Introduction
According to a previous study, ovarian cancer still
remains the fifth-leading cause of mortality in women, with
approximately 22,280 newly diagnosed cases and 14,240 mortalities
reported in 2016 worldwide (1).
With such a high recurrence rate, over three quarters of ovarian
cancer patients eventually relapse following primary platinum and
taxane-based chemotherapy (2).
Development of cytotoxic chemotherapy and novel targeted therapies
have enhanced progression-free survival, however failed to have
significant influence on overall survival (3,4).
Malignant tumours, including ovarian cancer, are characterized by
tumour metastasis from the primary site to other parts via the
lymphatic system, blood vessels, or body cavity (5). To better understand the underlying
molecular mechanism of carcinogenesis in order to develop more
effective therapeutic strategies, more research studies need to be
conducted.
Previously, the use of miRNAs as a novel effective
target in cancer therapy has been extensively reported (6-10).
miRNAs are small non-coding RNA molecules with a length varying
from 19-25 nt. Previous studies have demonstrated that miRNAs that
are generated in cells serve important roles in a variety of
biological processes; in particular, they negatively regulate gene
expression following transcription. A mature miRNA chain is able to
combine with the 3′ untranslated region (UTR) of target mRNA in an
RNA-induced silencing complex; in this way, complete complementary
miRNA and target mRNA result in inhibition or activation of
translation of the mRNA (11-13). miRNAs take part in various
physiological processes, including cell proliferation,
differentiation, and apoptosis, by regulating transcription of
their target genes. In neoplastic processes, aberrantly expressed
miRNAs affect proliferation, migration, and invasion of cancer
cells by mediating associated proteins or transcription factors so
as to exert promotive or inhibitory effects on tumorigenesis and
metastasis (14-16). Several lines of evidence have
verified that miRNAs may directly or indirectly control oncogenes
or suppressor genes to mediate protein expression of
cancer-associated pathways (17,18). Based on previous studies, miR-377,
a member of the large miRNA cluster on chromosome 14q32, is weakly
expressed in several human malignancies, including ependymoma,
osteosarcoma, neuroblastoma, gastro-intestinal stromal tumours, and
gliomas (6-10). To the best of the author's
knowledge, there is a lack of information about the association
between miR-377 and ovarian cancer; thus, the present study
investigated whether miR-377 serves an important role in the
control of proliferation and metastasis in ovarian cancer, and in
particular, if it exhibits inhibitory capabilities.
Cullin (CUL) 4A, which was demonstrated to be one of
the target genes of miR-377 in the present study, belongs to the
family of Cullin-Ring E3-ligases (CRLs) and serves a key role in
tumorigenesis and tumor progression (19). CUL4A constitutes a helical
N-terminal domain and a spherical C-terminal domain which combine
with Ras like without CAAX 1 (RIT1) and damage specific DNA binding
protein 1, respectively (20).
CUL4A, initially reported in breast cancer, was previously
demonstrated overexpressed in numerous cancers, including
squamous-cell carcinoma, adrenocortical carcinoma, medulloblastoma
and liver cancer (21).
Overexpression of CUL4A leads to promotion of cancer cell
proliferation, and is closely and positively associated with
overall survival and disease-free survival of cancer patients
(22). The present study focused
on the role of miR-377 in the control of CUL4A expression in
ovarian cancer cell lines.
Materials and methods
Tumor samples
A total of 44 ovarian cancer patients in the Third
Affiliated Hospital of Wenzhou Medical University were included in
the present study. During the period from February 2015 to March
2016, all cancer specimens were obtained from surgical tumor
resections, and their adjacent normal ovarian tissue specimens were
collected at the same time, as negative controls. The normal and
cancer tissues represented matched pairs from each patient. Basic
clinical and pathological data of these patients at the average age
of 51.12±16.35 were collected with their written informed consent
forms. The present study was approved by the ethics committee of
the Third Affiliated Hospital of Wenzhou Medical University
(Wenzhou, China).
Cell culture and grouping
Normal ovary cell line (IOSE80) and seven ovarian
cancer cells lines including CAOV3, SKOV3, A2780, OVCAR3, HO-8901,
3AO and TC-1 were all obtained from Shanghai Bogoo Biotechnology
Co. Ltd. (Shanghai, China). Cells were cultured in Dulbecco's
modified Eagle's medium (DMEM) supplemented with 10% fetal calf
serum (Thermo Fisher Scientific Inc, Waltham, MA, USA) at 37°C in
5% CO2 incubator. Medium was replaced once every two
days. Cell passage cultivation was performed when cells grew to 90%
confluency.
One cell line from six ovarian cancer cell lines was
selected based on the expression of the miR-377 and CUL4A levels
for the primary in vitro experiments of the effects of
miR-377 on CUL4A in ovarian cancer in the present study. Cells were
randomly allocated into three groups: Control group, mock group
(cells transfected with blank plasmids) and mimics (cells
transfected with miR-377 mimics). Recombinant plasmids were
purchased by Nanjing Cobioer Biotechnology Co. Ltd. (Jiangsu,
China). A total of 500 ng/µl plasmids were prepared.
Transfection processes were carried out using
Lipofectamine® 2000 reagent (Thermo Fisher Scientific
Inc.) according to the manufacturer's protocol. Prior to subsequent
experimentation, transfected cells were incubated at 37°C in a 5%
CO2 incubator for 48 h.
Bioinformatics analysis
To analyze the role of miRNA-377 in ovarian cancer,
prediction of its target genes was carried out using the prediction
software miRanda (www.microrna.org/microrna/home.do), miRDB (www.mirdb.org/), PicTar (pictar.mdc-berlin.de/) and
TargetScan (www.targetscan.org/vert_71/) and the function of
miRNA-377 was analyzed through the Database for Annotation,
Visualization and Integrated Discovery (www.david.ncifcrf.gov/).
Luciferase reporter assay
CUL4A 3′-UTR and mutated CUL4A 3′-UTR were prepared
by GeneCopoeia Inc. (Rockville, MD, USA). The 3′-UTR fragment of
CUL4A with the binding site for miR-377 mimic and inhibitor was
amplified via polymerase chain reaction (PCR) with the primer
sequences presented in Table I
and cloned into luciferase vectors (Promega Corporation, Madison,
WI, USA). SKOV3 cells were seeded into 96-well plates at the
density of 1×105 cells per well one day prior to
transfection. Control luciferase reporter plasmid, CUL4A 3′-UTR and
mutated CUL4A 3′-UTR were co-transfected with either miR-377 mimic
or miR-377 inhibitor using Lipofectamine® 3000 (Thermo
Fisher Scientific Inc., USA). A total of 48 h following
transfection, luciferase activity was determined with Secret-Pair™
Dual-Luciferase Reporter Assay (GeneCopoeia Inc.). The results were
normalized through comparison with Renilla luciferase
activity.
 | Table ISequence of miR-377 mimics and
inhibitor. |
Table I
Sequence of miR-377 mimics and
inhibitor.
| Name | Direction | Sequence
(5′-3′) |
|---|
| miR-377 mimics | Forward |
TGCTGATCACACAAAGGCAACTTTTGTGTTTTGGCCACTGACT |
|
GACACAAAAGTCCTTTGTGTGAT |
| Reverse CC |
TGTAGTGTGTTTCCGTTGAAAACACAAAACCGGTG |
|
ACTGACTGTGTTTTCAGGAAACACACTA |
| miR-377
inhibitor | Forward |
TGCTGGGAAGTCATACAATCCTACA |
|
TTGTTTTGGCCACTGACTGACAATGTAGGTGTATGACTTCC |
| Reverse CC |
TGCCTTCAGTATGTTAGGATGTAACAA |
|
AACCGGTGACTGACTGTTACATCCACATACTGAAGG |
| miR-377 UTR | |
AUCACACAAAGGCAACUUUUGU |
| CUL4A UTR | |
UGGUUUGUU-CUCGUGUGUGAU |
Cell Counting Kit (CCK)-8 assay
Cell viability in control, mock, and mimic groups
was measured using the CCK-8 assay (Beyotime Institute of
Biotechnology, Shanghai, China). Cells were seeded into a 96-well
plate (100 µl/well), and cultured at 37°C in a 5%
CO2 incubator. A total of four hours following
incubation, 10 µl CCK-8 reagent was added to each well and
incubated for 1-4 h. Optical density values were read by a iMark
microplate absorbance reader (Bio-Rad Laboratories, Inc., Hercules,
CA, USA) at a wavelength of 450 nm.
