Introduction
The majority of patients with type 2 diabetes
mellitus (T2DM) have the characteristics of high blood lipids in
addition to high glucose (1,2).
High plasma free fatty acids (FFAs) are associated with insulin
resistance, inflammation and oxidative stress, which are a result
of advanced diabetes mellitus (3). Therefore, high glucose combined with
high FFAs can contribute to the unfavorable development of T2DM. By
contrast, monocytes/macrophages are important in the occurrence and
development of T2DM, which is regarded as a type of low-grade
inflammation (4,5). In our previous study, it was
demonstrated that the increased expression of P2X7
receptor (P2X7R) in peripheral blood monocytes may alter
the innate immune system, contributing to the development of T2DM
(6). The high plasma FFA levels
in the inflammatory condition of T2DM may result in the continuous
secretion of intracellular ATP, which causes the accumulation of
extracellular ATP (7).
Overstimulation of the P2X7R by increasing levels of
extracellular ATP may induce uncontrolled Ca2+ influx
and activate the release of mature cytokines, including interleukin
(IL)-1β, IL-18 and tumor necrosis factor (TNF)-α (8,9).
Transcriptome analysis has revealed that only 1-2%
of the human gene encodes protein and the remaining 98% of the
human gene is non-coding RNA (10,11). Long non-coding RNAs (lncRNAs)
represent a subgroup of non-coding RNAs, which are >200
nucleotides in length and lack the ability to encode proteins
(10). Studies have shown that
lncRNAs regulate various biological processes, including the
expression of genesin epigenetics at the transcription and
post-transcription levels (12)
and they are involved in the regulation of apoptosis, proliferation
and differentiation (13). In
previous years, studies have identified several hundred lncRNAs
characterized with absolute conservation (100% homology with no
insertions or deletions) in the human, mouse and rat genomes
(14,15). These conserved lncRNAs have been
named ultra conserved RNA (ucRNA). The uc.48+ is one identified
lncRNA (http://genome.ucsc.edu). Although our
previous preliminary results revealed that lncRNA uc.48+ was
involved in diabetic neuropathic pain (16), the role of uc.48+ in
monocyte/macrophages has not been reported. Therefore, the present
study focused on the role of uc.48+ and its possible mechanism in
high glucose and FFA-induced immune and inflammatory responses
mediated by P2X7R in RAW264.7 macrophages.
Materials and methods
Individuals and serum RNA isolation
A total of 20 healthy subjects were enrolled from
the Blood Center of Jiangxi Province (Nanchang, China). In
addition, 38 patients fulfilling the American Diabetes Association
diagnostic criteria for T2DM, including 20 patients with normal
C-reactive protein (CRP) and 18 patients with high CRP, were
recruited from the First Affiliated Hospital of Nanchang University
(Nanchang, China). The present study was approved by the local
Ethics Committee of from the First Affiliated Hospital of Nanchang
University (approval no. 2018 16), and written informed consent was
obtained from each individual.
The participant recruitment/samples were collected
between June 1, 2014 and August 31, 2014. Blood samples obtained
from all individuals were collected and allowed to coagulate for 30
min at room temperature, followed by centrifugation for 10 min at
1,300 × g at 4°C. The collected sera were centrifuged for another
10 min at 3,000 × g at 4°C to remove any remaining cellular
components, divided into 500-µl aliquots, and stored
immediately at −80°C.
Cell culture exposure to high glucose and
FFAs
The RAW 264.7 murine macrophage-like cell line was
purchased from the Type Culture Collection of the Chinese Academy
of Sciences (Shanghai, China). The cells were cultured in DMEM
medium (low glucose; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA; cat. no. 11885092) containing 10% fetal bovine
serum (FBS; Hyclone; GE Healthcare Life Sciences, Logan, UT, USA),
100 U/ml penicillin, and 100 mg/ml streptomycin sulphate at 37°C in
a humidified atmosphere containing 5% CO2. Based on a
previous report, the treatment doses of high glucose (17) and high FFAs (18) were 44.4 and 1.0 mM, respectively,
throughout the study.
uc.48+ siRNA treatment
The siRNA specific for uc.48+ was purchased from
Invitrogen; Thermo Fisher Scientific, Inc., with the target
sequence of 5′-GGC ACT ACT ACT TGC AGA A-3′. The siRNA
oligonucleotides that specifically targeted uc.48+ were used in
this experiment (16). The uc.48+
was knocked down by RNA interference using Entranster™ RNA
transfection reagent. According to the manufacturer's protocol of
the transfection reagent (Engreen Biosystem Co., Ltd. Beijing,
China), the uc.48+ siRNA injection at a final concentration of 20
mM was used for subsequent experiments.
