Introduction
Induced pluripotent stem cells (iPSCs) provide a
promising resource for drug research and the investigation of
disease mechanisms and regenerative medicine (1). Nonhuman primates (NHPs) represent an
ideal preclinical model for human diseases. Among various NHPs, the
marmoset has several unique advantages, including its smaller size,
higher rate of breeding, and defined housing conditions, in
addition to their relatedness and similar physiology to humans,
which makes these animals ideal for developmental research and
clinical application (2).
Marmoset embryonic stem cell (ESC) lines have previously been
established (3–5). According to their requirement of
growth factors, including basic fibroblast growth factor (bFGF),
for maintaining pluripotency and their molecular signaling pathways
for self-renewal, marmoset ESCs are more similar to human ESCs than
mouse ESCs (mESCs) (6). To date,
marmoset iPSCs have been described in several reports (7–9).
In our previous study, it was found that marmoset iPSCs undergo
self-renewal and exhibit differentiation similar to that of ESCs
(7,10).
Leukemia inhibitory factor (LIF) is a member of the
interleukine-6 cytokine family, which utilizes a receptor
comprising LIF receptor β and the glycoprotein 130 subunit
(11). The functions of LIF,
including stimulating or inhibiting proliferation, differentiation,
apoptosis and inflammation, are multifaceted, even paradoxical, in
different cell types (12–14).
Upon LIF stimulation, three intracellular pathways are mainly
activated, comprising the Janus kinase (Jak)-signal transducer and
activator of transcription (Stat)3, mitogen-activated protein
kinase (MAPK) and phosphoinositide 3-kinase (PI3K)-Akt pathways
(15–17). Each of the three pathways has its
own critical function in mESCs. Among these pathways, the Jak-Stat3
pathway facilitates the self-renewal of mESCs, which are solely
regulated by LIF, whereas the PI3K-Akt pathway maintains
pluripotency, and the MAPK pathway promotes differentiation, both
of which are regulated by multiple pathways (18). Compared with those of mice, human
ESCs and iPSCs are primed pluripotent stem cells, which are
bFGF-dependent for self-renewal and LIF-independent (19,20). However, the growth factors used in
the culture medium of pluripotent stem cells differ among various
reports (6,7). Therefore, the most appropriate
growth factor and its downstream pathway for maintaining the
self-renewal of pluripotent stem cells remains to be
elucidated.
In our previous study, it was reported that bFGF was
essential and critical for maintaining marmoset iPSCs (7,10).
However, whether LIF has an effect in culturing marmoset iPSCs
remains to be fully elucidated. Accordingly, in the present study,
the functional effects of LIF on the pluripotent state of marmoset
iPSCs was investigated. To the best of our knowledge, the present
study is the first to apply LIF for the maintenance of marmoset
iPSCs. The results of the present study are likely to improve the
culture techniques for marmoset iPSCs and facilitate their
application as a preclinical experimental resource for human
regenerative medicine.
Materials and methods
Cell culture
Marmoset iPSCs were established according to the
methods in Dr Peter Hornsby's laboratory (Department of Physiology
and The Barshop Institute for Longevity and Aging Studies,
University of Texas Health Science Center, San Antonio, TX 78245,
USA) and maintained in our laboratory in the Department of
Biochemistry and Molecular Biology, College of Life Science,
Zhejiang Sci-Tech University (Hangzhou, China). All animal
procedures, husbandry and housing were in accordance with the
Animal Care and Use Committee requirements of Southwest National
Primate Research Center (San Antonio, TX, USA (7). The cells were cultured in 5%
CO2 with 95% humidity at 37°C in human ESC medium
TeSR-E8 (cat. no. 05840; StemCell Technologies, Inc., Vancouver,
BC, Canada) supplemented with 10% ESC qualified FBS (cat no.
SH30406.02E; Hyclone; GE Healthcare Life Sciences, Logan, UT, USA),
100 U/ml of penicillin, 100 mg/ml of streptomycin (cat. no.
15070-063; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), 100 ng/ml of bFGF (cat. no. F0291; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) and 10 µM Rho-associated kinase (ROCK)
inhibitor (Y-27632, cat no. Y0503-1MG, Sigma-Aldrich; Merck KGaA)
on Matrigel-coated dishes. The iPSCs were passaged when they
reached 80–90% confluency. For passaging, the cells were incubated
with Accutase (cat no. 07920, StemCell Technologies, Inc.) to a
single-cell mass and replated on Matrigel-coated dishes according
to the appropriate split ratio. The medium was replaced daily. To
investigate the effect of LIF, the treatment group was supplemented
with 1,000 U/ml of mouse LIF (cat no. ESG1106; EMD Millipore),
whereas the control cells were cultured in the absence of LIF.
Cell proliferation assay
An MTT assay (Sigma-Aldrich; Merck KGaA) was
performed to assess cell proliferation. Briefly, the cells were
seeded onto 96-well plates at a density of 5×103. After
24 h, 20 µl of MTT solution [0.5 mg/ml dissolved in
phosphate-buffered saline (PBS)] was added to each test well,
comprising the LIF group, the absence of LIF control group and a
blank control group. The cells were then incubated for 3 h at 37°C.
The produced formazan precipitate was dissolved in dimethyl
sulfoxide (DMSO; Sigma-Aldrich; Merck KGaA). The absorbance was
measured at 495 nm using a multi-mode microplate reader. The
optical density (OD) values represented the proliferation or the
survival of the marmoset iPSCs. Each experiment was repeated three
times, followed by calculation of the standard error of the
mean.
Flow cytometry
Cell apoptosis was measured according to the
manufacturer's protocol with the FITC Annexin V Apoptosis Detection
kit (BD Biosciences, Franklin Lakes, NJ, USA). Briefly, the cells
were harvested by Accutase, washed twice with PBS and then
resuspended in binding buffer. Subsequently, the cells were
incubated with Annexin V-fluorescein isothiocyanate (FITC) and
propidium iodide (PI) for 15 min at 37°C. The samples were analyzed
using the FACSAria system (BD Biosciences). Cell apoptosis was
analyzed using FlowJo version 10 software (BD Biosciences).
cDNA library construction, solexa
sequencing and digital gene expression (DGE) analysis
The marmoset iPSCs were seeded in a
35-cm2 cell culture dish (~104 cells) and
cultured in the presence or absence of LIF (100 U/ml) at 37°C, 5%
CO2 for 24 h. The samples were treated in triplicate.
For the DGE analysis (Beijing Genomics Institute, Shenzhen, China),
total RNA was extracted using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
Total RNA was treated with DNase I, and poly-(A) mRNA was enriched
with magnetic oligo (dT) beads. The RNA quality and quantity were
verified using an Agilent 2100 Bioanalyzer and the ABI StepOnePlus
Real-Time PCR system. The mRNA was mixed with fragmentation buffer
(10X Fragmentation Reagent, Ambion; Thermo Fisher Scientific, Inc.;
cat. no. AM8740) to create short fragments. These were used as
templates for first-strand cDNA synthesis with random hexamer
primers.
Second-strand cDNA was synthesized according to the
manufacturer's protocol using a reaction system of buffer, dNTPs,
RNaseH and DNA polymerase I included in the SuperScript®
Double-Stranded cDNA Synthesis kit (Invitrogen; Thermo Fisher
Scientific, Inc.; cat. no. 11917-010). Short fragments were
purified with the QiaQuick PCR extraction kit (Qiagen GmbH, Hilden,
Germany; cat. no. 28104) and resolved with EB buffer (Qiagen GmbH;
cat. no. 19086) for end reparation and poly (A) addition. These
were then connected with adapters and suitable fragments were
selected as templates for PCR amplification by using Platinum™ Pfx
DNA Polymerase (Invitrogen; Thermo Fisher Scientific, Inc.; cat.
no. 11708013), to create the final cDNA library. The following
thermocycling conditions were used for the PCR: 94°C for 2 min; 13
cycles of 94°C for 15 sec, 55°C for 30 sec, 68°C for 30 sec; and
final extension at 68°C for 5 min. Primer sequences were as
follows: Forward, 5-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGA-3;
reverse, 5-CAAGCAGAAGACGGCATACGAGAT-3.