Transwell assay
SKOV3 cells (5×105 cells/ml) were seeded
in the upper well of a Transwell migration system on ThinCerts™
inserts with 8-µm membranes (Huayue ReaCon Inc., Guangdong,
China) in DMEM supplemented with 0.1% fetal bovine serum (FBS). The
lower wells received the same medium supplemented with 10% FBS.
Following a 24-h incubation period, the contents of the upper
wells, which contained non-migrating cells were removed by cotton
swabs. For invasion detection, the upper chambers were specifically
coated with Matrigel. Cells that migrated or invaded through the
membranes were fixated with 70% cold ethanol, stained with 0.1%
crystal violet for 20 min at room temperature, and imaged using a
light microscope (Olympus Corporation, Tokyo, Japan). The quantity
of migrated cells was analysed from five randomly selected fields
under the light microscope at a magnification of ×100.
Reverse transcription-quantitative (RT-q)
PCR
Quantification of miR-377 was carried out using
TaqMan miRNAs assay (Thermo Fisher Scientific Inc). The PCR
conditions included activating the DNA polymerase at 95°C for 15
min, followed by 40 cycles of three-step PCR (94°C for 15 sec, 55°C
for 30 sec, and 70°C for 30 sec). Reverse transcription of 10 ng
template RNA was performed with a TaqMan MicroRNA Reverse
Transcription kit and miRNA-specific stem-loop primers (Table I). The expression of miR-377 was
normalized to U6 expression. The 2−ΔΔCq method was
performed for the quantification of gene expression data (23).
To determine mRNA levels of CUL4A, β-catenin, Wnt3a,
cyclin D1, matrix metalloproteinases (MMP)-2 and -9,
metastasis-associated protein (MTA)-1, and metallopeptidase
inhibitor (TIMP), total RNA was firstly reverse transcribed using
the Takara PrimeScript RT reagent kit (Takara Bio, Inc., Otsu,
Japan). Quantification of mRNA was carried out by TaqMan Gene
Expression Assay (Thermo Fisher Scientific Inc, USA). PCR was
carried out by activating the DNA polymerase at 95°C for 10 min,
followed by 40 cycles of two-step PCR (95°C for 15 sec and 60°C for
45 sec). Target gene expression was normalized to GAPDH expression.
Primer sequences were listed as follows: CUL4A forward,
5′CAAGAACTTCCGAGACAGACC3′ and reverse, 5′TGCTTGTAGAGCATTGGGGA3′;
β-catenin forward, 5′TATAAGAGCTCCTTGTGCGGC3′ and reverse,
5′CTGAAGCTGCTCCTCAGACC3′; Wnt3a forward, 5′CTGGAGCTAGTGTCTCCTCTCT3′
and reverse, 5′GGAAGAAGCCTCATCCACCA3′; cyclin D1 forward,
5′CAATGACCCCGCACGATTTC3′ and reverse, 5′AAGTTGTTGGGGCTCCTCAG3′;
MMP-2 forward, 5′TGTGTTGTCCAGAGGCAATG3′ and reverse,
5′ATCACTAGGCCAGCTGGTTG3′; MMP-9 forward, 5′TTTGAGTCCGGTGGACGATG3′
and reverse, 5′GCTCCTCAAAGACCGAGTCC3′; MTA1 forward,
5′AAACTGCCCTGAGTGTGGT3′ and reverse, 5′AAATATGTTGACCCAGCTCATCT3′;
TIMP1 forward, 5′GCCTGACGGTCATATGGTAGA3′ and reverse,
5′GAATGCGCCAAAAACCCCAT3′ and GAPDH forward,
5′GAATGGGCAGCCGTTAGGAA3′ and reverse, 5′AAAAGCATCACCCGGAGGAG3′. The
2−ΔΔCq method was performed for the quantification of
gene expression data (23).
Western blot analysis
Total protein of cells in each group was extracted
using the ProteoPrep® Total Extraction Sample kit
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Cell lysates were
prepared with the cell lysis buffer (Beyotime Institute of
Biotechnology). Following centrifugation (12,000 × g) at 4°C for 10
min, the supernatant was collected. Concentration of the protein
was determined by Bradford assay (Bio-Rad Laboratories, Inc.). A
total of 30 mg of each protein sample were separated on 10-15%
SDS-PAGE (Merck KGaA, Darmstadt, Germany). The membranes were
blocked using 5% fat-free milk in TBST at room temperature for 1 h.
Membranes were incubated with primary antibodies at 4°C for 6 h and
then at room temperature for 4 h. The primary specific antibodies
used were anti-CUL4A (catalog no. ab72548; 1:1,000; Abcam,
Cambridge, MA, USA), anti-β-catenin (catalog no. ab32572; 1:5,000;
Abcam), anti-Wnt3a (catalog no. ab19925; 1:1,000; Abcam),
anti-cyclin D1 (catalog no. ab134175; 1:10,000; Abcam), anti-MMP2
(catalog no. ab92536; 1:1,000; Abcam), anti-MMP9 (catalog no.
ab38898; 1:1,000; Abcam), anti-MTA1 (catalog no. ab71153; 1:2,000;
Abcam) and anti-GAPDH (catalog no. ab8245, 1:2,000; Abcam). The
membranes were incubated with the following secondary antibodies at
room temperature for 1 h: Goat anti-mouse IgG H&L (catalog no.
ab6789; 1:2,000; Abcam), goat anti-rabbit IgG H&L (catalog no.
ab6721; 1:2,000; Abcam) and donkey anti-goat IgG H&L (catalog
no. ab6885; 1:2,000; Abcam). Blots were visualized using enhanced
chemiluminescence (Thermo Fisher Scientific, Inc.). Densitometry of
the bands was performed using Quantity One software, version 4.6.9
(Bio-Rad Laboratories, Inc.).
Statistical analysis
Statistical analysis was performed using SPSS
software, version 22.0 (IBM Corp., Armonk, NY, USA). Each
experiment was repeated three times, and data are presented as the
mean ± standard deviation. Student's t-test and one-way analysis of
variance, followed by Tukey and Bonferroni post-hoc tests were used
in either two or multiple group comparisons, for statistical
significance. Spearman's nonparametric correlation test was used to
analyze the correlations between the miR-377 and CUL4A mRNA levels
in tissue samples. The Chi squared test was used for analysis of
the significance of CUL4A to clinicopathological characteristics.