Cell treatment and experimental
groups
The RAW264.7 macrophages were treated under control
[5.5 mM glucose +1% bovine serum albumin (BSA; Zhejiang Tianhang
Biotechnology Co., Ltd. Hangzhou, China], high glucose and high FFA
(44.4 and 1.0 mM, respectively) conditions for 3 days. RNA was
isolated following treatment in the presence of uc.48+ siRNA or
scramble siRNA for 48 h. The other experiment assays (western blot,
cell viability and apoptosis assays, and measurement of ROS) were
performed following treatment in the presence of uc.48+ siRNA or
scramble siRNA for 72 h. The supernatants were stored at −80°C for
the enzyme-linked immunosorbent assay. For experiments, the four
following groups were assigned: Control group, high glucose and FFA
treatment (HGHF) group, uc.48+ siRNA vector-treated HGHF group
(HGHF+ uc.48+ si), and scramble siRNA vector-treated HGHF group
(HGHF+ NC si).
Serum RNA and total RNA isolation, and
reverse transcription-polymerase chain reaction (RT-PCR)
analysis
Serum RNA was isolated from the serum using RNA pure
Circulating Reagent (CWBIO, Beijing, China; cat. no. CW2281), with
modifications. Briefly, 300 µl of serum was added to three
volumes of RNA pure Circulating Reagent, mixed thoroughly by
vortexing and left to stand at room temperature for 5 min.
Subsequently, one-fifth of the volume of chloroform was added and
shaken vigorously for 30 sec, incubated for 5 min at room
temperature, and then centrifuged for 20 min at 12,000 × g at 4°C.
The upper aqueous phase was transferred into a new collection tube
and one volume of isopropanol was added, mixed thoroughly for 30
min at room temperature, and centrifuged for 20 min at 12,000 × g
at 4°C. The precipitate was washed twice with 1 ml of 75% ethanol
(diluted with DEPC water). The RNA was eluted by 20 µl of
RNase-free water. The RAW264.7 macrophages were treated with the
control, high glucose and high FFA conditions in the presence of
uc.48+ siRNA or scramble siRNA for 48 h. Total RNA was isolated
using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. The quality of the RNA
was profiled using NanoDrop 2000 spectrophotometer (Thermo Fisher
Scientific, Inc.).
RNA was used to synthesize cDNA in 20 µl
reactions containing 500 ng of DNase-treated RNA, 200 ng of random
hexamers, 100 U of Revert Aid reverse transcriptase and 8 units of
RiboLock RNase Inhibitor using a Revert Aid First Strand cDNA
Synthesis kit (Fermentas; Thermo Fisher Scientific, Inc.). The
reaction samples were incubated for 10 min at 25°C and 60 min at
42°C, followed by enzyme inactivation for 10 min at 70°C and
storage at −20°C until use. In the serum experiments, PCR
amplification of the uc.48+ and GAPDH (internal standard for
quantification) was performed according to our previously described
method (19). A total of 2
µl cDNA was amplified in 12.5 µl of PCR mixture
(Tiangen Biotech Co., Ltd. Beijing, China), 2 µl primers (1
µl sense primer and 1 µl antisense primer) and 8.5
µl nuclease-free water. The oligonucleotides used to amplify
the uc.48+ and GAPDH were as follows: uc.48+, sense 5′-GCA AAC TGG
ATG AGG AT-3′ and antisense 5′-GTA GTG CCA CAA GGA GA-3′, and
GAPDH, sense 5′-CAG GGC TGC TTT TAA CTC TGG T-3′ and antisense
5′-GAT TTTGGA GGG ATC TCG CT-3′. The length of the PCR product was
231 bp for uc.48+ and 199 bp for GAPDH. The PCR conditions of
uc.48+ and GAPDH included a hot start at 94°C for 3 min, 45 sec
denaturation (94°C), 45 sec annealing (57°C), and 45 sec extension
(72°C) for 36 (uc.48+) or 32 (GAPDH) amplification cycles, and a
final 5-min extension at 72°C.
In the RAW264.7 cell experiment, the PCR
amplification of the P2X7R and β-actin (internal
standard for quantification) was performed according to our
previous method using oligonucleotides as described previously
(16). A total of 2 µl
cDNA was amplified in 12.5 µl PCR mixture (Tiangen Biotech
Co., Ltd.), 2 µl primers (1 µl sense primer and 1
µl antisense primer) and 8.5 µl nuclease-free water.