The fragments were purified by agarose gel
electrophoresis and enriched by PCR amplification. The library
products were then ready for solexa sequencing via the Illumina
HiSeq 2000 sequencing platform (21), followed by bioinformatics
analysis. The transcriptome sequence was assembled into distinct
contigs using the short reads with SOAPdenovo software (version
2.21; http://soap.genomics.org.cn). The genome
sequence of marmoset was obtained from the National Center for
Biotechnology Information (NCBI, www.ncbi.nlm.nih.gov) database. GO term annotation
(molecular function, biological process and cellular component) and
enrichment analysis was conducted using the Blast2GO software
(version 2.2.5) (22).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. For RT-qPCR analysis, 1 µg of RNA
was used for cDNA synthesis in a reverse transcription reaction
using the Prime Script Reagent kit (Takara Bio, Inc., Otsu, Japan).
The reaction was incubated at 37°C for 15 min anf at 85°C for 5 sec
and then amplified by using gene specific primers for qPCR. The
qPCR analysis was set up in duplicate with SYBR Premix Ex Taq
(Takara Bio, Inc.) and performed using the 7500 Real-Time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). qPCR
reactions of 20 µl for each sample were made and consisted
of cDNA (200 µg cDNA after dilution), 2X SYBR Premix Ex Taq,
50X ROX Reference Dye, dH2O and 10 µM of each
gene specific primer. After initial denaturation at 95°C for 30
sec, cDNA was amplified in 40 cycles. Amplification conditions
were: 5 sec of denaturation at 95°C, 34 sec of annealing at 60°C
and 30 sec of extension at 72°C, followed by a final extension step
at 72°C for 10 min. Using the 2−ΔΔCq method (23), the analysis of each sample was
repeated three times, and β-actin was used as the housekeeping gene
for internal normalization (24).
The primers were designed using Primer Premier 5.0 software
(Premier Biosoft International, Palo Alto, CA, USA). The sequences
of the primers are listed in Table
I.
 | Table IPrimers used in the present
study. |
Table I
Primers used in the present
study.
| Gene | Sense primer
(5′-3′) | Antisense primer
(5′-3′) |
|---|
| β-actin |
ATGGAAGAAGAAATTGCTGCG |
GCCCATGTAGGAGTCCTTCTG |
| TBX-3 |
GAGGCTAAAGAACTTTGGGATC |
TGGGCTATCTGGGTGAATG |
| PI3K |
GGCAGCAGTGGAGAGATTTGTTCCC |
AAGAATGTGCCCGAAG |
| NANOG |
ACCTTCCGGTATGGAACAA |
CCAAGTCACTGGCAGGAGA |
| OCT4 |
AAGGGCAAGCGATCAAGCA |
GGGAATGGGACCGAGGAGTA |
Western blot analysis
The cells were washed twice in PBS, collected, lysed
and subsequently boiled in loading buffer at 100°C for 10 min.
Protein concentration was determined by the Quick Start Bradford
Protein Assay kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Equal quantities of total protein (20 µg) were separated on
a 12% SDS-PAGE gel at 120 V for 1 h and subsequently blotted onto a
nitrocellulose membrane (EMD Millipore) at 100 V for 75 min at 4°C.
The membranes were then blocked in 5% nonfat dried milk in PBST [10
mM Tris-HCl (pH 7.5), 150 mM NaCl and 0.1% Tween-20] at room
temperature for 1 h according to the antibody manufacturer's
protocol. Following a brief wash in PBST (0.1% Tween 20 in PBS),
the membranes were incubated with primary antibodies at 4°C
overnight. The primary antibodies included anti-β-actin (cat. no.
bs-0061R, BIOSS, Beijing, China), anti-PI3K (85α) antibody (cat.
no. 60225-1-Ig; Full), anti-AKT (cat. no. 9272; Cell Signaling
Technology, Danvers. MA, USA), anti-p-AKT (cat. no. 9271; Cell
Signaling Technology), anti-STAT3 (cat. no. 10253-2-AP;
ProteinTech, Rosemont, IL, USA), anti-Tbx-3 (cat. no. 16741-1-AP;
ProteinTech) and anti-NANOG (cat. no. 14295-1-AP; ProteinTech)
primary antibodies at a dilution of 1:1,000. The immunoreactive
bands were then incubated with the appropriate horseradish
peroxidase (HRP)-conjugated secondary anti-rabbit IgG-HRP
antibodies (cat. no. A0208; 1:1,000; Beyotime Institute of
Biotechnology, Haimen, China), or anti-mouse IgG-HRP antibodies
(1:1,000, cat. no. A0216; Beyotime Institute of Biotechnology) for
1 h at room temperature. The protein bands were visualized using
enhanced chemiluminescence (ECL) reagent (Amersham; GE Healthcare
Life Sciences, Piscataway, NJ, USA) and images were captured by the
ECL Tanon 5500 system (Tanon Science and Technology Co., Ltd.,
Shanghai, China).
PI3K/Akt signaling inhibition
LY294002, a synthetic selective inhibitor of PI3K,
was dissolved in DMSO at a concentration of 10 mg/ml based on the
manufacturer's protocol. To confirm the involvement of PI3K/Akt
signaling on the effects of LIF on marmoset iPSCs, 2×106
cells were treated with the indicated concentration (20 µM)
of LY294002 or an equal volume of DMSO and cultured for 24 h at
37°C. Cell proliferation assays and western blot analysis were then
performed, as described above.