The method of survival analysis was used for testing divided
phases. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression levels of miR-377 and CUL4A in
ovarian cancer tissues and cell lines
The present study included a total of 44 patients at
the average age of 51.12±16.35 (Table II). The expression of miR-377 and
CUL4A in surgically resected ovarian cancer specimens and adjacent
normal ovarian tissues was assessed using RT-qPCR. Compared with
the normal ovarian tissues, the levels of miR-377 in ovarian cancer
specimens was decreased, and the expression of CUL4A was increased,
and the correlation analysis indicated a negative correlation
between miR-377 and CUL4A expression (Fig. 1A–C). It was also detected that the
survival rate of ovarian cancer patients with decreased levels of
CUL4A and increased miR-377 was greater compared with those that
demonstrated the opposing trends (P<0.05; Fig. 1D and E).
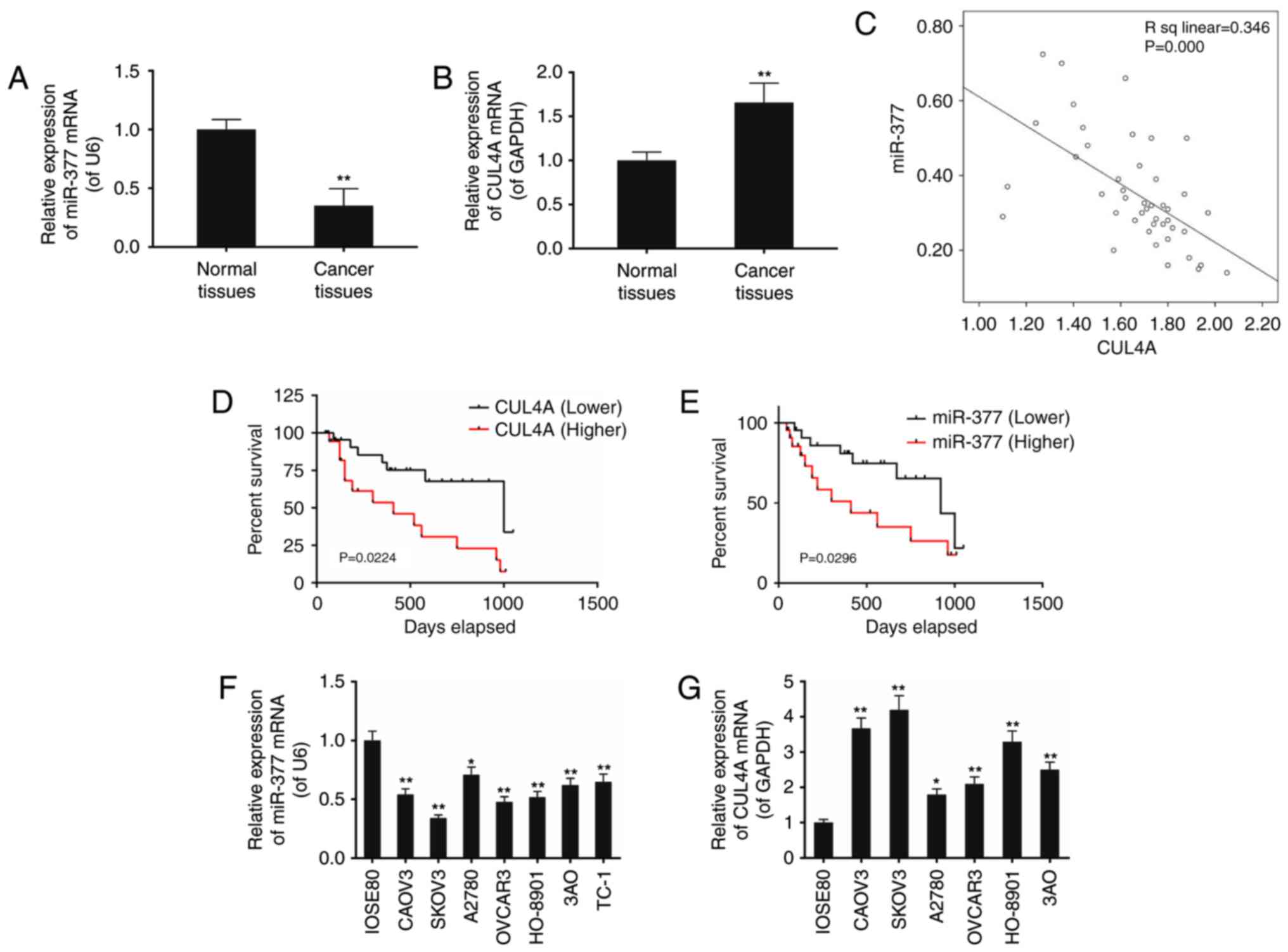 | Figure 1The expression levels of miR-377 and
CUL4A in ovarian cancer tissues and cell lines (CAOV3, SKOV3,
A2780, OVCAR3, HO-8901 and 3AO) were assessed. (A) miR-377 levels
were decreased in cancer tissues compared with their adjacent
normal tissues, whereas (B) mRNA levels of CUL4A were elevated. (C)
Linear correlation of mRNA levels between miR-377 and CUL4A were
assessed. Survival analysis demonstrated that ovarian cancer
patients with (D) increased level of CUL4A and (E) decreased level
of miR-377, lived shorter that the patients with decreased CUL4A
and increased miR-377 expression level, respectively. (F) The
miR-377 level was significantly lower in ovarian cancer cells
compared with normal cells (IOSE80), whereas (G) mRNA levels of
CUL4A were greater in ovarian cancer cells compared with normal
cells (IOSE80). Data are expressed as the mean ± standard deviation
from three independent experiments. **P<0.01 vs.
IOSE80 normal cells. CUL4, cullin 4A; miR, microRNA. |
 | Table IIAssociation between CUL4A and
clinical data of ovarian cancer patients. |
Table II
Association between CUL4A and
clinical data of ovarian cancer patients.
| Cancer stage | Age
(<45/≥45) | CUL4A
expression
(low/high) |
|---|
| TNM | | |
| I | 6/8 | 9/5 |
| II | 4/8 | 3/9 |
| III | 7/11 | 4/14 |
| P-value | 0.883 | 0.031a |
Decreased miR-377 expression along with increased
CUL4A expression was also detected in seven ovarian cancer cell
lines including CAOV3, SKOV3, A2780, OVCAR3, HO-8901, 3AO and TC-1,
compared with in the normal cell line IOSE80, however this
difference was most evident in SKOV3 cells (P<0.05; Fig. 1F and G).
