Introduction
During the process of bone regeneration, three main
phases, inflammation, new bone formation and bone remodeling take
place in an orchestrated fashion to repair the injury. Bone
remodeling is regulated by both the number and activity of
osteoblasts and osteoclasts. Osteoblast lineage commitment,
proliferation and differentiation are controlled by a well-defined
genetic program. Bone morphogenetic proteins (BMPs) are unique
extracellular multifunctional signaling cytokines belonging to the
large transforming growth factor-β (TGF-β) superfamily (1–6).
BMPs can induce heterogeneous oligomerization of type I and II
receptor serine/threonine kinases and Smad1/5/8 phosphorylation.
Subsequently, the phosphorylated Smad1/5/8 are associated with
Smad4 and translocate into the nucleus where they cooperate with
tissue-specific transcription factors to drive osteogenic target
gene expression (7). Or through
the non-canonical Smad-independent signaling [p38 mitogen-activated
protein kinase (MAPK)] pathway to active downstream transcription
factors to potentiate the osteoblastogenesis role (8). Bone morphogenetic protein 9 (BMP9),
a member of the BMP family, in previous studies was identified it
as one of the most osteogenic BMPs (1,4,6,9,10),
but the mechanisms involved remain largely unclear.
MicroRNAs (miRNAs or miRs) are a class of endogenous
noncoding RNAs, ~22 nucleotides in length that are involved in the
regulation of gene expression, coordinating a broad spectrum of
biological processes (11,12).
miRNAs influence the expression of the target gene by binding to
its 3′-untranslated region (3′-UTR) and inactivate it by promoting
its degradation or translational repression (13,14). Several studies indicated that
miRNAs act as key regulators in cell differentiation (15–20), cell growth (21,22), cell death (23), and lipid metabolism (24). Previous studies have shown that
miR-206, -204, -133, -135, -125b, -141, -200a and -214 can inhibit
osteogenic differentiation (12,16,25-30). On the contrary, miR-15b and
miR-29b have been demonstrated to promote osteogenesis (31,32). Apparently, miRNAs are important
regulators during the osteogenic differentiation. As a member of
the oncomir class of microRNAs, miR-155 is implicated in
lymphomagenesis (33-36) and a wide array of nonlymphoid
tumors including breast, colon, and lung (33,37-41).
In recent years, several studies have focused on
miRNAs being modulated by BMP signaling as a means to investigate
the role of miRNAs in osteoblasts (17,26,28,29,42). Another study indicated that
suppressing the expression and function of miR-155 contributes to
mitigate the inhibition of TNF-α on BMP-2-induced osteogenic
differentiation (43), indicating
that there was a link between miR-155 and BMP signaling. From the
early studies we found that during the process of osteogenic
differentiation induced by BMP9, the expression of miR-155 has
changed. Overexpression of miR-155 in C2C12 myoblasts and mouse
embryonic fibroblasts (MEF), induced osteogenesis by BMP9, the
expression of osteogenesis-related genes like runt-related
transcription factor 2 (Runx2) and alkaline phosphatase (ALP)
declined, so we reasonably hypothesized that miR-155 can inhibit
this process, but the relevant mechanisms are not clear. The aim of
this work is to explore the effect of miR-155 on the osteogenic
differentiation induced by BMP9, and investigate the correlation
mechanisms.
Materials and methods
Cell lines and culture
The HEK-293 (human embryonic kidney) cell line,
C2C12 (myoblasts) cell line, and MEF cell line were obtained from
American Type Culture Collection (ATCC, Manassas, VA, USA). The
HEK-293 cells, C2C12 cells and MEF cells were all immortalized and
maintained in DMEM, supplemented with 10% fetal bovine serum (FBS)
(Gibco, Grand Island, NY, USA), 100 U/ml penicillin and 100
µg/ml streptomycin. The cells were maintained at 37°C in a
humidified 5% CO2 atmosphere.
Transfection of the miR-155 mimic,
miR-155 inhibitor, and shBMPR2
miR-155 mimic, miR-155 inhibitor, and shBMPR2 were
purchased from GenePharma (Shanghai, China). The sequences for each
oligonucleotide are listed in Table
I. Entranster™ R4000 (Engreen Biosystems, Beijing, China) was
utilized to transfect miR-155 mimic, miR-155 inhibitor and miR-155
negative control (NC) according to the manufacturer's instructions.
To transfect shBMPR2 and it negative control shNC, Lipofectamine
2000 transfection reagent (Invitrogen, Shanghai, China) was used
according to the manufacturer's instructions. In this study, C2C12
cells and MEF cells were transfected with with 100 nM of miR-155
mimic, or miR-155 inhibitor or NC in a 24-well plate or 500 nM of
the above in a 6-well plate. In a 6-well plate, shNC or shBMPR2
were transfected into cells at 800 ng each well.
 | Table IOligoribonucleotides used in the
study. |
Table I
Oligoribonucleotides used in the
study.
| Name | Sequence
(5′→3′) |
|---|
| mmu-miR-155
mimic | F:
UUAAUGCUAAUUGUGAUAGGGGU |
| R:
CCCUAUCACAAUUAGCAUUAAUU |
| mmu-miR-155
inhibitor | F:
ACCCCUAUCACAAUUAGCAUUAA |
| Negative control
(NC) | F:
UUCUCCGAACGUGUCACGUTT |
| R:
ACGUGACACGUUCGGAGAATT |
| shBMPR2 | F:
CACCGCCAAGATGAATACAATCAATTTCAAGAGAATTGATTGTATTCATCTTGGCTTTTTTG |
| R:
GATCCAAAAAAGCCAAGATGAATACAATCAATTCTCTTGAAATTGATTGTATTCATCTTGGC |
| shNC | F:
CACCGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTTG |
| R:
GATCCAAAAAATTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAAC |
Transfection of the BMP9 and
si-Runx2
C2C12 cells and MEF cells were cultured in 24-well
plates, each well was added with 500 µl DMEM supplemented
with 10% FBS, and before using BMP9 and si-Runx2, we tested the
titer of these recombinant adenoviruses. Each well contained 3
µl of polybrene to contribute to the adenovirus entry into
cells, different volumes of adenovirus was added, until the
infection efficiency reached 30%, the volume of the adenovirus was
the titer for the corresponding cells, respectively. In a 6-well
plate, 12 µl of polybrene and four-fold titer volume of
adenovirus were added after that the cells attached.
RNA extraction, reverse transcription and
real-time quantitative PCR (RT-qPCR) analysis
Total RNA was extracted from C2C12 cells and MEF
cells at the indicated time points using TRIzol reagent (Ambion,
Aukland, New Zealand) according to the manufacturer's instructions
for the analysis of mRNA and miRNA expression. The concentration
and purity of RNA were quantified using spectrophotometer (NanoDrop
1000 Spectrophotometer; Thermo Fisher Scientific Inc., Waltham, MA,
USA). The extracted total RNA (2,000 ng) was reverse transcribed
using reverse transcription primers according to the manufacturer's
instructions (Reverse Transcriptase M-MLV, Takara code: D2639A;
Takara Bio Inc., Otsu, Japan). The primers used for reverse
transcription PCR (RT-PCR) and RT-qPCR are listed in Table II. RT-qPCR were performed on a
Bio-Rad iQ5 instrument (Bio-Rad, Hercules, CA, USA) used cDNA as
templates to amplify target genes, mixing with
SYBR®-Green PCR Master Mix (Takara Bio, Inc.),
Fold-changes in expression were calculated using the
2-∆∆Ct method, and each experiment was performed in
triplicate. Data were analyzed using Optical system software,
version 2.0. The expression levels of β-actin and the small U6 RNA
were used as internal controls for mRNA and miRNA,
respectively.
 | Table IIPrimers used in the study. |
Table II
Primers used in the study.
| Genes | Sequence
(5′→3′) |
|---|
| mmu-miR-155 | RT:
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACCCCT |
| U6 | RT:
AAAATATGGAACGCTTCACGAATTTG |
| mmu-miR-155 | F:
GGCGTTAATGCTAATTGTGAT | R:
GTGCAGGGTCCGAGGT |
| U6 | F:
CTCGCTTCGGCAGCACATATACT | R:
ACGCTTCACGAATTTGCGTGTC |
| Mouse Runx2 | F:
TCTGACAAAGCCTTCATGTCC | R:
AAATAGTGATACCGTAGATGCG |
| Mouse OSX | F:
GAAGTCCAATGGGGATCTGA | R:
GAATCCCTTTCCCTCTCCAG |
| Mouse ALP | F:
TGACCTTCTCTCCTCCATCC | R:
CTTCCTGGGAGTCTCATCCT |
| Mouse OCN | F:
CTGCTTGTGACGAGCTATCAG | R:
TGATACCGTAGATGCGTTTGT |
| Mouse β-actin | F:
ATGAAGGCGTGGCAACAT | R:
GCCATTGGCTCTGTCCTG |
| Mouse BMPR2 | F:
GCAGTGAGGTCACTCAAGGA | R:
CATCATGAGTTCAGCCATCC |
ALP staining and ALP activity
ALP staining was performed with BCIP/NBT Alkaline
Phosphatase Color Development kit (C3206; Beyotime, Shanghai,
China) according to the manufacturer's instructions. Briefly, we
seeded the C2C12 cells and MEF cells in 24-well plates followed by
treatment with NC, miR-155 mimic or miR-155 inhibitor and BMP9 for
7 days. The supernatant was discarded, cells were washed three
times using phosphate-buffered saline (PBS). Then 3 ml of ALP
developing buffer was mixed with 10 µl BCIP, and 20
µl NBT to form BCIP/NBT working liquid and 200 µl
BCIP/NBT was the working liquid in each well of 24-well plates, and
the plates were placed in the dark to undergo ALP staining for 30
min. Finally, BCIP/NBT working liquid was removed by suction and
the staining observed. Cells were scanned at both a lower
(ScanMaker 3600; Shanghai Microtek Technology Co., Ltd., Shanghai,
China) and higher magnification under a bright field microscope
(T-DH; Nikon Corp., Tokyo, Japan). ALP activity was measured using
an established enzymatic assay. ALP substrate was prepared from
solution A (8 ng naphtol-AS-TR phosphate/300 ml
N,N-dimethylformamide; both from Sigma-Aldrich, St. Louis, MO, USA)
and solution B (24 mg fast blue BB salt/30 ml of 100 mmol/l Tris
HCl, pH 9.6). The two solutions were combined, 10 mg of
MgCl2 was added, the resulting solution was used
immediately. Enzymatic activity was determined in cell lysates that
were solubilized with 0.1% Triton X-100. Aliquots (5 µl) of
each sample were incubated with 15 µl of AP substrate buffer
(100 mmol/l diethanolamine, 150 mmol/l NaCl, 2 mmol/l
MgCl2 p-nitrophenylphosphate at 2.5 mg/ml) and 5
µl of ALP substrate under the conditions of protection from
light for 30 to 40 min at room temperature. ALP activity was tested
using a luminometer (Promega, Madison, WI, USA).