Statistical analysis
The data are presented as the mean ± standard
deviation of triplicate experiments (n=3). Student's t-test
(two-tailed unpaired) was used to evaluate the significant
differences between the control and treated groups. All graphs were
plotted using GraphPad Prism 5 software (GraphPad Software, Inc.,
La Jolla, CA, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
LIF induces the proliferation of marmoset
iPSCs, but has minimal effect on apoptosis
In the present study, the effects of LIF on
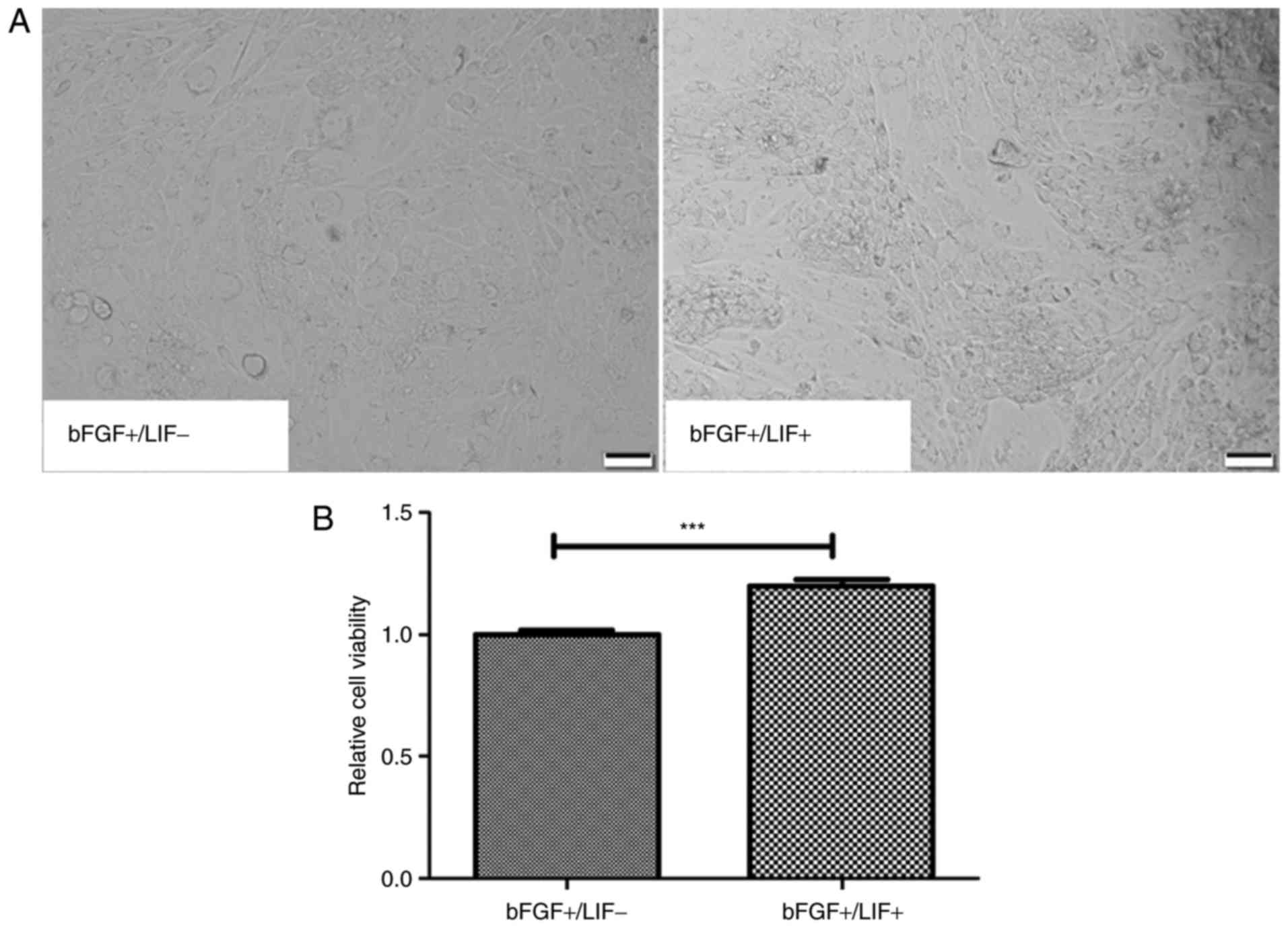
maintaining marmoset iPSCs were investigated. First, the morphology
and proliferation of marmoset iPSC clones were compared when
cultured in media containing different growth factor combinations
either in the presence of LIF (LIF+/bFGF+) or
in the absence of LIF (LIF−/bFGF+) for 24 h.
The morphology of the marmoset iPSCs remained unchanged, whereas
the cloning efficiency of the marmoset iPSCs was visibly increased
in the presence of LIF (Fig. 1A).
To further characterize the ability of LIF to support the
proliferation of marmoset iPSCs, an MTT assay was performed. The
results suggested that LIF significantly increased the cell
viability ratio, compared with that in the control (P<0.01;
Fig. 1B).
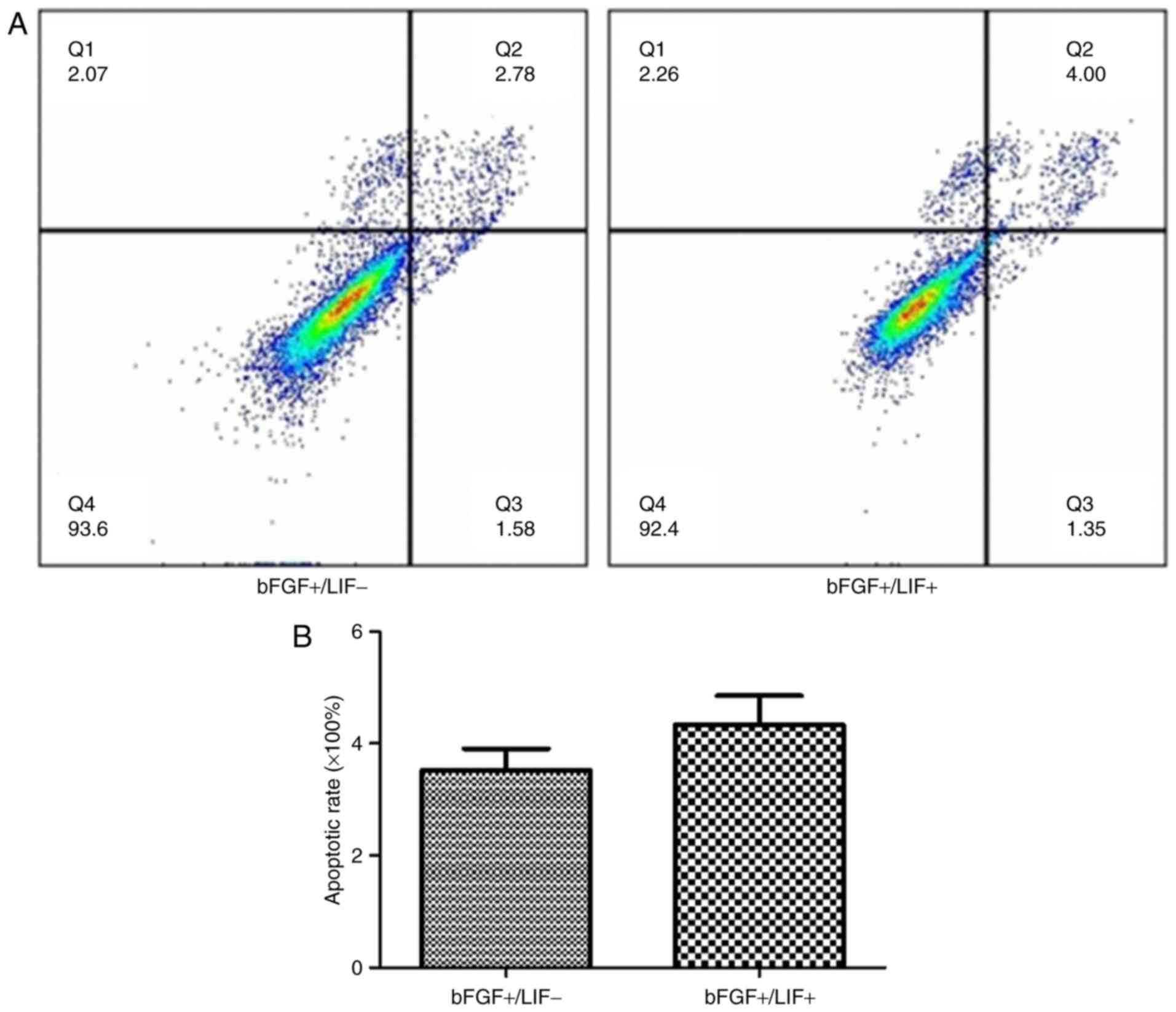
Subsequently, the effects of LIF on cell apoptosis
were evaluated. To assess cell apoptosis, the samples were
subjected to flow cytometric analysis. The results showed that the
apoptotic rate in the treatment group was 4.81±1.51% and that in
the control group was 4.25±1.01%. No significant difference was
found in the cell apoptotic rate between the treatment group and
control group (Fig. 2A and B).
These results revealed that the presence of LIF had no effect on
apoptosis.