Verification of CUL4A as the direct
target of miR-377
In silico analysis of human miR-377 began
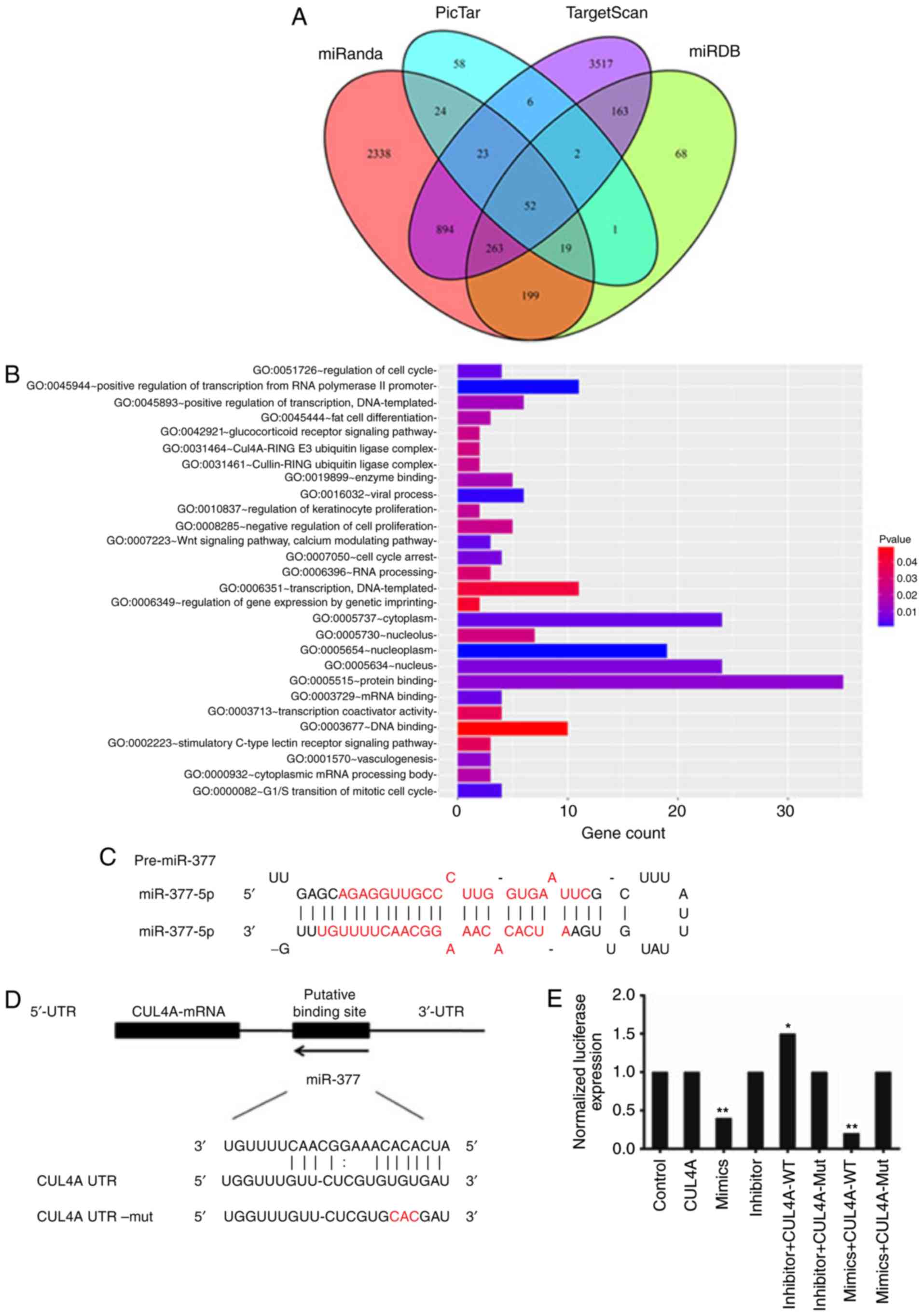
with surveys of its target-prediction. A total of four prediction
software programs; miRanda, miRDB, PicTar, and TargetScan, detected
3.812, 767, 185, and 4.920 target genes, respectively. Based on the
result of the Venn diagram, a total of 52 reliable target genes of
miR-377 were discovered through intersection calculation of
predicted target genes from the four online programs (Fig. 2A). Subsequent gene ontology (GO)
analysis of gene function revealed 28 annotations of biological
processes which were associated with miR-377 (P<0.05). The
target genes of miR-377 were primarily enriched during processes
regulating gene expression, cell proliferation, and signal
transduction (Fig. 2B).
Based on literature review, of the 52 predicted
target genes, CUL4A was selected as it has been previously
demonstrated to highly implicated in tumor progression and patient
survival (22,24). To investigate whether miR-377 is
involved in the regulation of CUL4A protein expression, the
alignment of miR-377/CUL4A was analysed (Fig. 2C). miR-377 is located in a miRNA
cluster on chromosome 14q32.2. The stem-loop for miR-377 includes
two different mature miRNA sequences, miR-377 (MIMAT0000730) from
positions 45-66 and miR-377* (MIMAT0004689) from positions 7-28
(25). miR-377 was predicted to
interact with a 7-mer seed match with the CUL4A-3′ UTR from
position 319-349 (Fig. 2D).
miR-377 interacts with CUL4A-3′ UTR
A luciferase reporter involving the human CUL4A-3′
UTR was applied to identify if miR-377 interacted directly with the
CUL4A-3′ UTR. Luciferase activity was tested 24 h following
transfection of SKOV3 cells with the reporter miR-377
overexpression and inhibition constructs. In the assay system, a
reduction in luciferase expression revealed a specific miR-377-3′
UTR interaction. It was demonstrated that luciferase activity was
significantly decreased in miR-377 overexpressed cells; in
particular, in the miR-377 mimics + CUL4A-WT group, the luciferase
expression was reduced to one fifth of the level in the control
group (P<0.01; Fig. 2E).
Mutation of the predicted binding sites in CUL4A-3′ UTR eliminated
this decrease in luciferase reporter activity. This result
suggested that miR-377 is involved in the control of CUL4A
expression.
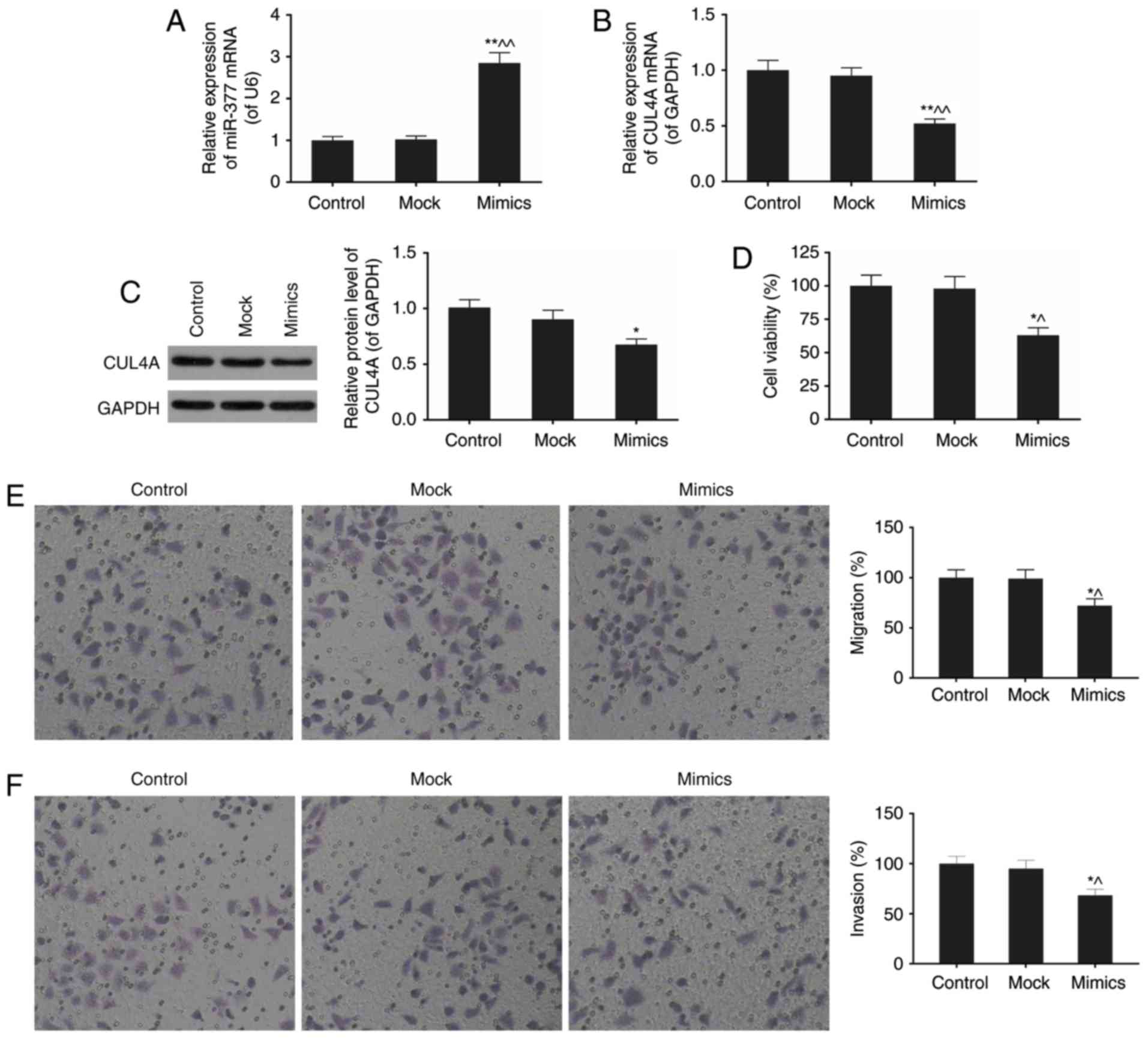
miR-377 downregulates CUL4A mRNA and
protein expression levels in SKOV3 cells
As presented in Fig.