Alizarin red S staining
C2C12 cells and MEF cells were seeded in 24-well
plates treated with NC, miR-155 mimic or miR-155 inhibitor and BMP9
for 14 days. The supernatant was discarded, and cells washed three
times with PBS. The cells were fixed with 0.05% ice-cold
glutaraldehyde (Chongqing Chuandong Chemical Group) for 10 min and
rinsed with double-distilled H2O. Then stained with 40
mM (2%) Alizarin red S (Sigma-Aldrich) at pH 4.0, rocked gently for
~5 min. The cells were rinsed three times with double-distilled
H2O and then rinsed for 15 min with PBS and gently
shaken. Finally cells were scanned at both a lower (ScanMaker 3600;
Shanghai Microtek Technology Co., Ltd.) and higher magnification
under a bright field microscope (T-DH; Nikon Corp.).
Western blot analysis
C2C12 cells and MEF cells were plated into a 10-cm
Petri dishes. After treating cells as before, and when cells
reached 80% confluence, cell lysate was collected and protein
concentrations were determined using the Bio-Rad protein assay kit.
The protein (50 µg) was boiled for 5 min in Laemmli buffer
[62.5 mM Tris (pH 6.8), 1% sodium dodecyl sulfate (SDS), 20%
glycerol, 0.01% bromophenol blue, and 100 mM DTT] before being
loaded onto a 12% SDS-polyacrylamide gel and then transferred to
PVDF membranes. Specific protein bands were detected with mouse
anti-BMPR2 polyclonal antibody (1:200 dilution, cat. no. BM2821;
BOSTER), rabbit anti-p-Smad1/5/8 monoclonal antibody (1:1,000
dilution, cat. no. sc-12353), rabbit anti-Smad1/5/8 monoclonal
antibody (1:1,000 dilution, cat. no. sc-6031-R, lot no. D1911),
rabbit anti-Runx2 monoclonal antibody (1:1,000 dilution, cat. no.
sc-10758, lot no. A0810) (all from Santa Cruz Biotechnology, Inc.,
Santa Cruz, CA, USA), rabbit anti-osteocalcin (OCN) polyclonal
antibody (1:200 dilution, cat. no. bs-4917R; Beijing Biosynthesis
Biotechnology Co., Ltd.), rabbit anti-osteopontin (OPN) polyclonal
antibody (1:500 dilution, cat. no. wl00691; Wanleibio, Shenyang,
China), or anti-β-actin mouse monoclonal antibody (1:1,000
dilution; Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing,
China) overnight at 4°C, and then washed with TBST three times,
each time for 15 min, the membranes were incubated with goat
anti-rabbit or goat anti-mouse horseradish peroxidase conjugated
secondary antibody (1:5,000 dilution; Zhongshan Golden Bridge
Biotechnology Co., Ltd.), for 1 h at 37°C. After that, membranes
were washed with TBST three times, each time for 15 min. The
secondary antibodies were detected with western chemiluminescence
reagent (Millipore Corp., Billerica, MA, USA). The results were
recorded using the Bio-Rad electrophoresis documentation system
(Gel Doc 1000) and Quantity One software, version 4.5.0.
Luciferase reporter assay
TargetScan (http://www.targetscan.org) and PicTar (http://pictar.mdc-berlin.de/) were utilized to
identify the possible target genes of miR-155 in BMP9-induced
osteogenic differentiation of C2C12 cells and MEF cells. The 3′-UTR
of Runx2 and BMPR2 containing miR-155-binding sequences, and the
sequences contained binding sites with miR-155 were synthesised
following the manufacturer's instructions (pMIR-REPORT™ system,
miRNA expression reporter vector; Ambion, Carlsbad, CA, USA). The
sequences contained binding sites with miR-155 are listed in
Table III. The annealing
fragments were then cloned into the SpeI/HindIII
backbone of the pMIR-REPORT microRNA (miRNA) expression reporter
(Ambion). Binding-region mutations were achieved using an oligo
synthesis following the manufacturer's instructions (Ambion).
Transient transfection into HEK-293 cells (1×104
cells/well) was carried out in 24-well plates with Lipofectamine
2000 transfection reagents (Invitrogen) following the
manufacturer's instructions. The cells were co-transfected with 800
ng of the luciferase construct plasmid and miR-155 mimic or NC, and
luciferase assays were performed using the dual-luciferase reporter
assay system (Ambion) according to the manufacturer's instructions.
Luminescent signals were quantified using a luminometer (Promega),
and each value of luciferase activity was normalized by the
luciferase activity of cells that transfected empty vector
only.
 | Table IIIThe sequences contained binding sites
with miR-155. |
Table III
The sequences contained binding sites
with miR-155.
| Name | Sequence
(5′→3′) |
|---|
| WT-Runx2 | F:
CTAGGATGTAGTTTGTTTTCACAATGTATGAAGGAGATGCTCTG |
| R:
AGCTCAGAGCATCTCCTTCATACATTGTGAAAACAAACTACATC |
| MT-Runx2 | F:
CTAGGATGTAGTTTGTTTTAGTAATGTATGAAGGAGATGCTCTG |
| R:
AGCTCAGAGCATCTCCTTCATACATTACTAAAACAAACTACATC |
| WT-BMPR2 | F:
CTAGCTTATGGGGTAATTAGCATTATAAGACTTTATAAAAGTGAGCTGATGGCTCTAGC |
| R:
AGCTGCTAGAGCCATCAGCTCACTTTTATAAAGTCTTATAATGCTAATTACCCCATAAG |
| MT-BMPR2 | F:
CTAGCTTATGGGGTAATTATTAGGATAAGACTTTATAAAAGTGAGCTGATGGCTCTAGC |
| R:
AGCTGCTAGAGCCATCAGCTCACTTTTATAAAGTCTTATCCTAATAATTACCCCATAAG |
Ectopic in vivo bone formation assay of
transfected MEF cells
All animal experiments were approved by
Institutional Animal Care and Use Committee (IACUC) of Chongqing
Medical University. The nude mice were purchased from Huafukang
(Beijing Hfk Bioscience Co., Ltd., Beijing, China). MEF cells were
transfected as described above, and the long-lasting mimic
(mmu-agomiR-155) and inhibitor (mmu-antagomiR-155) of miR-155 were
used, and the sequences are fully consistent with mmu-miR-155 or
mmu-anti-miR-155. The sequence of stable NC is the same as the NC
used before. In short, after treating cells like before, cells were
collected at 80% confluence and for subcutaneous injection
(5×106/injection) into the flanks of the athymic nude
mice (5 animals/group, 4–6-week old, female) (44). At 4 weeks after implantation,
heterotopic bones were harvested from mice after euthanasia, fixed
in 4% paraformaldehyde and scanned using a µCT 40 system
(Scanco Medical, Brüttisellen, Switzerland). Parameters computed
from these data include bone mineral density (BMD), bone volume
(BV/TV), trabecular thickness (Tb.Th), trabecular number (Tb.N) and
trabecular separation (Tb.Sp). For histological analysis of bones,
after scanning used µCT, decalcified in 10% EDTA for ~4
weeks, before being embedded in paraffin. Sections (5 µm)
were cut and stained with hematoxylin and eosin (H&E) or with
Masson's trichrome according to the manufacturer's instructions.
Bone histomorphometry was performed using a microscope (T-DH; Nikon
Corp.).
Statistical analysis
The quantitative experiments were performed in
triplicate and/or repeated three times. Data are represented as the
mean ± SD. Statistical significance between the control and
treatment groups were determined by one-way analysis of variance
and the Student's t-test. All statistical analyses were performed
using GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA,
USA). P<0.05 was considered statistically significant.
Results
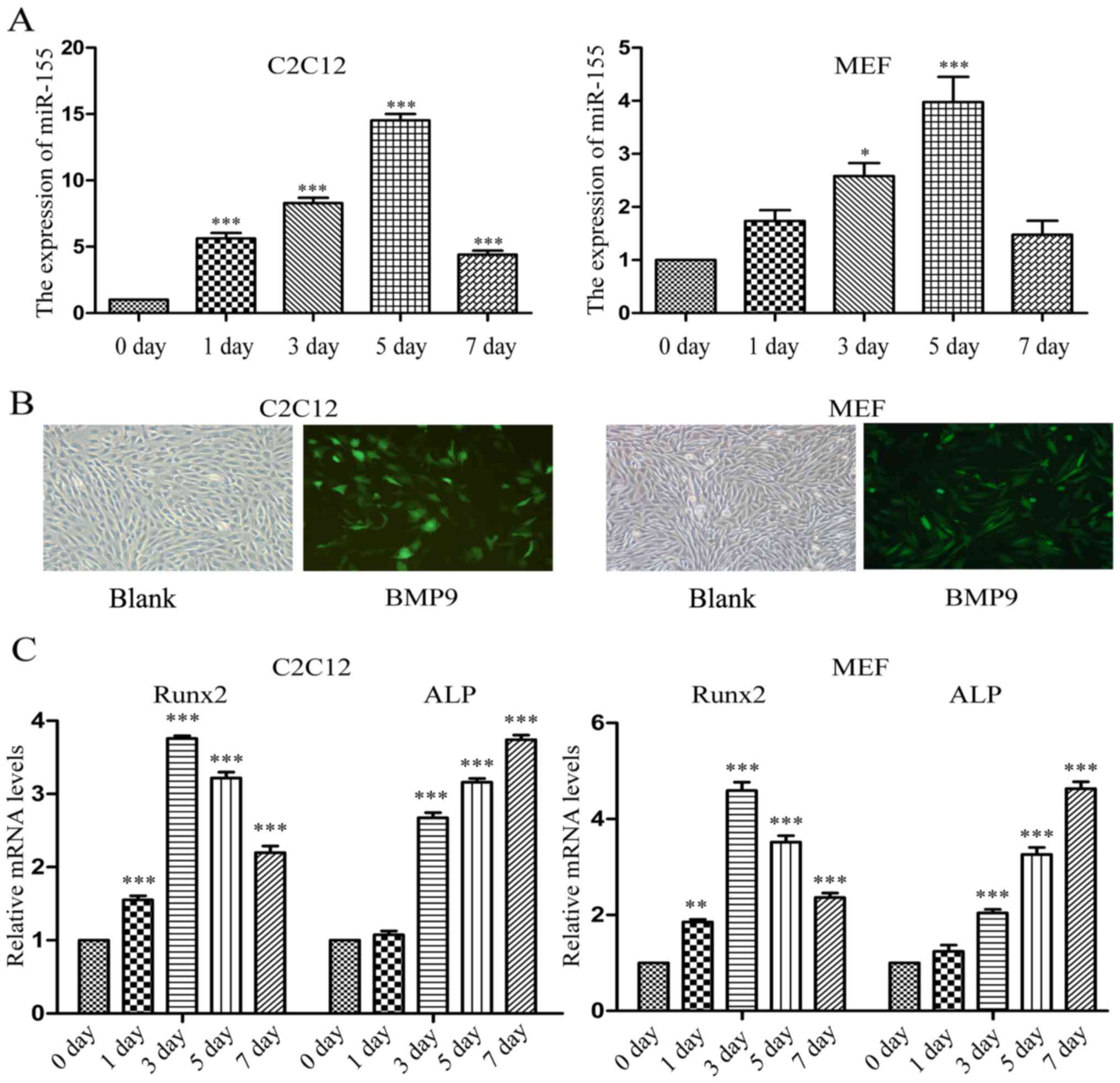
During the early stage of osteogenic
differentiation induced by BMP9, the expression of miR-155 was
increased at first and then decreased
Several differential expression miRNAs including
miR-21, miR-23b and miR-155 have been previously identified during
the early stage of mesenchymal stem cells (MSCs) osteogenesis
induced by BMP9 through microarray data analysis (data not shown).