Differentially expressed genes of
marmoset iPSCs in the presence or absence of LIF
The present study analyzed the differences between
the stages of pluripotency in the presence and absence of LIF
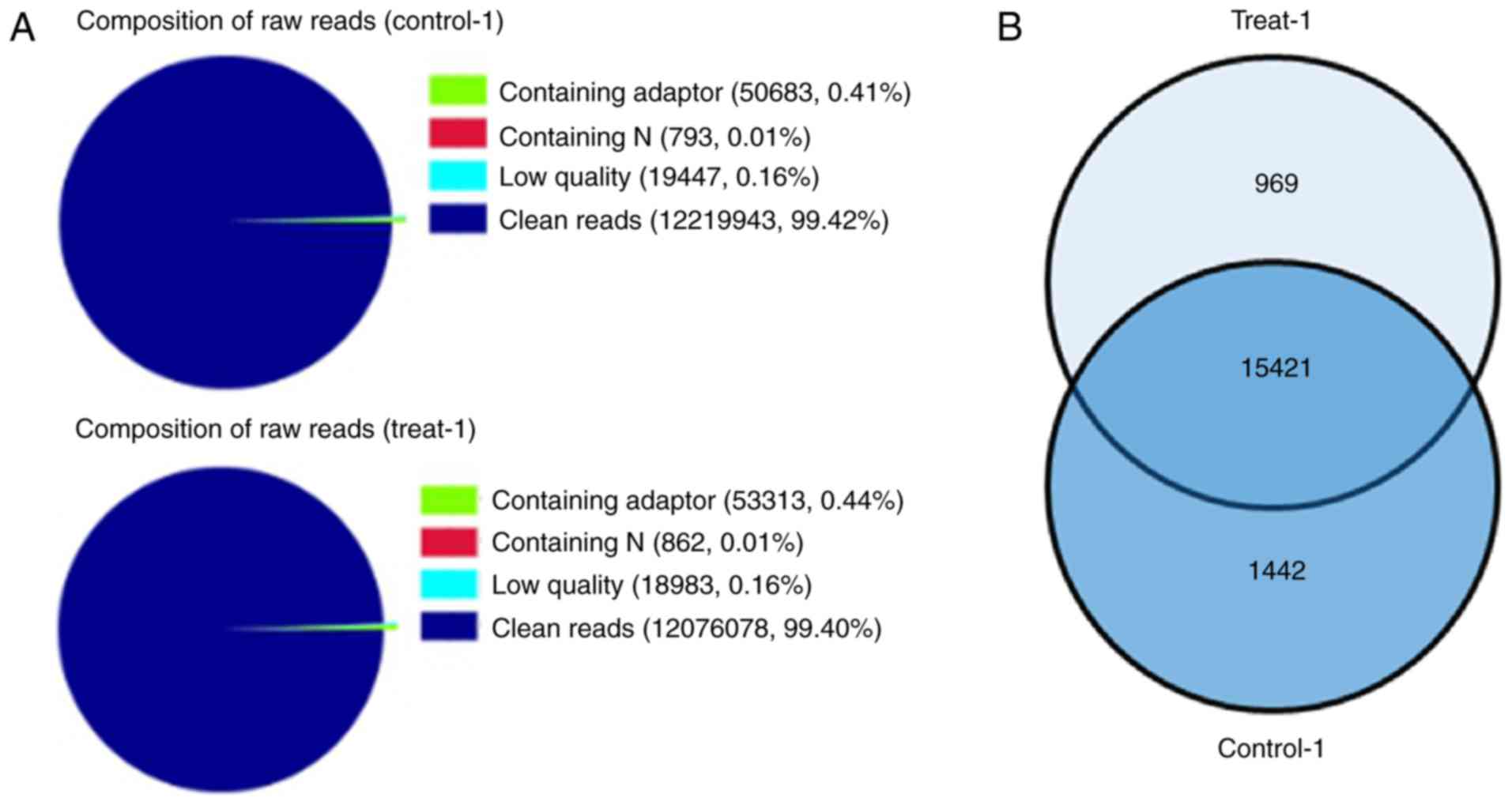
through DGE analysis. Two DGE libraries were constructed from the
control group and LIF-treated group using Solexa technology. Of the
total tags, 99.42 and 99.40% of the tags in each group,
respectively, were clean tags, and were used for the analysis
(Fig. 3A). The results suggested
that the sequences were sufficiently reliable to meet the
requirements for the experiment. A total of 15,421 genes were
shared between the two groups (Fig.
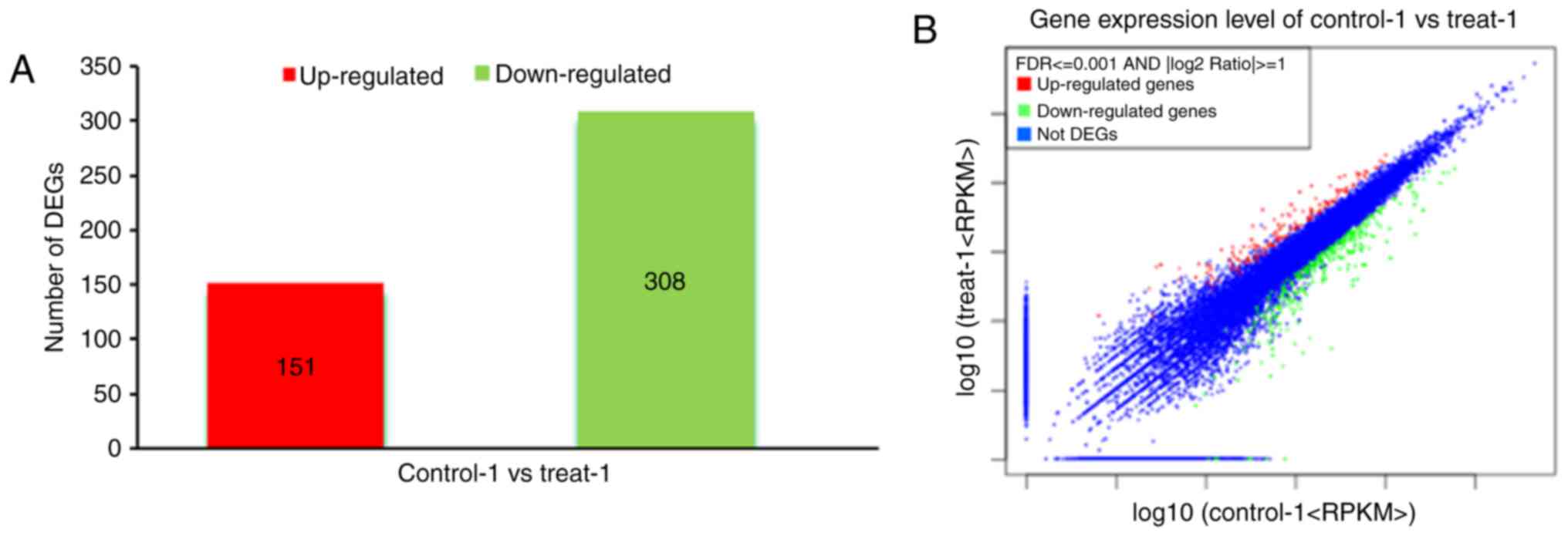
3B). In the common set of genes, a total of 459 differentially
expressed candidate genes were detected between the two groups,
among which 151 genes were upregulated and 308 genes were
downregulated (Fig. 4A and B). To
determine the functions of these differentially expressed genes in
inducing proliferation of marmoset iPSCs, a functional enrichment
analysis was performed, including all of the differentially
expressed pluripotency-associated genes. The 1.0-fold upregulated
genes in the samples were defined as significantly differentially
expressed genes or regulator genes following the addition of LIF.