The following primers were used for PCR analysis: P2X7R
(171 bp), sense 5′-GCA CGA ATT ATG GCA CCG TC-3′ and antisense
5′-CCC CAC CCT CTG TGA CAT TC-3′; uc.48+ (231 bp), sense 5′-GCA AAC
TGG ATG AGG AT-3′ and antisense 5′-GTA GTG CCA CAA GGA GA-3′;
β-actin, (240 bp), sense 5′-TAA AGA CCT CTA TGC CAA CAC AGT-3′ and
antisense 5′-CAC GAT GGA GGG GCC GGA CTC ATC-3′. The PCR conditions
of uc.48+, P2X7R and β-actin included a hot start at
94°C for 3 min, 45 sec denaturation (94°C), 45 sec annealing (57°C
for uc.48+ and β-actin; 59°C for P2X7R), and 45 sec
extension (72°C) for 36 (uc.48+ and P2X7R) or 32
amplification cycles (β-actin) and a 5-min final extension at
72°C.
The PCR products were run on 1.5% agarose gels with
EB using standard protocols. Densitometric analysis of three
different observations was performed using Image-Pro Plus software
(version 6.0, Media Cybernetics, Inc., Rockville, MD, USA). In the
experiments involving patient serum samples, the results are
expressed as the ratio of the uc.48+ band intensity/GAPDH band
intensity. In the RAW264.7 cell experiments, the results are showed
as the ratio of uc.48+ or P2X7R band intensity to
β-actin band intensity.
Western blot analysis
The cells were washed twice with ice-cold PBS and
harvested. The lysates were obtained using RIPA buffer containing a
protease/phosphatase inhibitor mixture (diluted 1:100, Vazyme
Biotech, Nanjing China) and then subjected to 10,000 × g
centrifugation at 4°C for 20 min. Total protein concentrations in
the supernatant were determined using a bicinchoninic acid assay
(Beyotime Institute of Biotechnology, Haimen, China). A total of 30
µg protein was separated by 6% sodium
deodecylsulfate-polyacrylamide gel electrophoresis, which was then
transferred onto polyvinyl difluoride membranes (Millipore,
Bedford, MA, USA) by electroblotting. Following blocking with 5%
BSA at room temperature for 2 h, the blots were incubated with the
following primary antibodies: P2X7R (diluted 1:800, cat.
no. ab229453, Abcam, Cambridge, MA, USA), extracellular
signal-regulated kinase (ERK)1/2 and phosphorylated (p-)ERK
(diluted 1:1,000, cat. no. 5013 and 8544, respectively, Cell
Signaling Technology, Inc., Danvers, MA, USA); β-actin (diluted
1:1,500, cat. no. TA09, Beijing Zhongshan Biotech Co, Ltd.,
Beijing, China) overnight at 4°C and developed with appropriate
horseradish peroxidase-conjugated secondary antibodies (diluted
1:2,000, cat. no. ZDR-5306 and ZDR-5307, Beijing Zhongshan Biotech
Co., Ltd.) at room temperature for 60 min. An enhanced
chemiluminescence kit (SuperSignal West Pico; Thermo Fisher
Scientific, Inc.) was used to produce chemiluminescent signals,
which were recorded by a Chemi Doc™ XRS Imaging system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Band intensity was
quantified using Image Pro-Plus software (version 6.0). Protein
expression levels are represented as densitometric ratios of the
targeted protein relative to β-actin or ERK1/2.
Analysis of cell proliferation and
apoptosis
Cell viability was determined using a
3-(4,5-dimethylthiazol-2-yl)-5-(3-car
boxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt
(MTS) assay. The RAW264.7 macrophages were plated in 48-well plates
at a concentration of 2×105 cells/well. At 72 h
post-transfection, the media from the control and high glucoseor
high FFA-treated cultures were discarded and 200 µl of MTS
(Promega Corporation, Madison, WI, USA) was added to each well. The
cells were incubated for 1-2 h at 37°C, following which the cell
solution was measured in a microplate reader (RT-6000; Rayto,
Shengzhen, China) at 490 nm. All experiments were performed in
triplicate.
At 72 h post-transfection, the cells were harvested
and washed three times with PBS, 5 µl of Annexin V-FITC and
5 µl of PI solution were added and the cells were stained
for 15 min in the dark using the Annexin V-FITC Apoptosis Detection
kit (KeyGEN Biotech Co., Ltd., Nanjing, China). The relative
percentage of Annexin V-FITC-positive/PI-negative cells was
analyzed by flow cytometry (Beckman Coulter, Inc., Brea, CA,
USA).
Measurement of ROS
Levels of ROS were measured with
2′,7′-dichlorofluorescin diacetate (DCFH-DA) according to the
manufacturer's protocol. Briefly, the RAW264.7 macrophages were
treated in 24-well dishes and then loaded with DCFH-DA and
incubated under the indicated conditions. Aliquots of cell
suspensions were centrifuged for 6 min at 6,000 × g at 4°C and
added to a cuvette on a spectrofluorometer (Perkin-Elmer, Inc.,
Waltham, MA, USA). The fluorescence generated by DCFH-DA was
monitored and recorded by ratio fluorometry with an emission
wavelength of 525 at 488 nm excitation wavelengths.