1D and E, the differential expression of miR-377 and CUL4A in
SKOV3 cell line compared with normal cells was the most significant
of the six ovarian cancer cell lines. In the present study, SKOV3
was selected for subsequent experimentation to probe into the
effects of miR-377 on ovarian cancer in vitro. The effect of
expression of the miR-377 mimics on the expression level of CUL4A
was subsequently assessed using RT-qPCR and western blotting
(Fig. 3A–C). The miR-377
expression levels in the mimics group exhibited an almost
three-fold increase compared with normal SKOV3 cells (P<0.05;
Fig. 3A). Overexpression of
miR-377 downregulated CUL4A mRNA and protein expression levels. As
presented in Fig. 3C, the
expression of miR-377 mimics resulted in an approximately 20%
decrease in CUL4A protein expression compared with expression of
the mock.
miR-377 mimics attenuate viability,
migration and invasion of SKOV3 cells
The effects of miR-377 mimics on biological
processes in SKOV3 cells were next investigated, including cell
viability, migration, and invasion (Fig. 3D-F). In the miR-377 mimic group,
cell viability was markedly inhibited by the high level of miR-377,
compared with normal cell group (P<0.05; Fig. 3D). Furthermore, with miR-377
overexpression, migration and invasion rates were attenuated
(P<0.01; Fig. 3E and F). These
results implied the important role of miR-377 in ovarian
cancer.
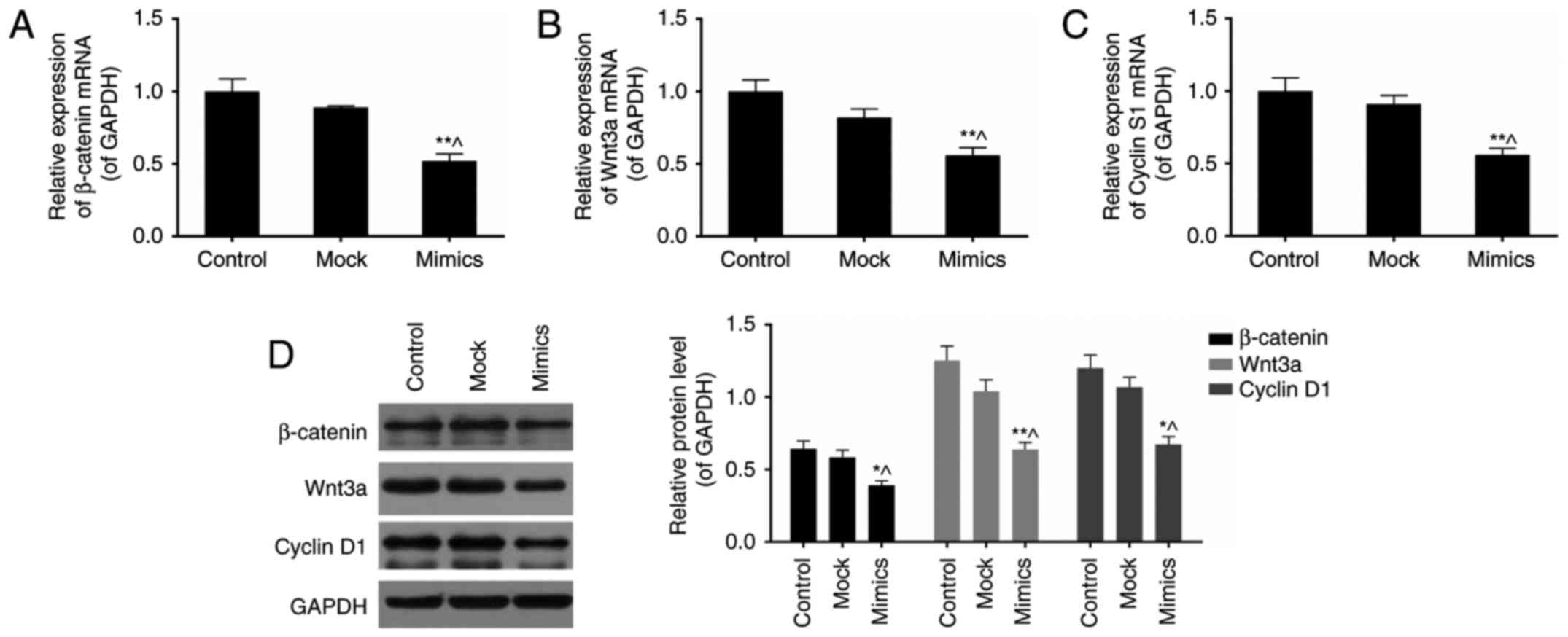
miR-377 mimics inhibit Wnt/β-catenin
signaling pathway in SKOV3 cells
β-catenin, Wnt3a, and cyclin D1 are three key
components in the Wnt/β-catenin signaling pathway (26). To investigate the effects of
miR-377 on this signalling pathway, expression levels of these
three genes were assessed in normal and miR-377 mimics conditions
(Fig. 4A–D). As a result,
overexpression of miR-377 downregulated the expression of
β-catenin, Wnt3a, and cyclin D1 (P<0.05).
miR-377 mimics affect the expression of
MMP-2, MMP-9, MTA1 and TIMP2 in SKOV3 cells
Subsequently, the influence of miR-377
overexpression on tumor metastasis-associated genes including MMP-2
and 9, MTA1 and TIMP2 was assessed. MMPs and TIMPs serve important
roles in extracellular matrix turnover (27). MTA1, a member of MTAs family, is
an integral component of the nucleosome remodeling and histone
deacetylation complexes (28). It
has been reported to be highly expressed in metastatic cells
compared with non-metastatic ones (29). Expression levels of MMP-2, MMP-9,
MTA1 and TIMP2 were influenced by miR-377 overexpression (Fig. 5A-E). Cells transfected with the
miR-377 overexpression construct expressed lower levels of MMP-2,
MMP-9, and MTA1, however increased levels of TIMP2 (P<0.05).