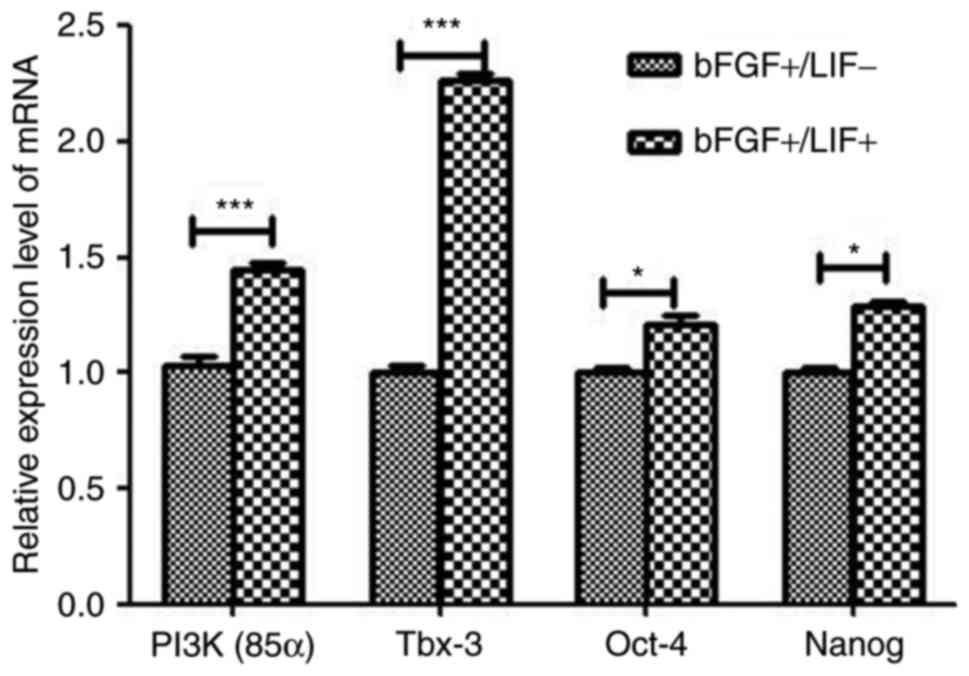
The analyses indicated that the expression levels of Tbx-3 and PI3K
were significantly higher in the LIF-treated group, compared with
those in the control group (Table
II). The expression levels of PI3K and Tbx-3 were significantly
increased, whereas those of others, including NANOG and SOX2, did
not show a significant increase. To confirm the DGE results,
RT-qPCR analysis was performed to confirm the expression levels of
pluripotency-associated genes. The results showed a marked
upregulation in the expression of Tbx-3 and PI3K (P<0.01;
Fig. 5), therefore, the results
were generally consistent with the DGE data.
 | Table IIDifferentially expressed
pluripotency-associated and signaling-related genes in marmoset
induced pluripotent stem cells cultured with leukemia inhibitory
factor. |
Table II
Differentially expressed
pluripotency-associated and signaling-related genes in marmoset
induced pluripotent stem cells cultured with leukemia inhibitory
factor.
| Gene | ID |
Control-Exp/RPKM | Treat-Exp/RPKM | log2 ratio
(treat/control) | P-value | FDR |
|---|
| OCT4 | NM_001265584.1 | 6,712/1,492.90 |
10,982/2,532.61 | 0.76 |
1.13×10−263 |
2.25×10−260 |
| NANOG | XM_002752302.1 | 85/22.29 | 104/28.29 | 0.34 | 0.1027 | 0.2932 |
| TBX-3 | XM_002753045.1 | 0/0.01 | 1/0.10 | 3.31 | 0.4818 | 0.7507 |
| PI3K (PIK3CG) | XM_002751727.1 | 61/3.83 | 185/12.06 | 1.65 |
6.68×10−17 |
3.12×10−15 |
LIF-induced upregulation of Tbx-3 is
mediated by the PI3K/Akt pathway
As LIF has previously been demonstrated to integrate
the Jak/Stat3 and PI3K/Akt signaling pathways to maintain the
pluripotency and survival of mESCs (15,16,18,25,26), it is reasonable to hypothesize
that one or both of these signaling processes may be key in the
activation of pluripotency-associated genes to maintain marmoset
iPSCs. To confirm this hypothesis, the cells were treated with or
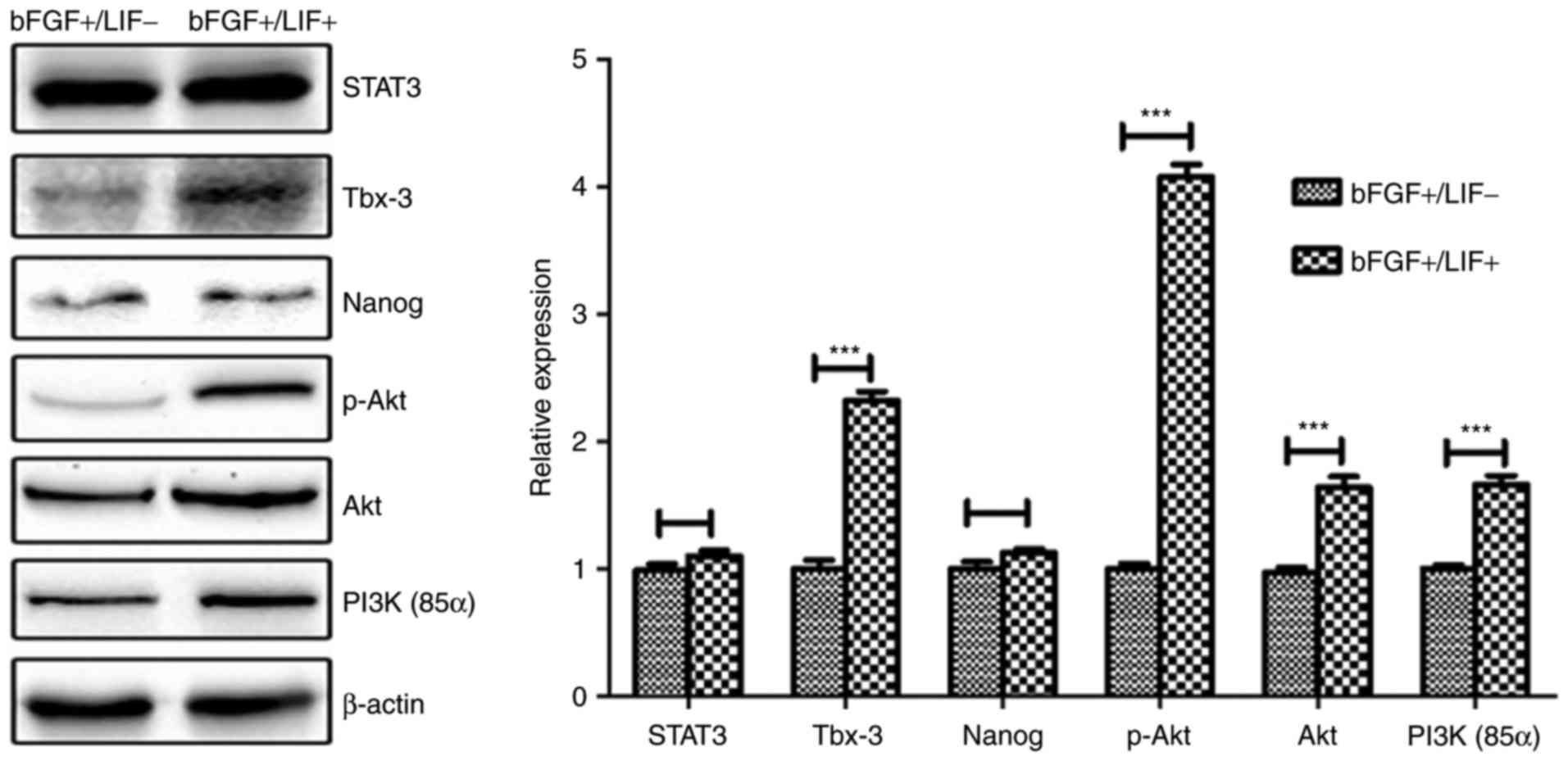
without LIF for 24 h, and the protein levels of STAT3, PI3K, AKT,
p-AKT, Tbx-3 and NANOG were assessed by western blot analysis.