Enzyme-linked immunosorbent assay
(ELISA)
The levels of IL-10, IL-1β, and TNF-α were assessed
in supernatants collected from RAW264.7 macrophages using specific
ELISA kits (Boster Biological Technology, Wuhan, China). The cat.
nos. of theIL-10, IL-1β and TNF-α kits were EK0417, EK0394, and
EK0527, respectively, which were used according to the
manufacturer's protocol.
Statistical analysis
All experiments were performed in triplicate to
ensure the accuracy of the results, and data are presented as the
mean ± standard deviation. The data from the cell experiments were
normalized by the control group. Statistical analyses were
performed using the Statistical Package for Social Sciences (SPSS)
11.5 software (SPSS, Inc., Chicago, IL, USA). Statistical
significance was determined by one-way analysis of variance.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Anthropometric data, serum biochemistry
parameters and serum expression of uc.48+ in healthy subjects and
patients with T2DM
The anthropometric data and biochemical parameters
obtained for the three groups are summarized in Table I. The serum expression of uc.48+
in the patients with T2DM was significantly higher, compared with
that in the healthy control subjects (0.84±0.12), and its
expression in patients with T2DM with high CRP levels (1.56±0.15)
was significantly higher compared with that in patients with T2DM
with normal CRP levels (1.33±0.13) (Fig. 1).
 | Table IAnthropometric data and serum
biochemical detection in healthy subjects and patients with
T2DM. |
Table I
Anthropometric data and serum
biochemical detection in healthy subjects and patients with
T2DM.
| Parameter | Healthy
subjects | Patients with T2DM
|
|---|
| Normal CRP | High CRP |
|---|
| Number of patients
(female/male) | 20 (10/10) | 20 (9/11) | 18 (8/10) |
| Weight (kg) | 65.30±8.36 | 68.17±5.63 | 61.29±7.23 |
| Age (years) | 41.70±8.27 | 47.67±7.06 | 52.43±5.34 |
| CRP (mg/l) | 1.57±0.43 | 2.46±0.28 |
8.17±2.95a,b |
| FPG (mmol/l) | 4.60±0.55 | 10.01±3.92a | 9.89±2.14a |
| PPG (mmol/l) | 6.52±0.59 | 14.18±4.06a | 13.86±2.62a |
| FPI (mIU/l) | 5.88±1.34 | 6.12±1.52 |
8.73±1.83a,b |
| HOMA-IR | 1.20±0.18 | 2.72±0.32a |
3.84±0.46a,b |
| Glycosylated serum