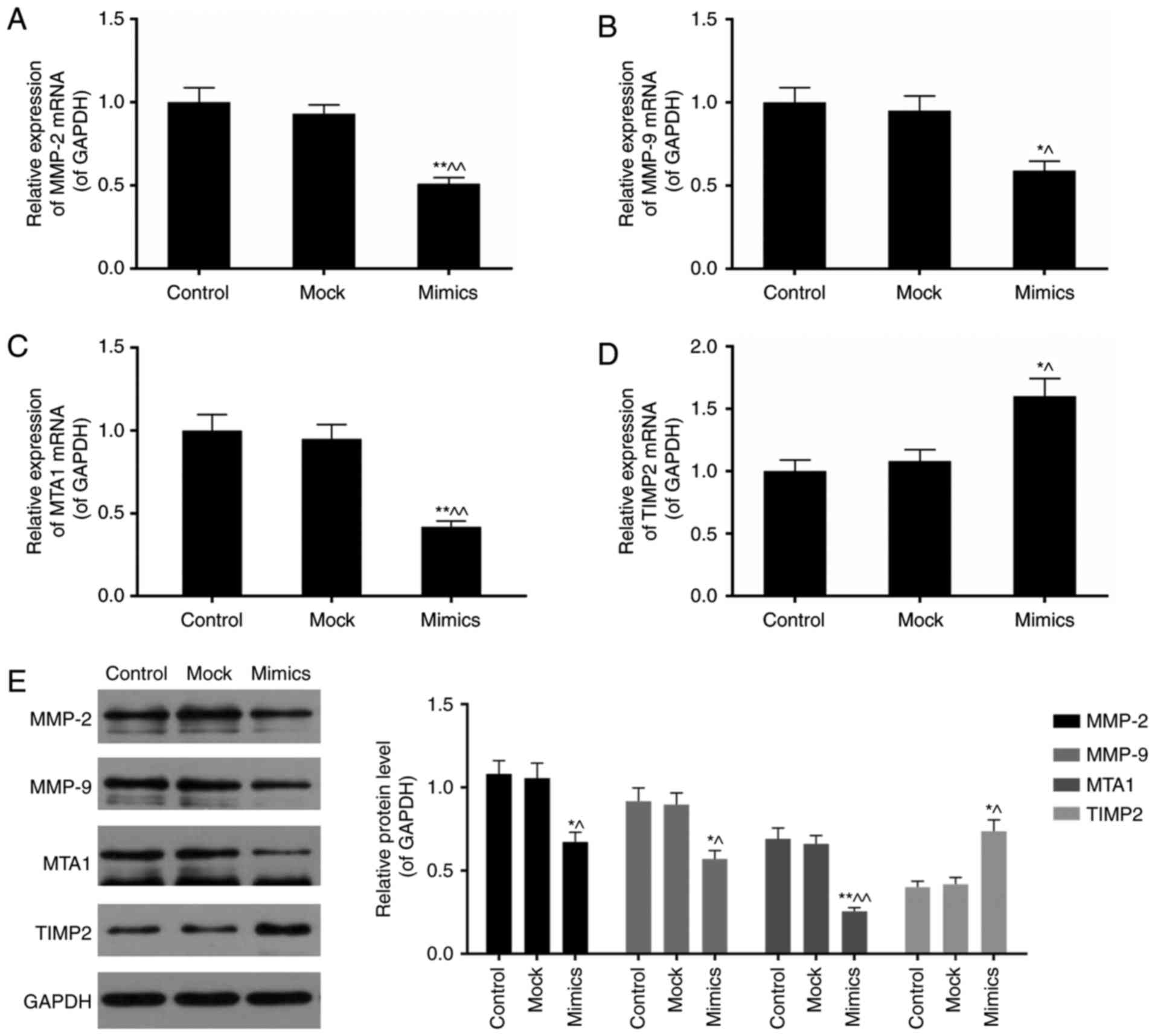 | Figure 5miR-377 overexpression influences the
expression of MMP-2, MMP-9, MTA1 and TIMP2. Aberrant miR-377 level
downregulated the expression of (A) MMP-2, (B) MMP-9 and (C) MTA1
mRNA, however upregulated (D) TIMP2 mRNA expression levels. (E)
MMP-2, MMP-9 and MTA1 protein expression levels were decreased
whereas TIMP2 protein expression was elevated in the miR-377 mimics
group. Data are expressed as the mean ± standard deviation from
three independent experiments. *P<0.05 and
**P<0.01 vs. control, ^P<0.05 and
^^P<0.01 vs. mock. MTA, metastasis-associated
protein; miR, microRNA.; MMP, matrix metalloproteinase; TIMP,
metallopeptidase inhibitor. |
Discussion
The expression of miR-377 is silenced in a number of
human malignancies, which has been verified in previous studies
(6-10). In fact, the large miRNA cluster on
chromosome 14q32 including miR-377 is termed as ʻthe largest miRNA
tumour suppressor clusterʼ (9).
The present analysis identified a total of 52 target genes of
miR-377, and demonstrated that these genes were enriched in primary
functions of regulation of gene expression, cell proliferation, and
signal transduction. Aberrant expression of miR-377 in the ovarian
cell line SKOV3 reduced cell viability, and resulted in inhibition
of migration and invasion of the cancer cells, which suggested that
miR-377 is likely to function as a suppressor in ovarian
cancer.
There is a general lack of information regarding
target genes of miR-377. A previous study demonstrated that miR-377
targets frizzled class receptor 4, which is required for
epithelial-mesenchymal transition (EMT) and metastasis in prostate
cancer cells (30). miR-377 has
additionally been demonstrated to target cyclin dependant kinase 6
and E2F transcription factor 3 (31). However, previously it was unknown
if miR-377 targeted CUL4A.
It was demonstrated that EMT is an essential process
for the metastasis of cancer cells (32-34). Through EMT, cancer cells obtain
stronger capability to move and migrate into the circulatory system
via the extracellular matrix and basement membrane of blood
vessels, and form secondary metastases as a consequence (35,36). A key factor which regulates the
occurrence of EMT in ovarian cancer may be not only an important
biomarker of prognostic prediction of ovarian cancer, however also
an important target in further studies for intervention treatments
to more effectively inhibit or interdict metastasis of ovarian
cancer. Ubiquitin ligase E3, which is classified into two
categories, termed, ring-finger E3 ubiquitin ligase family and
homologous to E6AP C-terminus (HECT) ubiquitin ligase family,
according to the difference in transfer methods of ubiquitin,
serves a role in ubiquitination-mediated degradation of proteins
(19,37,38). Belonging to the cullin-ringbased
E3-ligases (CRLs), a principal part of the ring-finger E3 ubiquitin
ligase family, CUL4A was first identified in breast cancer and its
amplification is strongly associated with tumorigenesis and tumour
migration of several cancers (39,40). CUL4A, which was demonstrated to be
overexpressed in ovarian cancer tissues and cell lines in the
present study, is a direct target of miR-377, and its activity is
regulated by miR-377. The present study demonstrated that the
decrease in CUL4A protein levels, which were detected following
overexpression of miR-377 resulted in inhibition of viability,
migration, and invasion of SKOV3 cells.
