor (VEGF)-induced
angiogenesis in herniated disc resorption. J Orthop Res.
20:409–415. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gawri R, Rosenzweig DH, Krock E, Ouellet
JA, Stone LS, Quinn TM and Haglund L: High mechanical strain of
primary intervertebral disc cells promotes secretion of
inflammatory factors associated with disc degeneration and pain.
Arthritis Res Ther. 16:R212014. View
Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lee JM, Song JY, Baek M, Jung HY, Kang H,
Han IB, Kwon YD and Shin DE: Interleukin-1β induces angiogenesis
and innervation in human intervertebral disc degeneration. J Orthop
Res. 29:265–269. 2011. View Article : Google Scholar
|
|
49
|
Sahoo S, Meijles DN and Pagano PJ: NADPH
oxidases: key modulators in aging and age-related cardiovascular
diseases? Clin Sci (Lond). 130:317–335. 2016. View Article : Google Scholar
|
|
50
|
Weyemi U, Lagente-Chevallier O, Boufraqech
M, Prenois F, Courtin F, Caillou B, Talbot M, Dardalhon M, Al
Ghuzlan A, Bidart JM, et al: ROS-generating NADPH oxidase NOX4 is a
critical mediator in oncogenic H-Ras-induced DNA damage and
subsequent senescence. Oncogene. 31:1117–1129. 2012. View Article : Google Scholar :
|