Allowing us to consider that there are some associations between
miR-155 and BMP9 in osteogenesis. During the early stage of
osteogenic differentiation induced by BMP9, we detected the
expression of miR-155 by using RT-qPCR on different days. It was
found that compared with the expression level of 0 day, the
expression level of miR-155 was upregulated in the early stage of
BMP9-induced C2C12 cells and MEF cells osteogenic differentiation,
and on the 5th day reached the maximum expression, then decreased
on the 7th day (Fig. 1A). In our
laboratory, BMP9 contains green fluorescence protein (GFP) tag, and
so the results of fluorescence microscopy proved that BMP9 was
transfected into C2C12 and MEF cells effectively (Fig. 1B). To further confirm the
osteoinduction effect of BMP9, RT-qPCR was used to detect the mRNA
expression level of osteoblast-specific genes over 7 days,
including Runx2 and ALP. The results revealed that successful
osteogenic differentiation of the C2C12 and MEF cells was induced
(Fig. 1C).
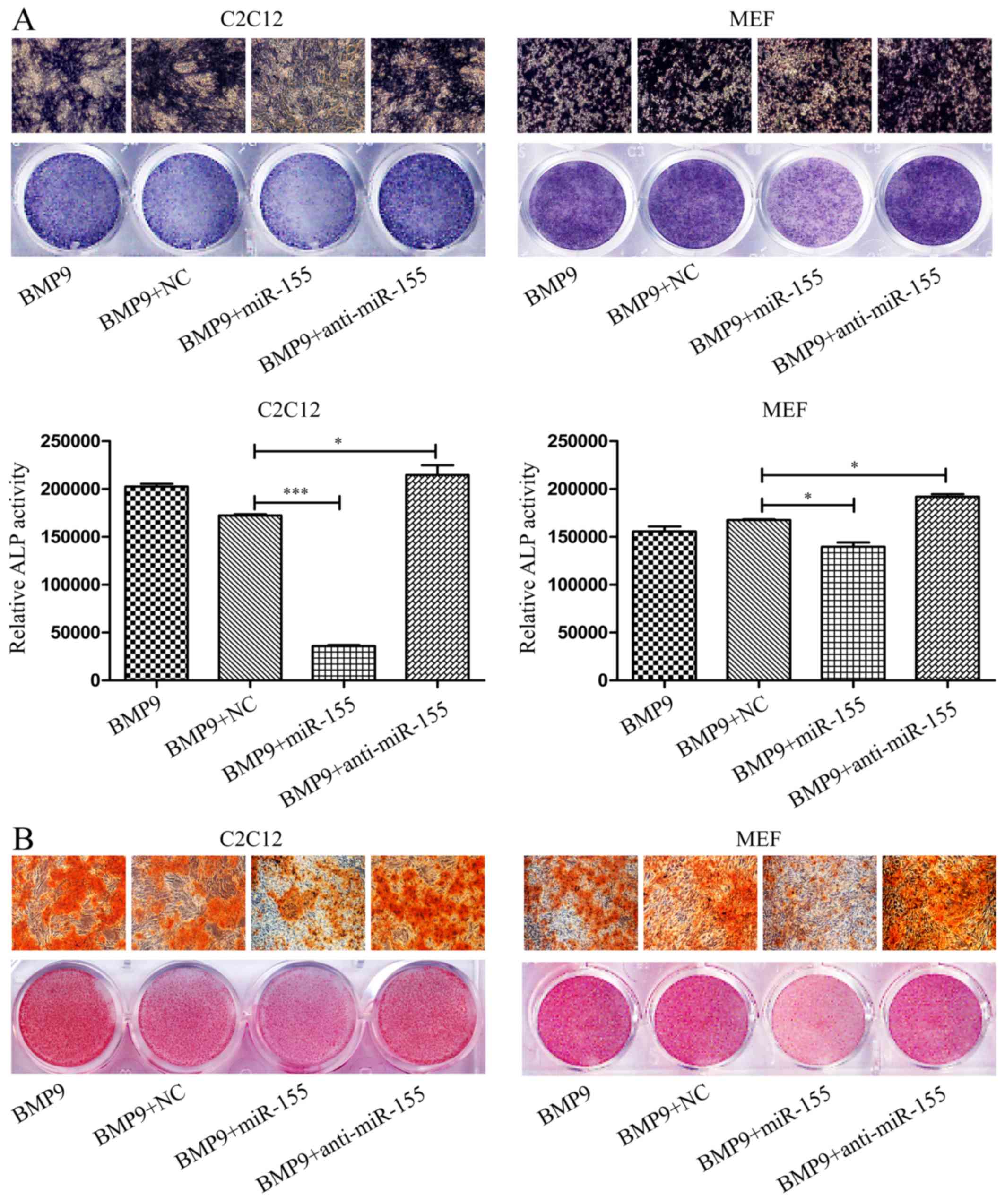
Overexpression of miR-155 suppresses
BMP9-induced ALP and calcium deposition in MSCs
Subsequently, to investigate the roles of miR-155 in
the osteogenic differentiation of C2C12 cells and MEF cells induced
by BMP9, the expression level of miR-155 following transfection
were tested by RT-qPCR, in the whole study, we set control groups
contain BMP9 group and BMP9 + miR-negative control group (BMP9 +
NC) (Fig. 3A). After transfecting
miR-155 or anti-miR-155 into C2C12 cells and MEF cells,
respectively, induced osteogenesis by BMP9 for 7 days. ALP staining
results showed that compared with the control groups, overexpressed
miR-155 (BMP9 + miR-155) downregulated the early osteogenesis
differentiation indicator-ALP activity, but inhibition of miR-155
(BMP9 + anti-miR-155) attenuated this inhibitive effect (Fig. 2A, top panel), quantitative
determination of ALP activity, the results were in accordance with
the ALP staining of C2C12 cells and MEF cells, respectively
(Fig. 2A, bottom panel).
We further analyzed the effect of miR-155 on late
osteogenic differentiation by Alizarin Red S staining of matrix
mineralization. The results were consistent with ALP staining,
overexpressed exogenous miR-155 repressed the BMP9-induced calcium
deposition of C2C12 cells and MEF cells, and decreased the
endogenous miR-155 adverse effect (Fig. 2B). In conclusion, the above
results indicated that miR-155 impairs the BMP9-induced osteoblast
lineage commitment of MSCs.
miR-155 downregulates the mRNA expression
level of osteogenesis-related genes
To detect the influence of miR-155 on the
osteogenesis induced by BMP9 on mRNA level, firstly, we used
RT-qPCR to measure the expression of miR-155 after transfection to
verified that our transfection was successful. The data showed that
during osteogenic differentiation induced by BMP9, compared with
control group, the intracellular miR-155 level was significantly
elevated by transfection with miR-155, whereas transfected with
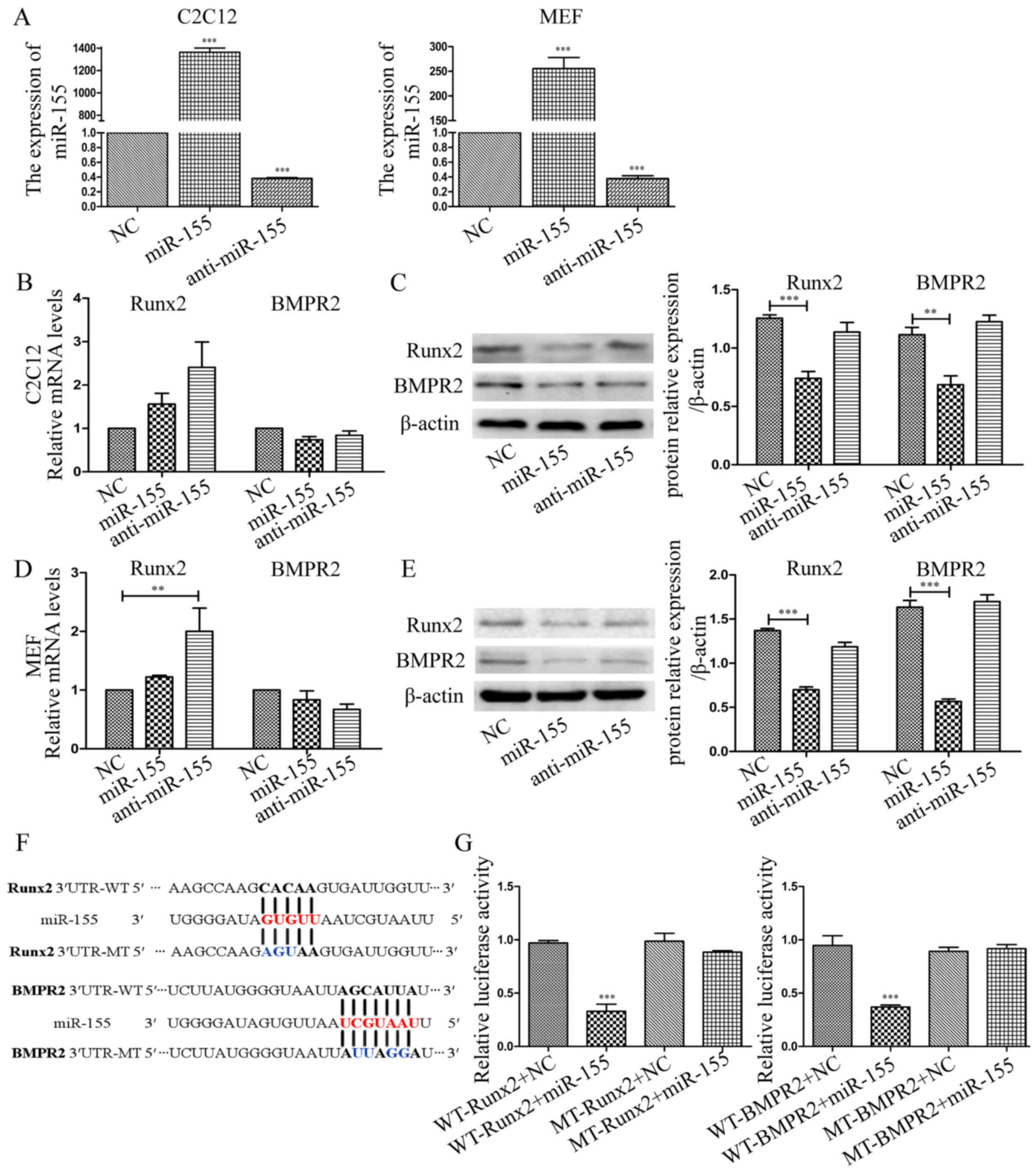
anti-miR-155 led to distinct reduction in miR-155 content (Fig. 3A). RT-qPCR was used to evaluate
the expression level of Runx2 and osterix (OSX) on the 3rd day, the
expression of ALP and OCN on the 7th day of BMP9-induced
osteogenesis. Compared with the control groups, the expression of
these osteogenesis markers were decreased by miR-155, conversely,
anti-miR-155 increased their expression in the process of
osteogenic differentiation induced by BMP9 in C2C12 cells, except
the expression of OSX and ALP (Fig.