Compared with the control group, treatment with LIF resulted in a
significant increase in the levels of p-AKT and Tbx-3, whereas
minimal change in the level of STAT3 was observed (Fig. 6), suggesting that the LIF-induced
upregulation of Tbx-3 was potentially mediated by the PI3K/Akt
pathway, but not the Jak/Stat3 pathway.
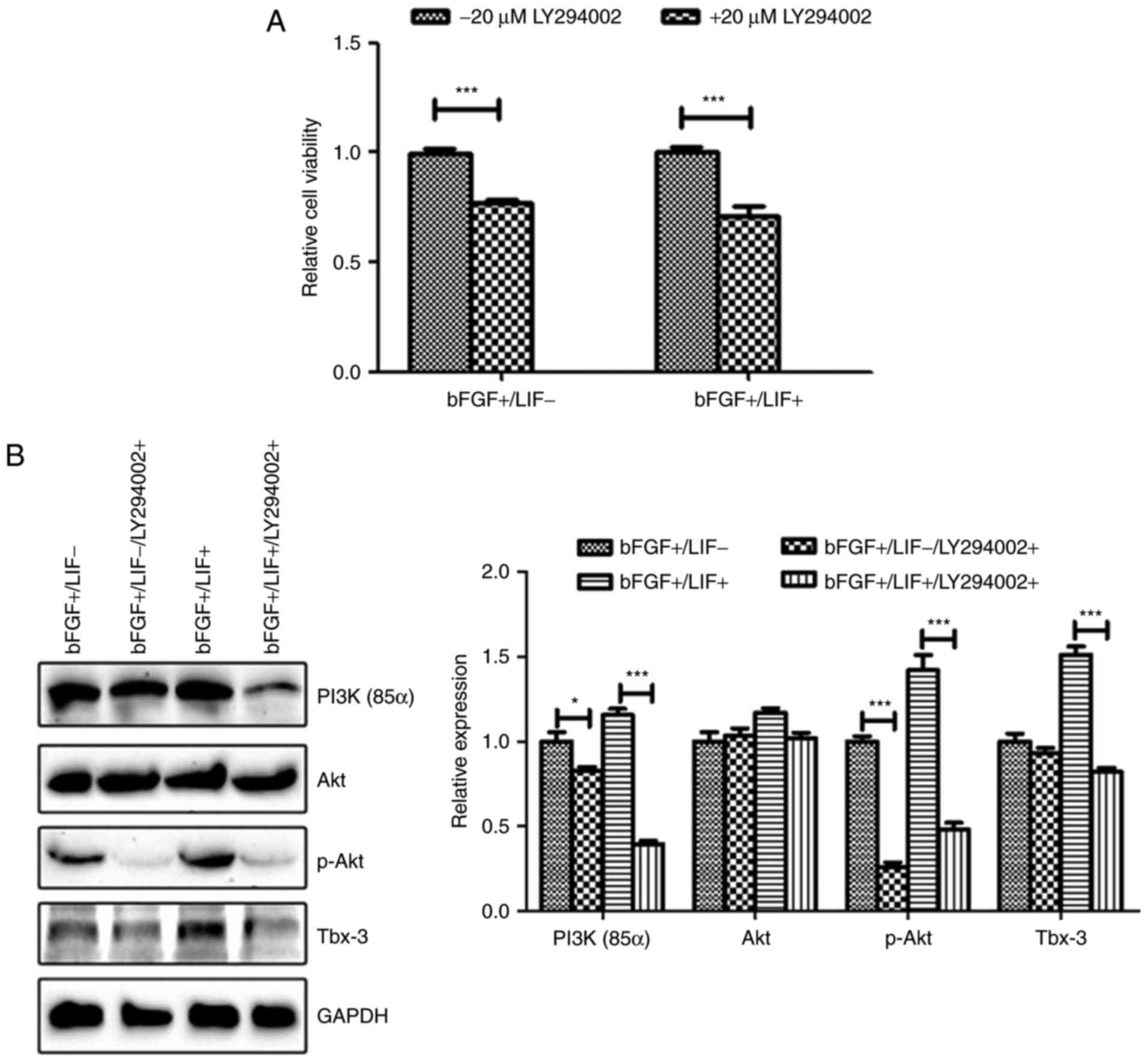
To confirm the above hypothesis, the present study
analyzed marmoset iPSCs cultured in the presence of LY294002, a
PI3K inhibitor. The results of the cell proliferation assay
demonstrated that inhibition of the PI3K/Akt pathway significantly
suppressed cell viability, even in the presence of LIF (P<0.01;
Fig. 7A). Additionally, western
blot analysis was performed to detect the levels of p-AKT and
Tbx-3. The results revealed that the upregulation of p-AKT and
Tbx-3 in marmoset iPSCs by LIF was significantly impaired following
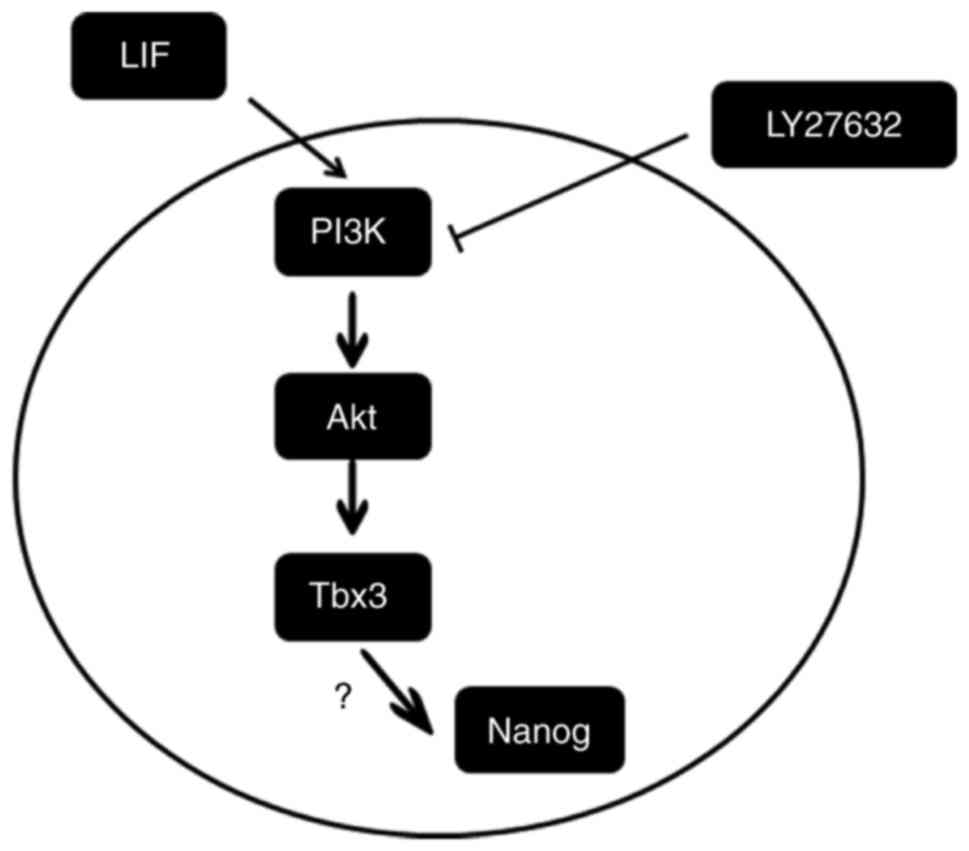
culture of the marmoset iPSCs in the presence of LY294002 (Fig. 7B). Taken together, these results
indicated that LIF promoted the expression of the
pluripotency-associated gene Tbx-3 by activating the PI3K/Akt
signaling pathway (Fig. 8).