A number of signal transduction pathways, including
transforming growth factor-β, mitogen activated protein kinase,
Notch and Wnt, participate in the process of EMT of tumour cells
(26). The Wnt signaling pathway
transmits signals from the cell surface into the cell nucleus by
secreting Wnt signalling proteins, transmembrane receptor proteins,
and other intracellular proteins (41). The canonical Wnt signaling
pathway, also named Wnt/β-catenin signaling pathway, serves a vital
role in proliferation, differentiation, movement, and morphology of
numerous cells (42-44). In the presence of Wnt signalling,
Wnt proteins combine with the cell surface receptor Frizzled and
members of the low-density lipoprotein receptor-related protein
(LRP) family, LRP-5, -6 to activate intracellular dishevelled
protein, inhibit phosphorylation of β-catenin, and protect
intracytoplasmic unphosphorylated β-catenin from degradation via
the ubiquitin-proteasome system. β-catenins in the cytoplasm gather
and shift toward the cell nucleus so as to interact with the
lymphoid enhancer factor/T cell factor to activate the
transcription of target genes, including cyclin D1 and c-myc. The
protein complex of β-catenin/E-cadherin combines with the actin
cytoskeleton to reduce cell-cell adhesion and induce the EMT
process of tumours, which strengthens the migration and invasion
abilities of tumour cells (45-49).
In the Wnt/β-catenin signaling pathway, Wnt3, a
secretory protein of the pathway, is the promoter, and β-catenin,
an important regulatory factor that conducts signals from the cell
membrane or cytoplasm to the cell nucleus, is the executor of the
regulatory function on its downstream targets. Cyclin D1, one of
its targets, is a key intranuclear transcription factor that
regulates the cell cycle (50).
The present study demonstrated that aberrant miR-377 expression,
attenuated the mRNA and protein levels of β-catenin, Wnt3a, and
cyclin D1, which indicated the reduced activation of the
Wnt/β-catenin signaling pathway by miR-377 mimics. The results of
the present study suggested that miR-377 reduces the migratory
ability of SKOV3 cells, potentially by inhibiting the EMT process
via the Wnt/β-catenin signalling pathway.
MMP2, together with MMP9, is able to degrade type IV
collagen, and degradation of the basement membrane allows cancer
cells to migrate out of the tumour, resulting in metastasis
(51). MTA1 suppresses the
expression of numerous tumour suppressor genes contributing to cell
migration and invasion (52). In
the present study, it was demonstrated that the expression levels
of MMP-2, MMP-9, MTA1, and TIMP2 were significantly influenced by
miR-377 overexpression at the mRNA and protein levels. Cells
transfected with the miR-377 overexpression construct expressed
lower levels of MMP-2, MMP-9, and MTA1, however increased levels of
TIMP2, which suggested that miR-377 is necessary for attenuating
migration-associated protein expression.
In conclusion, the results of the present study
suggested that miR-377 acted as a negative regulator of CUL4A,
which resulted in reduced cancer cell viability and migration in an
ovarian cancer cell line. It was then hypothesized that
downregulation of miR-377 in ovarian cancer may promote
tumorigenesis and metastasis through activation of CUL4A
expression, however further in vitro and in vivo
research is required to verify this.
Acknowledgments
Not applicable.
Notes
[1]
Funding
No funding was received.
[2] Availability
of data and materials
The analysed data sets generated during the study
are available from the corresponding author on reasonable
request.
[3] Authors'
contributions
RY and YC conceived and designed the study. XD
analyzed and interpreted the patient data regarding the
relationship of miR-377 and CUL4A expression in ovarian cancer
tissues. LC and XW performed the cell experiments to investigate
the role of miR-377 in metastatic capability regulation. RY was a
major contributor in writing the manuscript. All authors read and
approved the final manuscript.
[4] Ethics
approval and consent to participate
The present study was approved by the ethics
committee of the Third Affiliated Hospital of Wenzhou Medical
University (Wenzhou, China). Written informed consent was
obtained.
[5] Consent for
publication
Not applicable.
[6] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ozols RF, Bundy BN, Greer BE, Fowler JM,
Clarke-Pearson D, Burger RA, Mannel RS, DeGeest K, Hartenbach EM
and Baergen R; Gynecologic Oncology Group: Phase III trial of
carboplatin and paclitaxel compared with cisplatin and paclitaxel
in patients with optimally resected stage III ovarian cancer: A
Gynecologic Oncology Group study. J Clin Oncol. 21:3194–3200. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Smith HJ, Straughn JM, Buchsbaum DJ and
Arend RC: Epigenetic therapy for the treatment of epithelial
ovarian cancer: A clinical review. Gynecol Oncol Rep. 20:81–86.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Armstrong DK, Bundy B, Wenzel L, Huang HQ,
Baergen R, Lele S, Copeland LJ, Walker JL and Burger RA;
Gynecologic Oncology Group: Intraperitoneal cisplatin and
paclitaxel in ovarian cancer. N Engl J Med. 354:34–43. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chambers AF, Groom AC and MacDonald IC:
Dissemination and growth of cancer cells in metastatic sites. Nat
Rev Cancer. 2:563–572. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Costa FF, Bischof JM, Vanin EF, Lulla RR,
Wang M, Sredni ST, Rajaram V, Bonaldo Mde F, Wang D, Goldman S, et
al: Identification of microRNAs as potential prognostic markers in
ependymoma. PLoS One. 6:e251142011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gattolliat CH, Thomas L, Ciafre SA,
Meurice G, Le Teuff G, Job B, Richon C, Combaret V, Dessen P,
Valteau-Couanet D, et al: Expression of miR-487b and miR-410
encoded by 14q32.31 locus is a prognostic marker in neuroblastoma.
Br J Cancer. 105:1352–1361. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Haller F, von Heydebreck A, Zhang JD,
Gunawan B, Langer C, Ramadori G, Wiemann S and Sahin O:
Localization- and mutation-dependent microRNA (miRNA) expression
signatures in gastrointestinal stromal tumours (GISTs), with a
cluster of co-expressed miRNAs located at 14q32.31. J Pathol.
220:71–86. 2010. View Article : Google Scholar
|
|
9
|
Lavon I, Zrihan D, Granit A, Einstein O,
Fainstein N, Cohen MA, Cohen MA, Zelikovitch B, Shoshan Y, Spektor
S, et al: Gliomas display a microRNA expression profile reminiscent
of neural precursor cells. Neuro Oncol. 12:422–433. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Thayanithy V, Sarver AL, Kartha RV, Li L,
Angstadt AY, Breen M, Steer CJ, Modiano JF and Subramanian S:
Perturbation of 14q32 miRNAs-cMYC gene network in osteosarcoma.
Bone. 50:171–181. 2012. View Article : Google Scholar
|
|
11
|
Vasudevan S, Tong Y and Steitz JA:
Switching from repression to activation: microRNAs can up-regulate
translation. Science. 318:1931–1934. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gong J, Zhang JP, Li B, Zeng C, You K,
Chen MX, Yuan Y and Zhuang SM: MicroRNA-125b promotes apoptosis by
regulating the expression of Mcl-1, Bcl-w and IL-6R. Oncogene.
32:3071–3079. 2013. View Article : Google Scholar
|
|
13
|
Henke JI, Goergen D, Zheng J, Song Y,
Schüttler CG, Fehr C, Jünemann C and Niepmann M: microRNA-122
stimulates translation of hepatitis C virus RNA. EMBO J.
27:3300–3310. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Banelli B, Forlani A, Allemanni G,
Morabito A, Pistillo MP and Romani M: MicroRNA in glioblastoma: An
overview. Int J Genomics. 2017:76390842017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Honardoost M and Rad SMAH: Triangle of
AKT2, miRNA, and tumorigenesis in different cancers. Appl Biochem
Biotechnol. 2017.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Slaby O, Laga R and Sedlacek O:
Therapeutic targeting of non-coding RNAs in cancer. Biochem J.
474:4219–4251. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu N, Zhao X, Liu M, Liu H, Yao W, Zhang
Y, Cao S and Lin X: Role of microRNA-26b in glioma development and
its mediated regulation on EphA2. PLoS One. 6:e162642011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kuninty PR, Schnittert J, Storm G and
Prakash J: MicroRNA targeting to modulate tumor microenvironment.