3B). The results of MEF cells were consis tent with C2C12 cells
(Fig. 3C). Taken together,
miR-155 could suppress the expression of Runx2, OSX, ALP and OCN,
which are bone differentiation related markers.
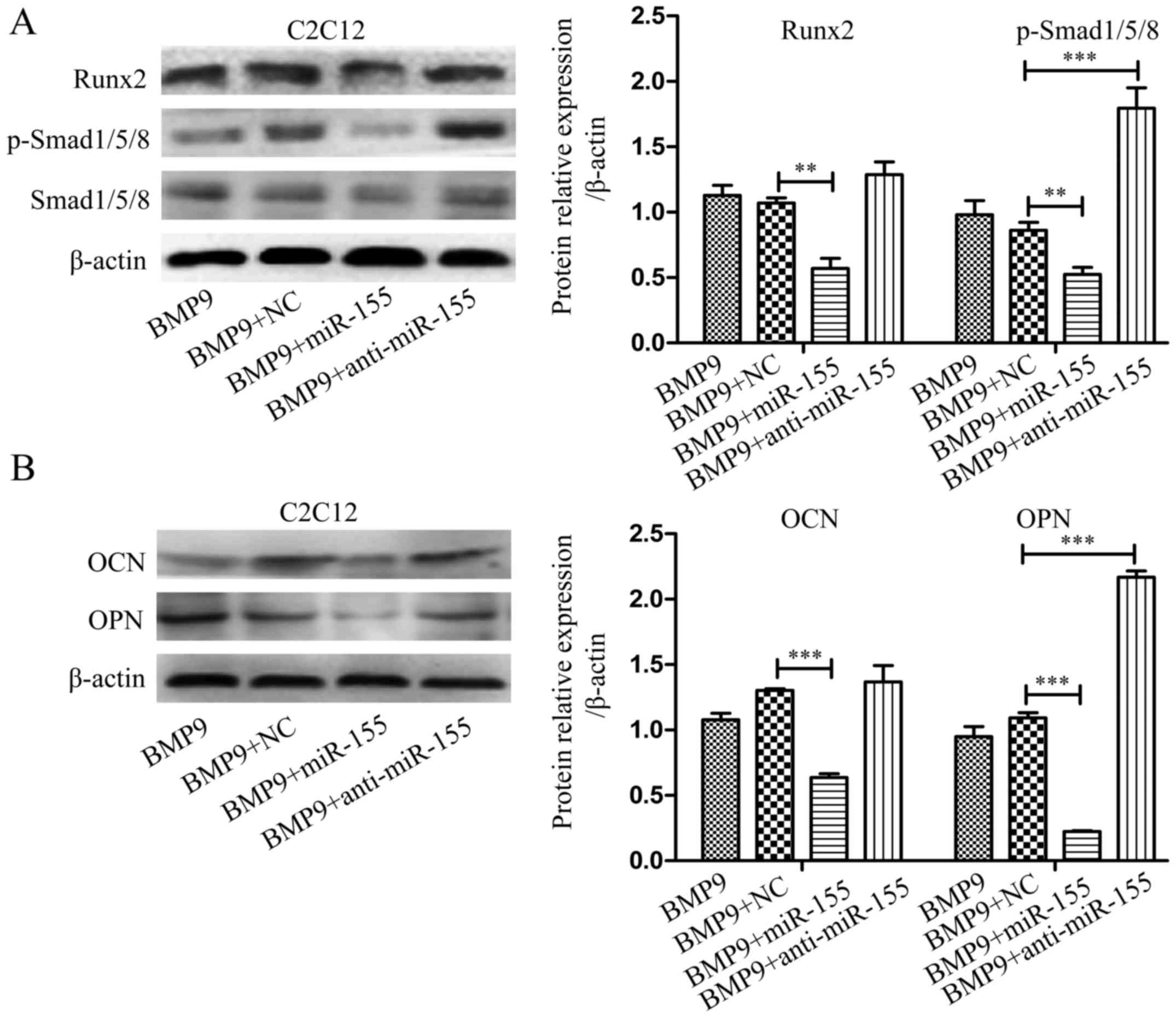
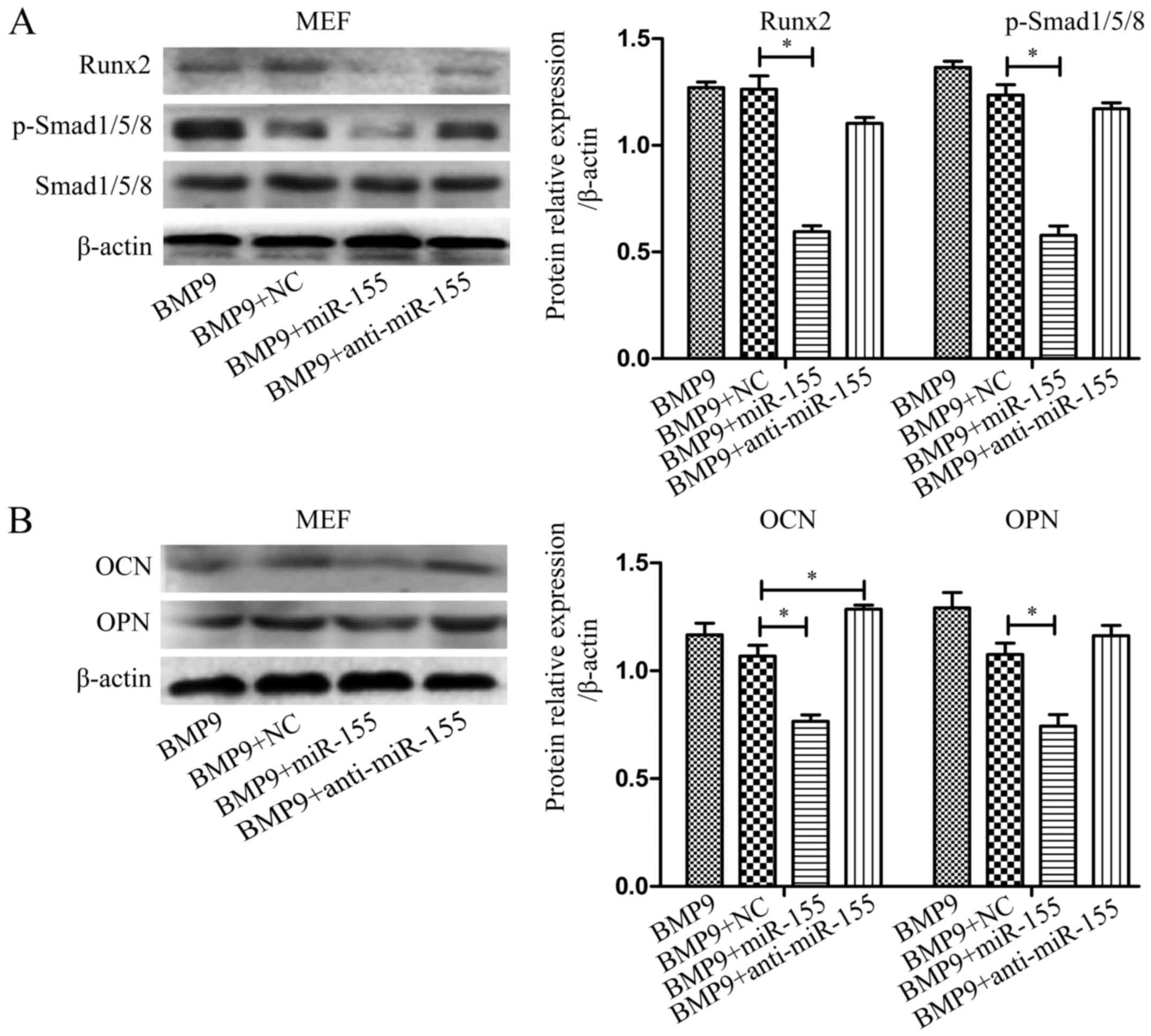
miR-155 suppresses BMP signaling during
the osteogenesis induced by BMP9
Previous studies revealed that Smads, p38 and ERK1/2
are involved in BMP9-induced osteogenic differentiation (45). We hypothesized that miR-155 may
through inhibiting canonical Smad-dependent signaling repress the
activity of downstream transcription factors to achieve suppressed
osteoblastogenesis in MSCs. The expression levels of p-Smad1/5/8
and Runx2 were measured by western blot analysis after treatment of
cells as above. Compared with the control group, overexpressed
miR-155 group (BMP9 + miR-155) diminished the expression of
p-Smad1/5/8 and Runx2, however, downregulated miR-155 group (BMP9 +
anti-miR-155) eliminated this inhibitory effect in C2C12 cells
(Fig. 4A) and MEF cells (Fig. 5A). Based on these data, we
concluded that the negative effect of miR-155 on osteogenic
differentiation may be through suppressing canonical BMP (BMP/Smad)
signaling.