Discussion
Marmosets have been widely recognized as vital,
non-human primate models for disease research and preclinical
assessment (27). Therefore,
marmoset iPSCs offer potential not only for basic mechanism
evaluation, but also for therapeutic applications, including
regenerative medicine (2).
Increasing advances in the field of basic investigations, including
the generation of marmoset iPSCs and development of culture
conditions, have provided realistic possibilities for human
regenerative medicine (7–9). To maintain the self-renewal and
pluripotency of marmoset iPSCs in vitro, establishment of a
simple and effective culture condition, containing a series of
cytokines, is significant and urgent. Therefore, the present study
investigated the effects of LIF on maintaining marmoset iPSCs, in
addition to the underlying mechanisms.
In general, mouse ESCs are naïve pluripotent stem
cells, whereas human ESCs are primed pluripotent stem cells,
depending on their patterns of gene expression and the cytokines
required to maintain self-renewal (19,20,28). An experimental culture of human
ESCs requires bFGF supplementation, whereas mESCs are LIF-dependent
in vitro (29). The
application of LIF to establish and maintain marmoset ESCs has long
been controversial (3–6). In our previous study, it was found
that bFGF is essential and critical for maintaining marmoset iPSCs
(7,10). In the present study, the
self-renewal ability of marmoset iPSCs in a feeder-free culture was
markedly promoted by LIF, in contrast to the characteristic of
human iPSCs (20). Based on the
morphology of marmoset iPSCs, it was determined that these cells
progressed towards a naïve pluripotency stage in the presence of
LIF and that marmoset iPSCs have the potential for differentiation
(data not shown).
To clarify the molecular mechanisms by which LIF
sustains the self-renewal and pluripotency of marmoset iPSCs, DGE
analysis was performed. The results showed that the expression
levels of Tbx-3 and PI3K were significantly upregulated. Tbx-3, a
known transcriptional repressor, is a member of the T-box
transcription factor family and is important in embryonic
development and cell fate determination (30,31). Previous studies have demonstrated
that the expression of Tbx-3 is associated with the maintenance of
pluripotency and self-renewal of ES cells, in addition to the
facilitation of reprogramming and establishment of iPSCs (32–36). The present data showed that the
effect of LIF on the core circuitry of proliferation and
pluripotency in marmoset iPSCs was mediated by the activation of
Tbx-3. A previous study showed that the overexpression of Tbx-3
promoted human ES cell proliferation; however, Tbx-3 knockdown
resulted in decreased neuroepithelial differentiation (37). Furthermore, the knockdown of Tbx-3
resulted in the loss of pluripotency and differentiation of mESCs
(38) and also attenuated the
self-renewal ability of mESCs (39), suggesting that Tbx-3 is necessary
for maintaining self-renewal ability. Notably, the overexpression
of Tbx-3 has been found to be sufficient to maintain mESCs in an
undifferentiated state in the absence of LIF (18). The knockdown of Tbx-3 has been
shown to prevent extra-embryonic endoderm differentiation, but
enhance ectoderm and trophectoderm differentiation (39). It has been reported that the
expression of Tbx-3 is downregulated for several days following LIF
withdrawal (18,39,40). In addition, the downregulation of
Tbx-3 has been shown to attenuate the proliferation of mESCs in the
presence of LIF (39).
In mESCs, three LIF signal pathways are involved via
different transcription factors (15–17). Briefly, LIF engagement of its
receptor results in a cascade of tyrosine phosphorylation, which
stimulates three distinct signaling pathways: The Jak/Stat3 pathway
primarily activates Kruppel-like factor 4, whereas the PI3K-Akt and
MAPK pathways regulate Tbx-3 (17,18). In marmoset ESCs, Nii et al
reported that LIF activated the Jak-Stat3 pathway, but did not
affect the capacity of self-renewal (6). In the present study, it was
identified that LIF activated Tbx-3 through the PI3K-Akt pathway to
maintain marmoset iPSC pluripotency and self-renewal. Activation of
the PI3K pathway following LIF stimulation was driven by the
phosphorylation of the p85 subunit, a regulatory subunit of PI3K,
consistent with previous studies (17,41). In addition, the effect of LIF was
independent of the Jak-Stat3 pathway. Notably, the DGE data showed
a marginal shift in the expression of NANOG and octamer-binding
protein 4 (OCT4) in the LIF-treated group, but subsequent
validation assessment indicated that the expression levels of NANOG
and OCT4 were significantly increased under the same conditions. In
the pluripotency network, Tbx-3 interacts with the pluripotency
factors NANOG and OCT4 to maintain the stem cell state and inhibit
differentiation (42). Niwa et
al reported that Tbx-3 preferentially activates Nanog and
subsequently maintains the expression of Oct3/4 in mESCs (18). In marmoset iPSCs, the expression
level of NANOG and OCT4 may be upregulated by the activation of
Tbx-3, which requires further investigation.
In conclusion, the present study is the first, to
the best of our knowledge, to show that LIF is critical in
maintaining the pluripotency and viability of marmoset iPSCs.
Furthermore, the effect and mechanism of LIF on marmoset iPSCs was
found to involve the activation of pluripotency factor Tbx-3 by the
PI3K/Akt signaling pathway. Therefore, the application of LIF
provided a simple and effective culture condition for maintaining
the self-renewal and pluripotency of marmoset iPSCs.
Acknowledgments
The authors would like to thank Dr Peter Hornsby
(Department of Physiology and The Barshop Institute for Longevity
and Aging Studies, University of Texas Health Science Center, San
Antonio, TX 78245, USA) for kindly providing the marmoset iPS
cells.
References
|
1
|
De Vos J, Bouckenheimer J, Sansac C,
Lemaître JM and Assou S: Human induced pluripotent stem cells: A
disruptive innovation. Curr Res Transl Med. 64:91–96. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu Y, Mishra A, Qiu Z, Farnsworth S,
Tardif SD and Hornsby PJ: Nonhuman primate induced pluripotent stem
cells in regenerative medicine. Stem Cells Int. 2012:7671952012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thomson JA, Kalishman J, Golos TG, Durning
M, Harris CP and Hearn JP: Pluripotent cell lines derived from
common marmoset (Callithrix jacchus) blastocysts. Biol Reprod.
55:254–259. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sasaki E, Hanazawa K, Kurita R, Akatsuka
A, Yoshizaki T, Ishii H, Tanioka Y, Ohnishi Y, Suemizu H, Sugawara
A, et al: Establishment of novel embryonic stem cell lines derived
from the common marmoset (Callithrix jacchus). Stem Cells.
23:1304–1313. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Müller T, Fleischmann G, Eildermann K,
Mätz-Rensing K, Horn PA, Sasaki E and Behr R: A novel embryonic
stem cell line derived from the common marmoset monkey (Callithrix
jacchus) exhibiting germ cell-like characteristics. Hum Reprod.
24:1359–1372. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nii T, Marumoto T, Kawano H, Yamaguchi S,
Liao J, Okada M, Sasaki E, Miura Y and Tani K: Analysis of
essential pathways for self-renewal in common marmoset embryonic
stem cells. FEBS Open Bio. 4:213–219. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu Y, Zhang Y, Mishra A, Tardif SD and
Hornsby PJ: Generation of induced pluripotent stem cells from
newborn marmoset skin fibroblasts. Stem Cell Res. 4:180–188. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Debowski K, Warthemann R, Lentes J,
Salinas-Riester G, Dressel R, Langenstroth D, Gromoll J, Sasaki E
and Behr R: Non-viral generation of marmoset monkey iPS cells by a
six-factor-in-one-vector approach. PloS One. 10:e01184242015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tomioka I, Maeda T, Shimada H, Kawai K,
Okada Y, Igarashi H, Oiwa R, Iwasaki T, Aoki M, Kimura T, et al:
Generating induced pluripotent stem cells from common marmoset
(Callithrix jacchus) fetal liver cells using defined factors,
including Lin28. Genes Cells. 15:959–969. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu Y, Shu J, He C, Li M, Wang Y, Ou W and
He Y: ROCK inhibitor Y27632 promotes proliferation and diminishes
apoptosis of marmoset induced pluripotent stem cells by suppressing
expression and activity of caspase 3. Theriogenology. 85:302–314.