protein (mmol/l) | 1.47±0.38 | 3.36±3.43a | 3.93±3.38a |
| TG (mmol/l) | 1.13±0.38 | 1.67±0.61a | 1.72±0.89a |
| TC (mmol/l) | 3.96±0.68 | 5.83±1.38a | 5.71±0.91a |
| HDL (mmol/l) | 1.28±0.13 | 1.14±0.48 | 1.13±0.12 |
| LDL (mmol/l) | 2.71±1.12 | 3.38±1.81 | 3.07±1.52 |
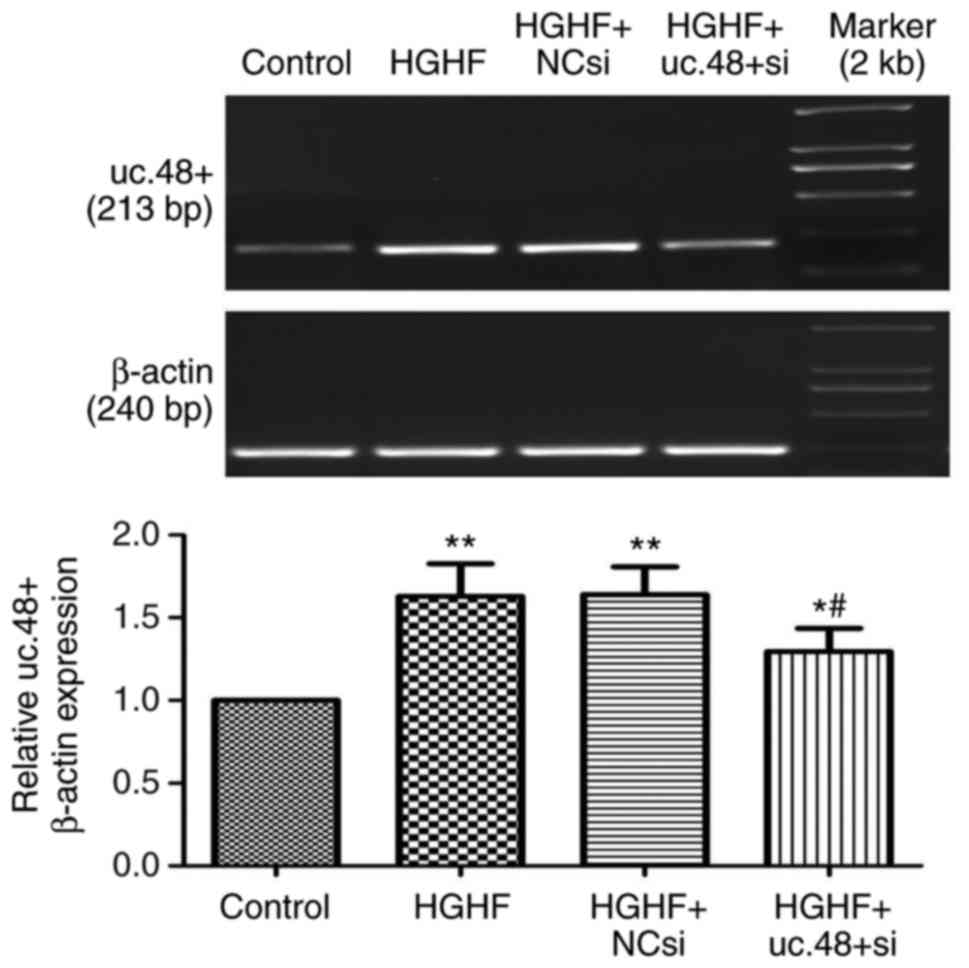
Effects of uc.48+ siRNA on the increased
expression of uc.48+ in RAW264.7 macrophages following exposure to
high glucose and high FFAs
The expression of uc.48+ was significantly
downregulated following treatment with uc.48+ siRNA (1.30±0.14),
whereas no significant difference was found between the HGHF group
(1.63±0.20) and the HGHF+NC siRNA group (1.64±0.18) (Fig. 2). The results showed that the
siRNA targeting uc.48+ effectively suppressed the expression of
uc.48+ in RAW264.7 macrophages.
Effects of uc.48+ siRNA on the increased
mRNA and protein expression of P2X7R in RAW264.7
macrophages following exposure with high glucose and high FFAs
As shown in Fig.
3, the upregulated mRNA (above) and protein (below) levels of
P2X7R were markedly decreased following uc.48+ siRNA
knockdown by transfection with uc.48+ siRNA (1.16±0.09 and
2.75±0.47, respectively), compared with those in the HGHF group
(1.42±0.21 and 4.66±0.58, respectively) and HGHF +NC siRNA group
(1.39±0.17 and 4.58±0.55, respectively). No significant difference
was found between the HGHF group and the HGHF +NC siRNA group.
 | Figure 3Effects of uc.48+ siRNA on the
increased mRNA and protein expression of P2X7R in
RAW264.7 macrophages following exposure to high glucose and high
FFAs. The mRNA (above) and protein (below) levels of
P2X7R were significantly decreased in the uc.48+ siRNA
group (1.16±0.09 and 2.75±0.47, respectively), as determined using
reverse transcription-polymerase chain reactionanalysis at 48 h and
western blot analysis at 72 h following transfection of the
RAW264.7 macrophages. No significant difference was found between
the HGHF group (1.42±0.21 and 4.66±0.58, respectively) and the
HGHF+NC siRNA group (1.39±0.17 and 4.58±0.55, respectively).
P2X7R, P2X7 receptor; siRNA, small
interference RNA; NC, negative control; FFAs, high plasma free
fatty acids; HGHF, high glucose and high FFAs.