To further examine the role of miR-155 in late stage
of osteogenic differentiation, we tested the expression of OCN and
OPN by western blot analysis after treating C2C12 cells and MEF
cells indicated above for 7 days. The results showed that compared
with NC group (BMP9 + NC), exogenous overexpression of miR-155
(BMP9 + miR-155) decreased the expression of OCN and OPN, and
downregulated miR-155 group (BMP9 + anti-miR-155) eliminated this
inhibitory effect in C2C12 cells (Fig. 4B) and MEF cells (Fig. 5B). The above data demonstrated
that inhibition of miR-155 led to BMP signaling that coincided with
increased bone formation, strongly suggested that miR-155
suppressed BMP9-induced osteogenic differentiation by repressing
BMP signaling.
miR-155 directly targets Runx2 and BMPR2
in BMP signaling
To further study the mechanisms of miR-155 inhibited
osteogenic differentiation induced by BMP9, we used biological
information analysis to find the potential targets of miR-155 in
BMP signaling. From the target prediction tools (TargetScan and
PicTar), we found the target binding sites of miR-155 on the 3′-UTR
of Runx2 and BMPR2, which can complementary bind with miR-155 seed
sequence. In order to evaluate whether Runx2 and BMPR2 are direct
targets of miR-155, we measured the mRNA and protein expression
levels following the regulation of miR-155 expression by
transfecting the MSCs with NC, miR-155, or anti-miR-155. After
determining the transfection efficiency (Fig. 6A), we discovered that in
overexpressed-miR-155 C2C12 cells, the mRNA expression level of
Runx2 and BMPR2 was not significantly altered compared with NC. In
anti-miR-155 group, the expression levels had no significant
changes as miR-155 group compared with NC (Fig. 6B). While, in MEF cells, the
expression of Runx2 was increased in anti-miR-155 group compared
with NC group, the expression level was not significantly altered
in miR-155 group. The mRNA expression of BMPR2 had no markedly
significant differences in different groups (Fig. 6D). However, compared with NC, the
protein expression level of Runx2 and BMPR2 was decreased in
miR-155 group in C2C12 cells and MEF cells, respectively (Fig. 6C and E). These results
demonstrated that miR-155 significantly decreased the protein
expression level of Runx2 and BMPR2, but did not markedly altered
the mRNA expression levels, as compared with NC, respectively.
To test whether miR-155 directly binds to the 3′-UTR
of Runx2 and BMPR2, luciferase reporter assays were performed in
HEK-293 cells. We directly designed and synthesized sequences
matching miR-155 target sites and cloned these wild-type and
mutant-type into the multiple cloning sites of pMIR-REPORT
luciferase, termed WT-Runx2, MT-Runx2, WT-BMPR2 and MT-BMPR2
(Fig. 6F). Luciferase levels were
significantly reduced when cells were co-transfected with WT-Runx2
and miR-155 compared with the cells that co-transfected with
WT-Runx2 and NC. However, this difference was abolished when the
binding site was mutated suggesting a specific Runx2∷miR-155 direct
interaction (Fig. 6G). For BMPR2,
the result was the same as Runx2, when co-transfected with WT-BMPR2
and miR-155 into HEK-293 cells, luciferase activity was decreased
remarkable, mutated binding site could offset this depressant
effect demonstrating a specific BMPR2∷miR-155 direct interaction
(Fig. 6G). In conclusion, the
results suggested that Runx2 and BMPR2 are direct targets of
miR-155.
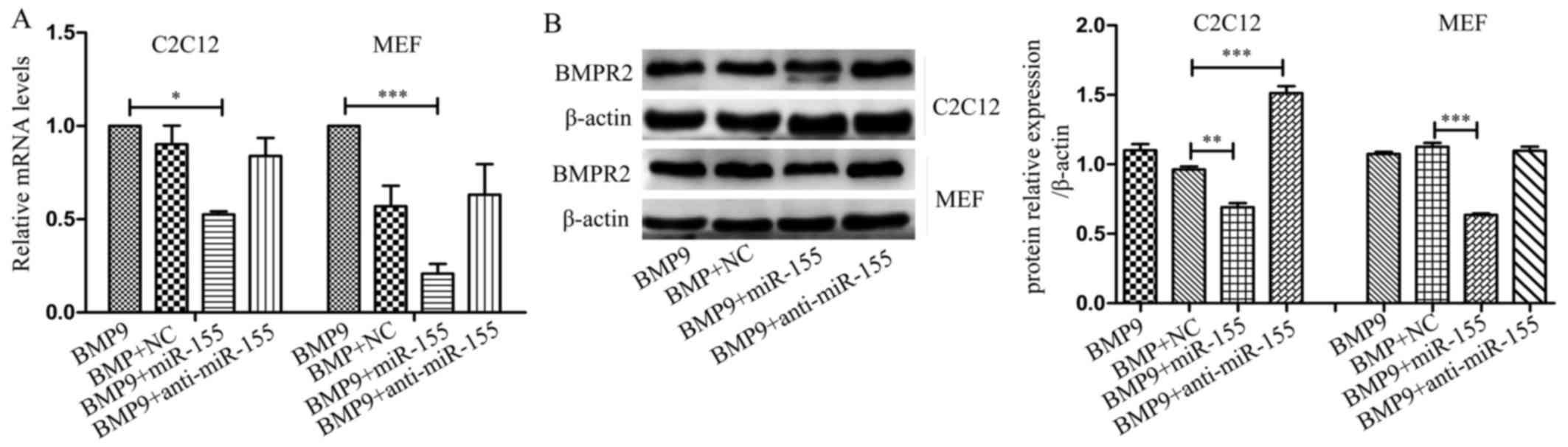
miR-155 decreases the the expression of
Runx2 and BMPR2 in the osteogenic differentiation induced by
BMP9
To validate the role of Runx2 and BMPR2 in miR-155
inhibited osteogenic differentiation of MSCs induced by BMP9, we
tested the expression changes of Runx2 and BMPR2 under the
influence of miR-155 in osteogenesis. In C2C12 cells and MEF cells,
in the process of differentiation, compared with NC, miR-155
repressed the expression of Runx2 at mRNA level (Fig. 3B and C) and protein level
(Figs. 4A and 5A), and anti-miR-155 reduced the
inhibitory effect of miR-155 on the expression of Runx2 and BMPR2.
In C2C12 and MEF cells, miR-155 had no obvious influence at the
expression of mRNA level, compared with NC during the
differentiation (Fig. 7A).
However, it reduced the protein expression level of BMPR2, and
anti-miR-155 promoted the protein level in C2C12 cells compared
with NC (Fig. 7B).
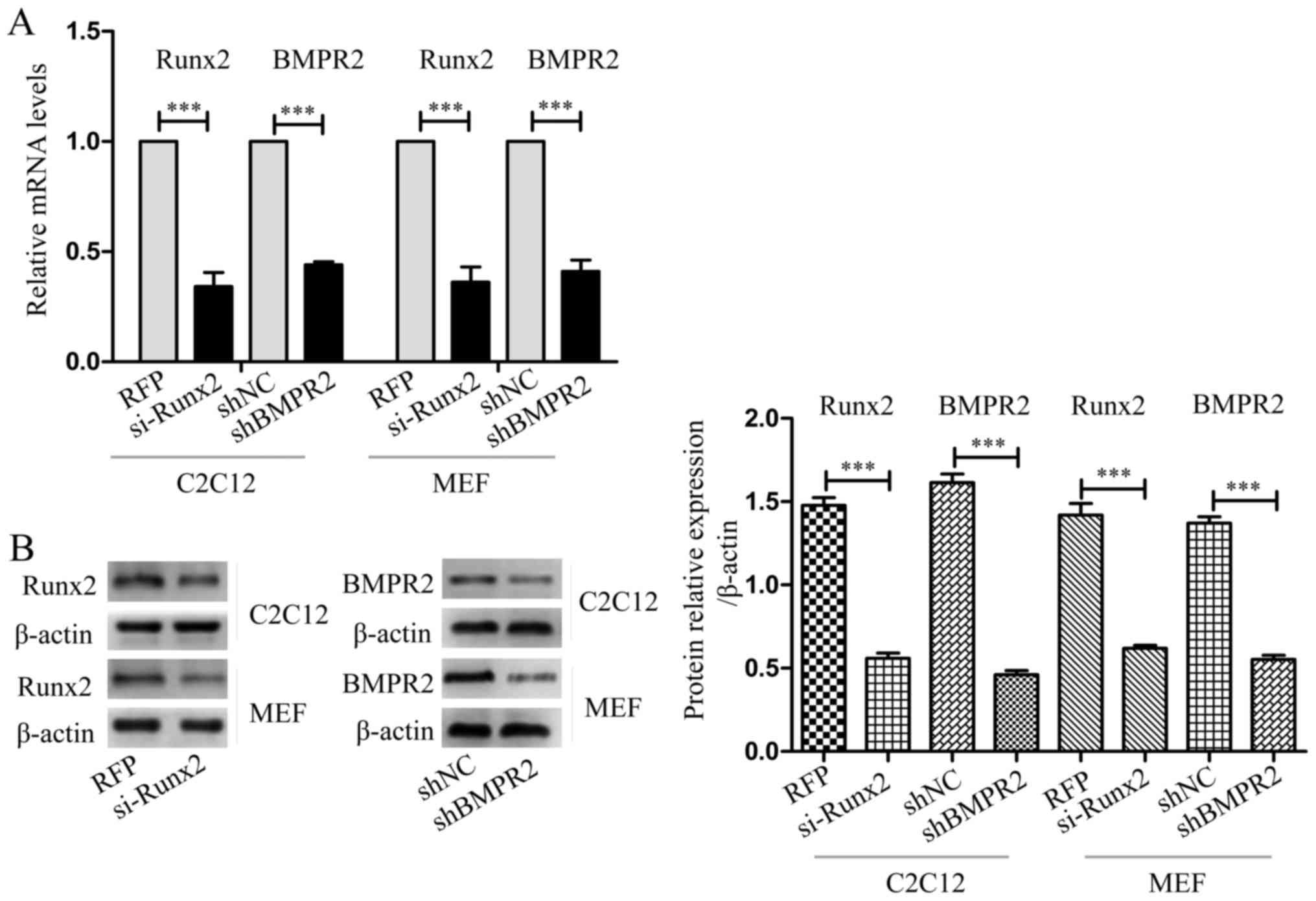
Knockdown of Runx2 and BMPR2 reduce the
inhibitory effect of miR-155 on the osteogenic differentiation
induced by BMP9
To further verify the association between miR-155
and its target genes, Runx2, and BMPR2, we blocked the expression
of Runx2 by si-Runx2, and blocked the expression of BMPR2 by
shBMPR2. Through blocking the target genes of miR-155, the effect
of miR-155 on osteogenesis of MSCs was evaluated, to speculate on
the function of the target genes of miR-155. After transfecting
si-Runx2, shBMPR2 or their controls into C2C12 cells and MEF cells,
respectively, used RT-qPCR to examine the interfering efficiency,
data showed that compared with the control groups, the expression
of Runx2 and BMPR2 was significantly decreased (Fig. 8A). The results of western blot
analysis shown that the protein expression of Runx2 and BMPR2 was
significantly reduced compared with their control group,
respectively (Fig. 8B).