2016. View Article : Google Scholar
|
|
11
|
Cullinan EB, Abbondanzo SJ, Anderson PS,
Pollard JW, Lessey BA and Stewart CL: Leukemia inhibitory factor
(LIF) and LIF receptor expression in human endometrium suggests a
potential autocrine/paracrine function in regulating embryo
implantation. Proc Natl Acad Sci USA. 93:3115–3120. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mathieu ME, Saucourt C, Mournetas V,
Gauthereau X, Thézé N, Praloran V, Thiébaud P and Boeuf H:
LIF-dependent signaling: New pieces in the Lego. Stem Cell Rev.
8:1–15. 2012. View Article : Google Scholar :
|
|
13
|
McKenzie RC and Szepietowski J: Cutaneous
leukemia inhibitory factor and its potential role in the
development of skin tumors. Dermatol Surg. 30:279–290.
2004.PubMed/NCBI
|
|
14
|
Gadient RA and Patterson PH: Leukemia
inhibitory factor, Interleukin 6, and other cytokines using the
GP130 transducing receptor: Roles in inflammation and injury. Stem
Cells. 17:127–137. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zouein FA, Kurdi M and Booz GW: LIF and
the heart: Just another brick in the wall. Eur Cytokine Netw.
24:11–19. 2013.PubMed/NCBI
|
|
16
|
Ohtsuka S, Nakai-Futatsugi Y and Niwa H:
LIF signal in mouse embryonic stem cells. JAKSTAT.
4:e10865202015.PubMed/NCBI
|
|
17
|
Nicola NA and Babon JJ: Leukemia
inhibitory factor (LIF). Cytokine Growth Factor Rev. 26:533–544.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Niwa H, Ogawa K, Shimosato D and Adachi K:
A parallel circuit of LIF signalling pathways maintains
pluripotency of mouse ES cells. Nature. 460:118–122. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hanna JH, Saha K and Jaenisch R:
Pluripotency and cellular reprogramming: Facts, hypotheses,
unresolved issues. Cell. 143:508–525. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hirai H, Firpo M and Kikyo N:
Establishment of LIF-dependent human iPS cells closely related to
basic FGF-dependent authentic iPS cells. PloS One. 7:e390222012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tang LD, Wang XM, Jin FL, Qiu BL, Wu JH
and Ren SX: De novo sequencing-based transcriptome and digital gene
expression analysis reveals insecticide resistance-relevant genes
in De novo sequencing-based transcriptome and digital gene
expression analysis reveals insecticide resistance-relevant genes
in Propylaea japonica (Thunberg) (Coleoptea: Coccinellidae)
(Thunberg) (Coleoptea: Coccinellidae). PloS One. 9:e1009462014.
View Article : Google Scholar
|
|
22
|
Götz S, García-Gómez JM, Terol J, Williams
TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J and Conesa A:
High-throughput functional annotation and data mining with the
Blast2GO suite. Nucleic Acids Res. 36:3420–3435. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Yi F, Yang F, Liu X, Chen H, Ji T, Jiang
L, Wang X, Yang Z, Zhang LH, Ding X, et al: RNA-seq identified a
super-long intergenic transcript functioning in adipogenesis. RNA
Biol. 10:991–1001. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tan G, Cheng L, Chen T, Yu L and Tan Y:
Foxm1 mediates LIF/Stat3-dependent self-renewal in mouse embryonic
stem cells and is essential for the generation of induced
pluripotent stem cells. PloS One. 9:e923042014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu N, Lu M, Feng XM, Ma FX, Fang ZH, Tian
XM, Ren Q, Zhang L, Liu B, Huang PP, et al: Exogenous Nanog
alleviates but is insufficient to reverse embryonic stem cells
differentiation induced by PI3K signaling inhibition. J Cell
Biochem. 106:1041–1047. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Saito A: The marmoset as a model for the
study of primate parental behavior. Neurosci Res. 93:99–109. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Buecker C and Geijsen N: Different flavors
of pluripotency, molecular mechanisms, and practical implications.
Cell Stem Cell. 7:559–564. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nichols J and Smith A: Naive and primed
pluripotent states. Cell Stem Cell. 4:487–492. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Govoni KE, Linares GR, Chen ST,
Pourteymoor S and Mohan S: T-box 3 negatively regulates osteoblast
differentiation by inhibiting expression of osterix and runx2. J
Cell Biochem. 106:482–490. 2009. View Article : Google Scholar
|
|
31
|
Dan J, Li M, Yang J, Li J, Okuka M, Ye X
and Liu L: Roles for Tbx3 in regulation of two-cell state and
telomere elongation in mouse ES cells. Sci Rep. 3:34922013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao D, Wu Y and Chen K: Tbx3 isoforms are
involved in pluripotency maintaining through distinct regulation of
Nanog transcriptional activity. Biochem Biophys Res Commun.
444:411–414. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Willmer T, Peres J, Mowla S, Abrahams A
and Prince S: The T-Box factor TBX3 is important in S-phase and is
regulated by c-Myc and cyclin A-CDK2. Cell Cycle. 14:3173–3183.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Willmer T, Hare S, Peres J and Prince S:
The T-box transcription factor TBX3 drives proliferation by direct
repression of the p21 (WAF1) cyclin-dependent kinase inhibitor.
Cell Div. 11:62016. View Article : Google Scholar
|
|
35
|
Han J, Yuan P, Yang H, Zhang J, Soh BS, Li
P, Lim SL, Cao S, Tay J, Orlov YL, et al: Tbx3 improves the
germ-line competency of induced pluripotent stem cells. Nature.
463:1096–1100. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Russell R, Ilg M, Lin Q, Wu G, Lechel A,
Bergmann W, Eiseler T, Linta L, Kumar PP, Klingenstein M, et al: A
dynamic role of TBX3 in the pluripotency circuitry. Stem Cell
Reports. 5:1155–1170. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Esmailpour T and Huang T: TBX3 promotes
human embryonic stem cell proliferation and neuroepithelial
differentiation in a differentiation stage-dependent manner. Stem
Cells. 30:2152–2163. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ivanova N, Dobrin R, Lu R, Kotenko I,
Levorse J, DeCoste C, Schafer X, Lun Y and Lemischka IR: Dissecting
self-renewal in stem cells with RNA interference. Nature.
442:533–538. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lu R, Yang A and Jin Y: Dual functions of
T-box 3 (Tbx3) in the control of self-renewal and extraembryonic
endoderm differentiation in mouse embryonic stem cells. J Biol
Chem. 286:8425–8436. 2011. View Article : Google Scholar :
|
|
40
|
Galan-Caridad JM, Harel S, Arenzana TL,
Hou ZE, Doetsch FK, Mirny LA and Reizis B: Zfx controls the
self-renewal of embryonic and hematopoietic stem cells. Cell.
129:345–357. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Storm MP, Kumpfmueller B, Thompson B,
Kolde R, Vilo J, Hummel O, Schulz H and Welham MJ: Characterization
of the phosphoinositide 3-kinase-dependent transcriptome in murine
embryonic stem cells: Identificatio n of novel regulators of
pluripotency. Stem Cells. 27:764–775. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lee DF, Su J, Sevilla A, Gingold J,
Schaniel C and Lemischka IR: Combining competition assays with
genetic complementation strategies to dissect mouse embryonic stem
cell self-renewal and pluripotency. Nat Protoc. 7:729–748. 2012.
View Article : Google Scholar : PubMed/NCBI
|