*P<0.05 compared with control group,
**P<0.01 compared with control group,
#P<0.05 compared with HGHF group and
##P<0.01 compared with HGHF group. |
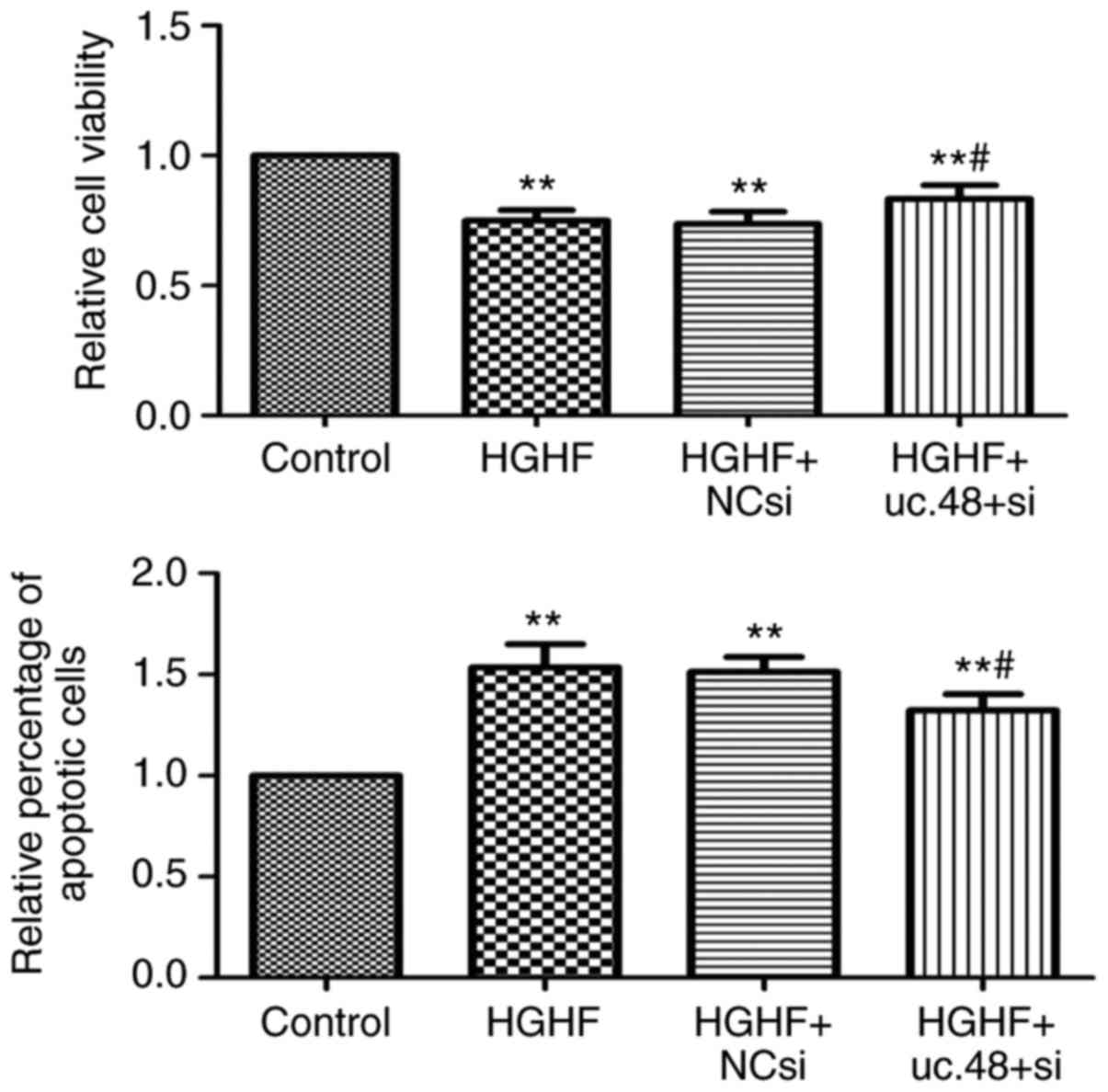
uc.48+ siRNA can partially reduce the
apoptosis and proliferation inhibition of RAW264.7 macrophages
induced by exposure of high glucose and high FFAs
The results indicated that the loss of uc.48+
partially reversed the inhibition of proliferation induced by high
glucose and high FFAs using the MTS assay (Fig. 4, above). The results also showed
that the uc.48+ siRNA significantly decreased the percentage of
apoptotic cells following high glucose and high FFA treatment by
flow cytometric analysis (Fig. 4,
below). Together, these results demonstrated that the uc.48+ siRNA
protected the RAW264.7 macrophages from the inhibition of
proliferation and induction of apoptosis induced by high glucose
and high FFAs.