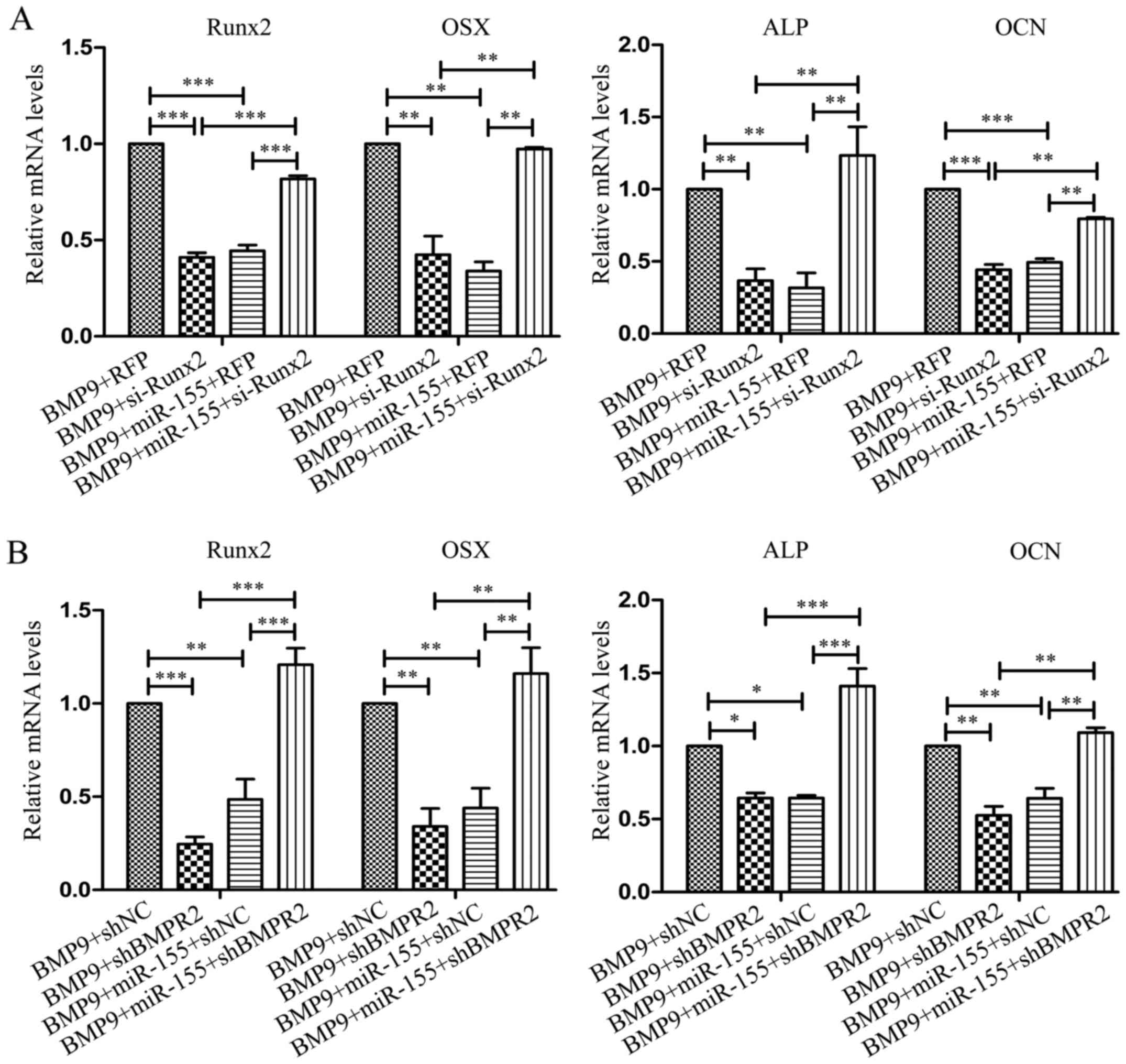
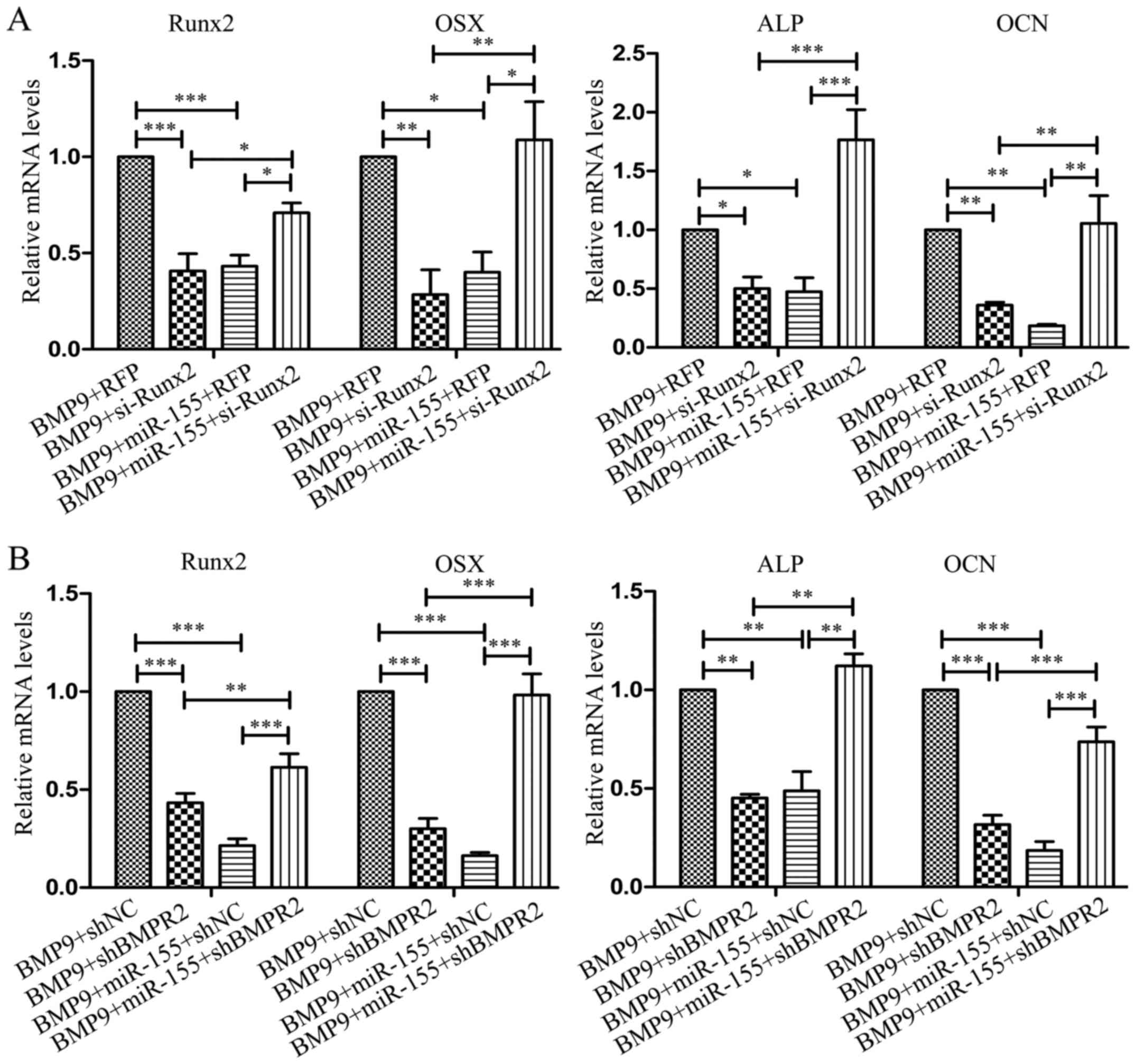
In C2C12 cells, the expression of Runx2, OSX, ALP
and OCN were distinctly decreased when transfected with si-Runx2
(BMP9 + si-Runx2) compared with RFP (BMP9 + RFP); and in
co-transfection with miR-155 and si-Runx2 group (BMP9 + miR-155 +
si-Runx2), the expression of these four bone-related genes were
significantly increased compared with co-transfection with miR-155
and RFP group (BMP9 + miR-155 + RFP) (Fig. 9A). In the process of osteogenesis,
after transfecting shBMPR2 (BMP9 + shBMPR2), the expression of
Runx2, OSX, ALP and OCN were markedly decreased compared with
control group (BMP9 + shNC); in co-transfected miR-155 and sh-BMPR2
group (BMP9 + miR-155 + sh-BMPR2), the expression of these four
bone differentiation related genes was notably increased compared
with co-transfected with miR-155 and shNC group (BMP9 + miR-155 +
shNC) (Fig. 9B). The results
obtained from MEF cells coincided with the results of C2C12 cells
(Fig. 10), when co-transfected
with miR-155 and si-Runx2 or sh-BMPR2, the expression of Runx2,
OSX, ALP and OCN was remarkably increased compared with
co-transfected with miR-155 and RFP or shNC, respectively. These
data indicated that knockdown of Runx2 and BMPR2, respectively, the
effect of miR-155 on the osteogenic differentiation induced by BMP9
was reduced. In conclusion, miR-155 inhibited osteogenic
differentiation induced by BMP9 may be through repression of the
expression of its target genes-Runx2 and BMPR2, which are, at least
partially, quite important components in BMP signaling.