Front Oncol. 6:32016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sarikas A, Hartmann T and Pan ZQ: The
cullin protein family. Genome Biol. 12:2202011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jackson S and Xiong Y: CRL4s: The
CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 34:562–570.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen X, Zhang Y, Douglas L and Zhou P:
UV-damaged DNA-binding proteins are targets of CUL-4A-mediated
ubiquitination and degradation. J Biol Chem. 276:48175–48182. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gupta A, Yang LX and Chen Lc: Study of the
G2/M cell cycle checkpoint in irradiated mammary epithelial cells
overexpressing Cul-4A gene. Int J Radiat Oncol Biol Phys.
52:822–830. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Birner P, Schoppmann A, Schindl M, Dinhof
C, Jesch B, Berghoff AS and Schoppmann SF: Human homologue for
Caenorhabditis elegans CUL-4 protein overexpression is associated
with malignant potential of epithelial ovarian tumours and poor
outcome in carcinoma. J Clin Pathol. 65:507–511. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Beckman JD, Chen C, Nguyen J, Thayanithy
V, Subramanian S, Steer CJ and Vercellotti GM: Regulation of heme
oxygenase-1 protein expression by miR-377 in combination with
miR-217. J Biol Chem. 286:3194–3202. 2011. View Article : Google Scholar :
|
|
26
|
Savagner P: The epithelial-mesenchymal
transition (EMT) phenomenon. Ann Oncol. 21(Suppl 7): vii89–vii92.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brun JL, Cortez A, Lesieur B, Uzan S,
Rouzier R and Daraï E: Expression of MMP-2, -7, -9, MT1-MMP and
TIMP-1 and -2 has no prognostic relevance in patients with advanced
epithelial ovarian cancer. Oncol Rep. 27:1049–1057. 2012.
View Article : Google Scholar
|
|
28
|
Malisetty VL, Penugurti V, Panta P, Chitta
SK and Manavathi B: MTA1 expression in human cancers-Clinical and
pharmacological significance. Biomed Pharmacother. 95:956–964.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xue Y, Wong J, Moreno GT, Young MK, Côté J
and Wang W: NURD, a novel complex with both ATP-dependent
chromatin-remodeling and histone deacetylase activities. Mol Cell.
2:851–861. 1998. View Article : Google Scholar
|
|
30
|
Formosa A, Markert EK, Lena AM, Italiano
D, Finazzi-Agro E, Levine AJ, Bernardini S, Garabadgiu AV, Melino G
and Candi E: MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c,
miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p
mapped to the 14q32.31 locus, regulate proliferation, apoptosis,
migration and invasion in metastatic prostate cancer cells.
Oncogene. 33:5173–5182. 2014. View Article : Google Scholar
|
|
31
|
Zehavi L, Schayek H, Jacob-Hirsch J, Sidi
Y, Leibowitz-Amit R and Avni D: miR-377 targets E2F3 and alters the
NF-kB signaling pathway through MAP3K7 in malignant melanoma. Mol
Cancer. 14:682015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sabe H: Cancer early dissemination:
Cancerous epithelial- mesenchymal transdifferentiation and
transforming growth factor β signalling. J Biochem. 149:633–639.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Micalizzi DS, Farabaugh SM and Ford HL:
Epithelial- mesenchymal transition in cancer: Parallels between
normal development and tumor progression. J Mammary Gland Biol
Neoplasia. 15:117–134. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Taylor MA, Parvani JG and Schiemann WP:
The pathophysiology of epithelial-mesenchymal transition induced by
transforming growth factor-beta in normal and malignant mammary
epithelial cells. J Mammary Gland Biol Neoplasia. 15:169–190. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial- mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chapman HA: Epithelial-mesenchymal
interactions in pulmonary fibrosis. Ann Rev Physiol. 73:413–435.
2011. View Article : Google Scholar
|
|
37
|
Zhao Y and Sun Y: Cullin-RING Ligases as
attractive anti-cancer targets. Curr Pharm Des. 19:3215–3225. 2013.
View Article : Google Scholar
|
|
38
|
Jia L and Sun Y: SCF E3 ubiquitin ligases
as anticancer targets. Curr Cancer Drug Targets. 11:347–356. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen LC, Manjeshwar S, Lu Y, Moore D,
Ljung BM, Kuo WL, Dairkee SH, Wernick M, Collins C and Smith HS:
The human homologue for the Caenorhabditis elegans cul-4 gene is
amplified and overexpressed in primary breast cancers. Cancer Res.
58:3677–3683. 1998.PubMed/NCBI
|
|
40
|
Schindl M, Gnant M, Schoppmann SF, Horvat
R and Birner P: Overexpression of the human homologue for
Caenorhabditis elegans cul-4 gene is associated with poor outcome
in node-negative breast cancer. Anticancer Res. 27:949–952.
2007.PubMed/NCBI
|
|
41
|
Toda H, Boku S, Nakagawa S, Inoue T, Kato
A, Takamura N, Song N, Nibuya M, Koyama T and Kusumi I: Maternal
separation enhances conditioned fear and decreases the mRNA levels
of the neurotensin receptor 1 gene with hypermethylation of this
gene in the rat amygdala. PLoS One. 9:e974212014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Moon RT, Bowerman B, Boutros M and
Perrimon N: The promise and perils of Wnt signaling through
beta-catenin. Science. 296:1644–1646. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Moon RT, Kohn AD, De Ferrari GV and Kaykas
A: WNT and beta-catenin signalling: Diseases and therapies. Nat Rev
Genet. 5:691–701. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang L, Yang X, Yang S and Zhang J: The
Wnt/β-catenin signaling pathway in the adult neurogenesis. Eur J
Neurosci. 33:1–8. 2011. View Article : Google Scholar
|
|
45
|
Nelson WJ and Nusse R: Convergence of Wnt,
beta-catenin, and cadherin pathways. Science. 303:1483–1487. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Toledo EM, Colombres M and Inestrosa NC:
Wnt signaling in neuroprotection and stem cell differentiation.
Prog Neurobiol. 86:281–296. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu F and Millar SE: Wnt/beta-catenin
signaling in oral tissue development and disease. J Dent Res.
89:318–330. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Michaelidis TM and Lie DC: Wnt signaling
and neural stem cells: Caught in the Wnt web. Cell Tissue Res.
331:193–210. 2008. View Article : Google Scholar
|
|
49
|
Liu J, Wang Z, Tang J, Tang R, Shan X,
Zhang W, Chen Q, Zhou F, Chen K, Huang A, et al: Hepatitis C virus
core protein activates Wnt/β-catenin signaling through multiple
regulation of upstream molecules in the SMMC-7721 cell line. Arch
Virol. 156:1013–1023. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Vallée A, Lecarpentier Y, Guillevin R and
Vallee JN: Thermodynamics in gliomas: Interactions between the
Canonical WNT/Beta-catenin pathway and PPAR Gamma. Front Physiol.
8:3522017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Mook OR, Frederiks WM and Van Noorden CJ:
The role of gelatinases in colorectal cancer progression and
metastasis. Biochim Biophys Acta. 1705:69–89. 2004.PubMed/NCBI
|
|
52
|
Zhang Y and Wang XF: Post-transcriptional
regulation of MTA family by microRNAs in the context of cancer.
Cancer Metastasis Rev. 33:1011–1016. 2014. View Article : Google Scholar : PubMed/NCBI
|