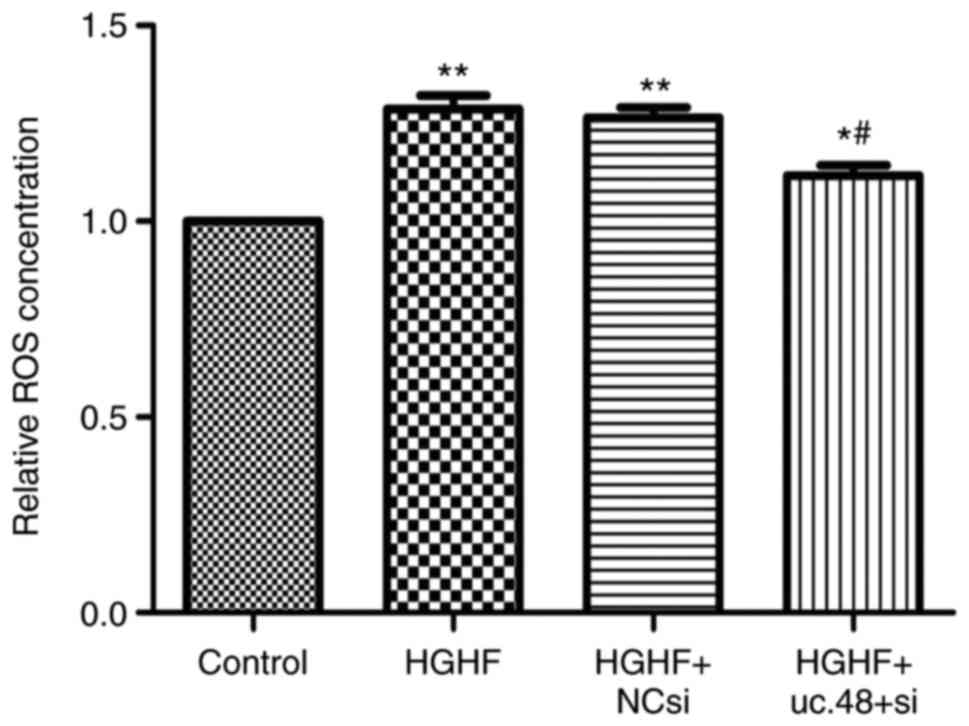
Effects of uc.48+ siRNA on the
upregulated ROS levels in RAW264.7 macrophages following exposure
to high glucose and high FFAs
The results showed that the ROS level was increased
in the HGHF group (1.29±0.06), compared with that in the control
(1.00±0.00), whereas uc.48+ siRNA (1.12±0.05) markedly reduced the
ROS concentration (Fig. 5).
Effects of uc.48+ siRNA on changes of
cytokines levels in RAW264.7 macrophages following exposure to high
glucose and high FFAs
Compared with the control (1.00±0.00), the levels of
IL-1β and TNF-α were elevated in the cells exposed to HGHF
(2.83±0.25 and 2.46±0.35, respectively) whereas uc.48+ siRNA
significantly decreased their levels (1.57±0.17 and 1.50±0.20,
respectively) (Fig. 6). Compared
with the results of IL-1β and TNF-α, the level of IL-10 was
significantly decreased in the HGHF group (0.65±0.08), compared
with that in the control (1.00±0.00) (P<0.05), whereas uc.48+
siRNA significantly increased the levels of IL-10 (0.79±0.06)
(Fig. 6).
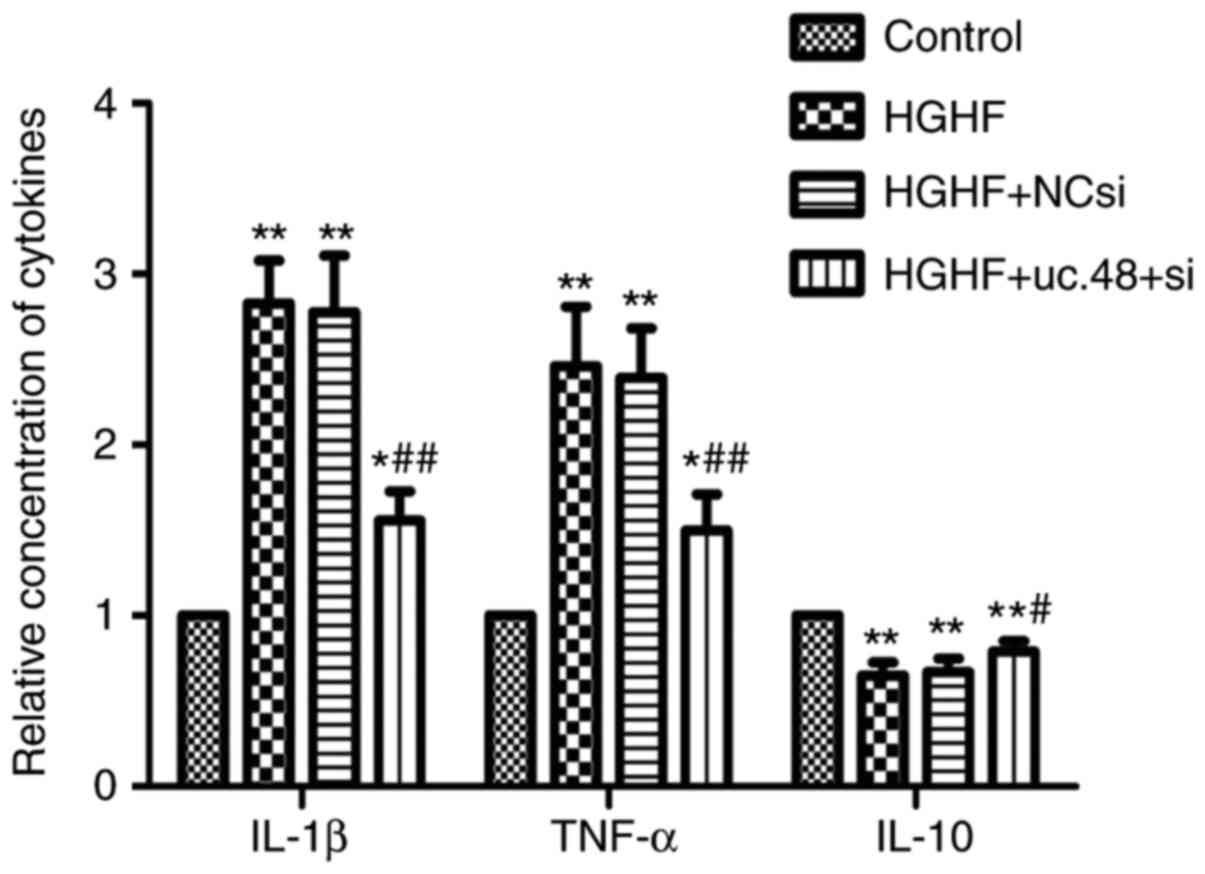 | Figure 6Effects of uc.48+ siRNA on changes of
cytokines levels in RAW264.7 macrophages following exposure to high
glucose and high FFAs. Cytokine levels in RAW264.7 macrophages,
including IL-1β, TNF-α and IL-10, were determined by enzyme-linked
immunosorbent assays. The results showed that levels of IL-1β and
TNF-α were increased in the HGHF group (2.83±0.25, 2.46±0.35,
respectively), compared with those in the control (1.00±0.00),
whereas the levels of IL-1β and TNF-α in the uc.48+ group
(1.57±0.17 and 1.50±0.20, respectively) were markedly decreased. In
contrast to the results of TNF-α and IL-1β, the level of IL-10
showed the opposite trend. siRNA, small interference RNA; NC,
negative control; FFAs, high plasma free fatty acids; HGHF, high
glucose and high FFAs; IL, interleukin; TNF-α, tumor necrosis
factor-α. *P<0.05 compared with control group,
**P<0.01 compared with control group,
#P<0.05 compared with HGHF group and
##P<0.01 compared with HGHF group. |
Effects of uc.48+siRNA on changes in the