Effect of miR-155 on in vivo ectopic bone
formation of BMP9
To determine the consequences of miR-155 on
osteogenic differentiation induced by BMP9 in vivo, we
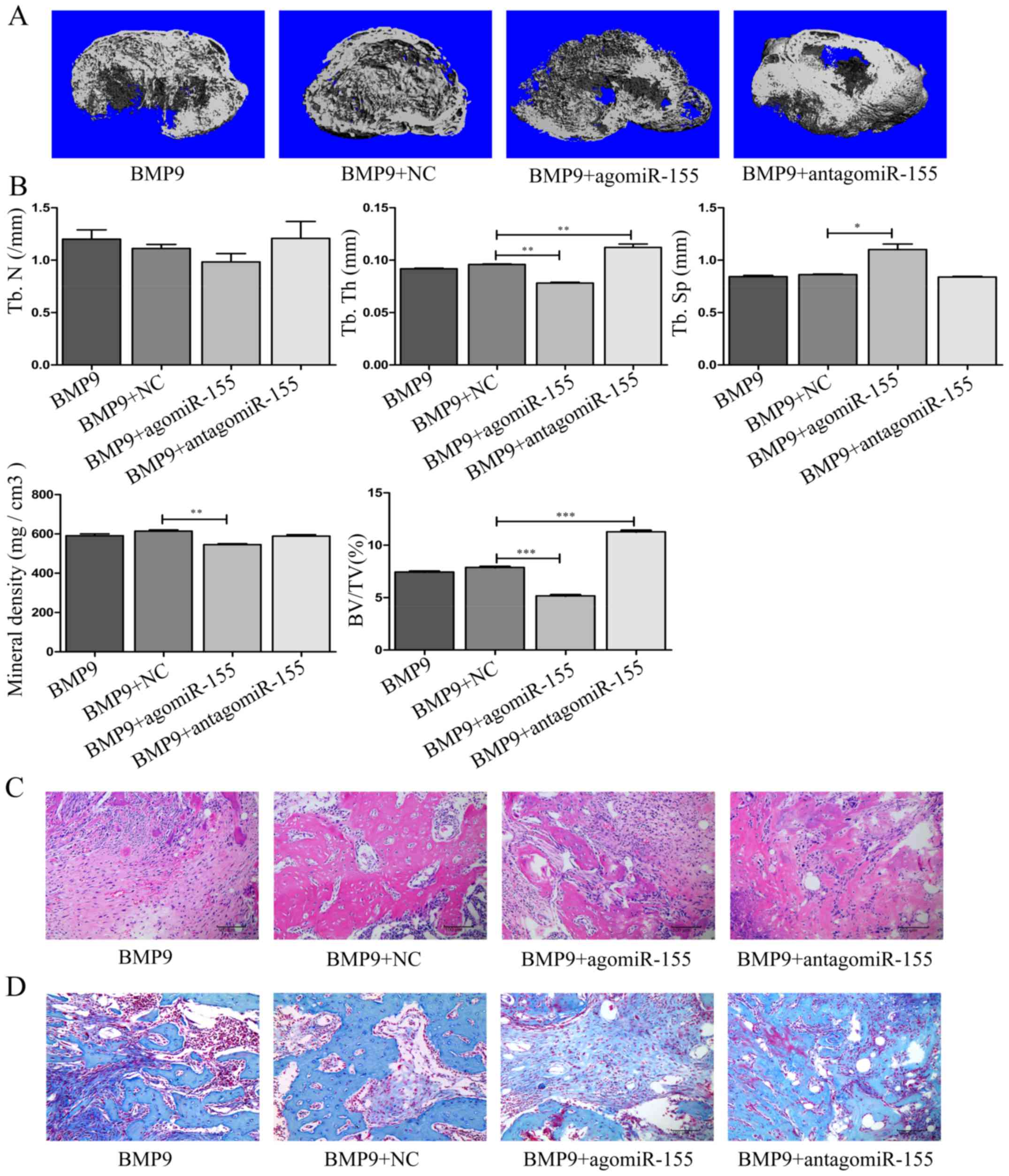
performed µCT analysis of heterotopic bones from 10-week-old
female nude mice. The microarchitecture of bones were in accordance
with data from cell culture experiments, µCT images of
ectopic bones showed decreased trabecular bone volume in BMP9 +
agomiR-155 group compared to BMP9 + NC group, while in BMP9 +
antagomiR-155 group, the trabecular bone volume was increased
compared with BMP9 + NC group (Fig.
11A). In µCT analyses, BMP9 + agomiR-155 group exhibited
significant decrease in BMD, BV/TV and Tb.Th, but a significant
increase in Tb.SP, there were no variances in Tb.N compared with
BMP9 + NC group, respectively. On the contrary, antago-miR-155
elevated BV/TV and Tb.Th of the bones consequently, and had no
effect on Tb.N, Tb.SP and BMD in the ectopic bone induction of BMP9
as well (Fig. 11B). After the
µCT scan, all specimens were subjected to a histological
study involving H&E staining and Masson's trichrome staining.
In histological analyses, the results correspond to the results
that we gained from cell culture in vitro. agomiR-155
inhibited the osteogenic differentiation of MEF cells induced by
BMP9 in vivo, decreased trabecular bone mass, whereas
antagomiR-155 reversed this phenomenon (Fig. 11C). The results of Masson's
trichrome staining were very similar to the H&E staining in all
the groups, showing that the chondrocytes were stained in BMP9 +
antagomir-155 group more, but in BMP9 + agomiR-155 group, the
staining was lighter compared with BMP9 + NC, respectively
(Fig. 11D). Given the above,
in vivo, agomiR-155 depressed the ectopic bone formation of
MEF cells induced by BMP9, antagomiR-155 facilitated ectopic bone
formation, conversely.
Discussion
Osteoblasts are a class of bone-forming cells that
originate from bone marrow stromal (skeletal or mesenchymal) stem
cells and are responsible for bone growth during development and
bone formation during remodeling of the post-natal skeleton.
Previous studies identified BMP9 as one of the most powerful
osteogenic BMPs, both in vitro and in vivo (2,4,9,29).
As one of the most osteogenic BMPs, BMP9 can induce MSCs to
differentiate into osteoblasts (46–48). But the specific mechanisms of BMP9
osteoinduction are not very clear, it is necessary to investigate
the complete regulatory mechanisms of BMP9-induced osteogenic
differentiation.
In this study, during the early stage of
BMP9-induced osteogenic differentiation of MSCs, the expression of
miR-155 presented early increased and later decreased trend,
indicated that there are some relationship between miR-155 and
BMP9. A one study indicated that miR-155 could modulate the
inhibitory effect of TNF-α on BMP-2-induced osteogenic
differentiation in MC3T3-E1 cells (43), suggested potential relationships
among them. Based on the staining experiments, we speculated
miR-155 may be a negative regulator in osteogenic differentiation.
The results of in vitro supported our hypothesis suggesting
that miR-155 inhibits the osteogenic differentiation of MSCs
induced by BMP9 may be via suppressing Smad/BMP signaling activity
and repressing the expression of Runx2 and BMPR2. Although, in some
cases, anti-miR-155 had a modest effect on the expression of
bone-related genes compared with NC, suggested that anti-miR-155
could reverse the inhibitory effect of miR-155 on the osteogenic
differentiation of MSCs induced by BMP9, sufficiently. In
vivo, agomiR-155 repressed the ectopic bone formation,
providing more believable evidence to prove our hypothesis.
Early studies revealed that canonical Smad-dependent
signaling pathway and non-canonical Smad-independent signaling (p38
MAPKs) pathway are involved in BMP9-induced osteogenic
differentiation of MSCs. MAPKs are a group of well-described
serine/threonine-specific protein kinases generally expressed in
all cell types (9,29,48). At least four subfamilies of
mammalian MAPKs have been identified, including ERK1/2, ERK5 (also
known as MAPK7 or BMK1), the Jun amino-terminal kinases (JNKs) and
p38 MAPKs. ERK1/2, JNKs, and p38 are involved in BMP9-induced
osteogenic differentiation of MSCs (44,45,49,50). Whether miR-155 influence the
non-canonical Smad-independent signaling (p38 mitogen-activated
protein kinase, MAPK) pathway has not been investigated, to study
the mechanisms further, we can focus on this aspect. As Runx2 and
BMPR2 were verified as the target genes of miR-155 in our study,
they were interfered by siRNA, or shRNA to block the targets of
miR-155. The results showed that miR-155 failed to inhibit
osteogenic differentiation, suggesting miR-155 may play negative
regulatory roles in BMP9-induced osteogenic differentiation through
repressed expression of Runx2 and BMPR2. More regulatory factors
and the corresponding mechanisms in the process of BMP9-induced
osteogenic differentiation need to be explored. Finding more target
genes of miR-155 in BMP signaling is beneficial to understand the
mechanism of action of miR-155, promote the clinic application of
BMP9 finally.
Acknowledgments
The authors would like to thank T.C. He (Medical
Center, the University of Chicago) for his kind provision of the
recombinant adenovirus (BMP9 and si-Runx2).
Notes
[1]
Funding
This study was supported by the National Natural
Science Foundation of China (NSFC 81672103), the National Natural
Science Foundation of China (NSFC 31200971), the National Ministry
of Education Foundation of China (20115503110009) and the Program
of the Ministry of Science and Technology of Yu-zhong District, CQ
(20130136).
[2] Availability
of data and material
All data generated or analyzed during this study are
included in this published article.
[3] Authors'
contributions
HL designed the experiments, controlled the progress
of the research, and was a major contributor in writing the
manuscript. LZ, TY, YZ, YG, MF, YL, YS, WL and SC critically
revised the manuscript for important intellectual content. LA was
involved in animal experiments of the nude mice preliminary
treatment. YW and QS analyzed the data and revised the manuscript.
All authors read and approved the final manuscript.
[4] Ethics
approval and consent to participate
All animal experiments were approved by the
Institutional Animal Care and Use Committee (IACUC) of Chongqing
Medical University (reference no. 2016009).
[5] Consent for
publication
Not applicable.
[6] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Luu HH, Song WX, Luo X, Manning D, Luo J,
Deng ZL, Sharff KA, Montag AG, Haydon RC and He TC: Distinct roles
of bone morphogenetic proteins in osteogenic differentiation of
mesenchymal stem cells. J Orthop Res. 25:665–677. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Beederman M, Lamplot JD, Nan G, Wang J,
Liu X, Yin L, Li R, Shui W, Zhang H, Kim SH, et al: BMP signaling
in mesenchymal stem cell differentiation and bone formation. J
Biomed Sci Eng. 6:32–52. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bidart M, Ricard N, Levet S, Samson M,
Mallet C, David L, Subileau M, Tillet E, Feige JJ and Bailly S:
BMP9 is produced by hepatocytes and circulates mainly in an active
mature form complexed to its prodomain. Cell Mol Life Sci.
69:313–324. 2012. View Article : Google Scholar
|
|
4
|
Brown MA, Zhao Q, Baker KA, Naik C, Chen
C, Pukac L, Singh M, Tsareva T, Parice Y, Mahoney A, et al: Crystal
structure of BMP-9 and functional interactions with pro-region and
receptors. J Biol Chem. 280:25111–25118. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang RN, Green J, Wang Z, Deng Y, Qiao M,
Peabody M, Zhang Q, Ye J, Yan Z, Denduluri S, et al: Bone
Morphogenetic Protein (BMP) signaling in development and human
diseases. Genes Dis. 1:87–105. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yan F, Luo S, Jiao Y, Deng Y, Du X, Huang
R, Wang Q and Chen W: Molecular characterization of the BMP7 gene
and its potential role in shell formation in Pinctada martensii.
Int J Mol Sci. 15:21215–21228. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Miyazono K, Kamiya Y and Morikawa M: Bone
morphogenetic protein receptors and signal transduction. J Biochem.
147:35–51. 2010. View Article : Google Scholar
|
|
8
|
Rahman MS, Akhtar N, Jamil HM, Banik RS
and Asaduzzaman SM: TGF-β/BMP signaling and other molecular events:
Regulation of osteoblastogenesis and bone formation. Bone Res.
3:150052015. View Article : Google Scholar
|
|
9
|
Chen L, Jiang W, Huang J, He BC, Zuo GW,
Zhang W, Luo Q, Shi Q, Zhang BQ, Wagner ER, et al: Insulin-like
growth factor 2 (IGF-2) potentiates BMP-9-induced osteogenic
differentiation and bone formation. J Bone Miner Res. 25:2447–2459.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kang Q, Song WX, Luo Q, Tang N, Luo J, Luo
X, Chen J, Bi Y, He BC, Park JK, et al: A comprehensive analysis of
the dual roles of BMPs in regulating adipogenic and osteogenic
differentiation of mesenchymal progenitor cells. Stem Cells Dev.
18:545–559. 2009. View Article : Google Scholar
|
|
11
|
Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D,
Ji Y, Zhao C, Wang J, Yang BB, et al: MiRNA-directed regulation of
VEGF and other angiogenic factors under hypoxia. PLoS One.
1:e1162006. View Article : Google Scholar
|
|
12
|
Wang X, Guo B, Li Q, Peng J, Yang Z, Wang
A, Li D, Hou Z, Lv K, Kan G, et al: miR-214 targets ATF4 to inhibit
bone formation. Nat Med. 19:93–100. 2013. View Article : Google Scholar
|
|
13
|
Sevli S, Uzumcu A, Solak M, Ittmann M and
Ozen M: The function of microRNAs, small but potent molecules, in
human prostate cancer. Prostate Cancer Prostatic Dis. 13:208–217.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Alshalalfa M: MicroRNA response
elements-mediated miRNA-miRNA interactions in prostate cancer. Adv
Bioinforma. 2012:8398372012. View Article : Google Scholar
|
|
15
|
Eskildsen T, Taipaleenmäki H, Stenvang J,
Abdallah BM, Ditzel N, Nossent AY, Bak M, Kauppinen S and Kassem M:
MicroRNA-138 regulates osteogenic differentiation of human stromal
(mesenchymal) stem cells in vivo. Proc Natl Acad Sci USA.
108:6139–6144. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Itoh T, Nozawa Y and Akao Y: MicroRNA-141
and -200a are involved in bone morphogenetic protein-2-induced
mouse pre-osteoblast differentiation by targeting distal-less
homeobox 5. J Biol Chem. 284:19272–19279. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kureel J, Dixit M, Tyagi AM, Mansoori MN,
Srivastava K, Raghuvanshi A, Maurya R, Trivedi R, Goel A and Singh
D: miR-542-3p suppresses osteoblast cell proliferation and
differentiation, targets BMP-7 signaling and inhibits bone
formation. Cell Death Dis. 5:e10502014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Luo W, Nie Q and Zhang X: MicroRNAs
involved in skeletal muscle differentiation. J Genet Genomics.
40:107–116. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qi R, Long D, Wang J, Wang Q, Huang X, Cao
C, Gao G and Huang J: MicroRNA-199a targets the fatty acid
transport protein 1 gene and inhibits the adipogenic
trans-differentiation of C2C12 myoblasts. Cell Physiol Biochem.
39:1087–1097. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu JY, Chung KH, Deo M, Thompson RC and
Turner DL: MicroRNA miR-124 regulates neurite outgrowth during
neuronal differentiation. Exp Cell Res. 314:2618–2633. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang J, Du YY, Lin YF, Chen YT, Yang L,
Wang HJ and Ma D: The cell growth suppressor, mir-126, targets
IRS-1. Biochem Biophys Res Commun. 377:136–140. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu B, Tao T, Wang Y, Fang F, Huang Y, Chen
S, Zhu W and Chen M: hsa-miR-135a-1 inhibits prostate cancer cell
growth and migration by targeting EGFR. Tumour Biol.
37:14141–14151. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shin S, Moon KC, Park KU and Ha E:
MicroRNA-513a-5p mediates TNF-α and LPS induced apoptosis via
downregulation of X-linked inhibitor of apoptotic protein in
endothelial cells. Biochimie. 94:1431–1436. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Esau C, Kang X, Peralta E, Hanson E,
Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, et
al: MicroRNA-143 regulates adipocyte differentiation. J Biol Chem.
279:52361–52365. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang J, Zhao L, Xing L and Chen D:
MicroRNA-204 regulates Runx2 protein expression and mesenchymal
progenitor cell differentiation. Stem Cells. 28:357–364. 2010.
|
|
26
|
Li Z, Hassan MQ, Volinia S, van Wijnen AJ,
Stein JL, Croce CM, Lian JB and Stein GS: A microRNA signature for
a BMP2-induced osteoblast lineage commitment program. Proc Natl
Acad Sci USA. 105:13906–13911. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mizuno Y, Yagi K, Tokuzawa Y,
Kanesaki-Yatsuka Y, Suda T, Katagiri T, Fukuda T, Maruyama M, Okuda
A, Amemiya T, et al: miR-125b inhibits osteoblastic differentiation
by downregulation of cell proliferation. Biochem Biophys Res
Commun. 368:267–272. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sato MM, Nashimoto M, Katagiri T, Yawaka Y
and Tamura M: Bone morphogenetic protein-2 downregulates miR-206
expression by blocking its maturation process. Biochem Biophys Res
Commun. 383:125–129. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang W, Zhang L, Zhou Y, Ji X, Liu J, Liu
D, Yin P, Peng Y, Hao M, Zhang L, et al: Synergistic effects of
BMP9 and miR-548d-5p on promoting osteogenic differentiation of
mesenchymal stem cells. BioMed Res Int. 2015:3097472015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Y, Xie RL, Croce CM, Stein JL, Lian
JB, van Wijnen AJ and Stein GS: A program of microRNAs controls
osteogenic lineage progression by targeting transcription factor
Runx2. Proc Natl Acad Sci USA. 108:9863–9868. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Z, Hassan MQ, Jafferji M, Aqeilan RI,
Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS and Lian JB:
Biological functions of miR-29b contribute to positive regulation
of osteoblast differentiation. J Biol Chem. 284:15676–15684. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vimalraj S, Partridge NC and Selvamurugan
N: A positive role of microRNA-15b on regulation of osteoblast
differentiation. J Cell Physiol. 229:1236–1244. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Elton TS, Selemon H, Elton SM and
Parinandi NL: Regulation of the MIR155 host gene in physiological
and pathological processes. Gene. 532:1–12. 2013. View Article : Google Scholar
|
|
34
|
Kluiver J, Poppema S, de Jong D, Blokzijl
T, Harms G, Jacobs S, Kroesen BJ and van den Berg A: BIC and
miR-155 are highly expressed in Hodgkin, primary mediastinal and
diffuse large B cell lymphomas. J Pathol. 207:243–249. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lawrie CH: MicroRNAs and lymphomagenesis:
A functional review. Br J Haematol. 160:571–581. 2013. View Article : Google Scholar
|
|
36
|
Wang M, Tan LP, Dijkstra MK, van Lom K,
Robertus JL, Harms G, Blokzijl T, Kooistra K, van T'veer MB, Rosati
S, et al: miRNA analysis in B-cell chronic lymphocytic leukaemia:
Proliferation centres characterized by low miR-150 and high
BIC/miR-155 expression. J Pathol. 215:13–20. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Faraoni I, Antonetti FR, Cardone J and
Bonmassar E: miR-155 gene: A typical multifunctional microRNA.
Biochim Biophys Acta. 1792:497–505. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu
H, Liu MF and Wang ED: MicroRNA-155 functions as an OncomiR in
breast cancer by targeting the suppressor of cytokine signaling 1
gene. Cancer Res. 70:3119–3127. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu J, Mao Q, Liu Y, Hao X, Zhang S and
Zhang J: Analysis of miR-205 and miR-155 expression in the blood of
breast cancer patients. Chin J Cancer Res. 25:46–54.
2013.PubMed/NCBI
|
|
40
|
Vigorito E, Perks KL, Abreu-Goodger C,
Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A,
Bradley A, et al: microRNA-155 regulates the generation of
immunoglobulin class-switched plasma cells. Immunity. 27:847–859.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen C, Tang Z, Song Q, Yang M, Shi Q and
Weng Y: Downregulated microRNA-23b promotes BMP9-mediated
osteogenesis in C2C12 myoblast cells by targeting Runx2. Mol Med
Rep. 13:2492–2498. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu T, Xie M, Wang X, Jiang X, Li J and
Huang H: miR-155 modulates TNF-α-inhibited osteogenic
differentiation by targeting SOCS1 expression. Bone. 51:498–505.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhao Y, Song T, Wang W, Wang J, He J, Wu
N, Tang M, He B and Luo J: P38 and ERK1/2 MAPKs act in opposition
to regulate BMP9-induced osteogenic differentiation of mesenchymal
progenitor cells. PLoS One. 7:e433832012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Miyazono K, Maeda S and Imamura T: BMP
receptor signaling: Transcriptional targets, regulation of signals,
and signaling cross-talk. Cytokine Growth Factor Rev. 16:251–263.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen L, Zou X, Zhang R-X, Pi CJ, Wu N, Yin
LJ and Deng ZL: IGF1 potentiates BMP9-induced osteogenic
differentiation in mesenchymal stem cells through the enhancement
of BMP/Smad signaling. BMB Rep. 49:122–127. 2016. View Article : Google Scholar :
|
|
47
|
Li R, Yan Z, Ye J, Huang H, Wang Z, Wei Q,
Wang J, Zhao L, Lu S, Wang X, et al: The prodomain-containing BMP9
produced from a stable line effectively regulates the
differentiation of mesenchymal stem cells. Int J Med Sci. 13:8–18.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Peng Y, Kang Q, Cheng H, Li X, Sun MH,
Jiang W, Luu HH, Park JY, Haydon RC and He TC: Transcriptional
characterization of bone morphogenetic proteins (BMPs)-mediated
osteogenic signaling. J Cell Biochem. 90:1149–1165. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xu DJ, Zhao YZ, Wang J, He JW, Weng YG and
Luo JY: Smads, p38 and ERK1/2 are involved in BMP9-induced
osteogenic differentiation of C3H10T1/2 mesenchymal stem cells. BMB
Rep. 45:247–252. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhao YF, Xu J, Wang WJ, Wang J, He JW, Li
L, Dong Q, Xiao Y, Duan XL, Yang X, et al: Activation of JNKs is
essential for BMP9-induced osteogenic differentiation of
mesenchymal stem cells. BMB Rep. 46:422–427. 2013. View Article : Google Scholar : PubMed/NCBI
|