expression of p-ERK1/2 in RAW264.7 macrophages following exposure
to high glucose and high FFAs
The levels of p-ERK1/2 and total ERK1/2 were
quantified by densitometry, and phosphorylated protein levels were
normalized to total protein levels. As shown in Fig. 7, the expression of p-ERK1/2 was
significantly increased in the HGHF group (3.28±0.34), compared
with that in the control (1.00±0.00). The inhibition of uc.48+ with
uc.48+ siRNA (2.17±0.24) markedly inhibited the p-ERK1/2 kinases;
scrambled (NC) siRNA (3.01±0.32) had no significant effect on the
expression of p-ERK (Fig. 7).
Discussion
The events leading to T2DM are complex, however,
emerging evidence has shown that various purinergic signaling
components may be important regulatory factors (20,21). Our previous preliminary study
demonstrated that P2X7R in peripheral blood monocytes
may be involved in the pathological changes of T2DM, particularly
affecting patients with high CRP levels (6). In the previous study, it was found
that the expression of uc.48+ in the serum of patients with T2DM
and in RAW264.7 macrophages treated with high glucose and high FFAs
was significantly increased, compared with the expression in the
healthy individuals and control group, respectively. This result
suggested that lncRNA uc.48+ was involved in the pathophysiological
process of T2DM.
Previous studies have shown that certain lncRNAs may
be involved in the pathological processes of diabetes mellitus
(22,23). Previous results obtained from rat
lncRNA array profiling revealed that the expression of the uc.48+
was significantly increased in the diabetic rat (24). Treatment of RAW264.7 macrophages
with uc.48+ siRNA significantly decreased the expression of uc.48+,
compared with that in the HGHF group. In the present study, it was
found that treatment with uc.48+ siRNA significantly improved the
cell survival status through a reduction in apoptosis and reduced
proliferation inhibition of RAW264.7 macrophages. Although the
present study was limited in that it was only a preliminary study
and did not investigate whether which caspases were involved in the
apoptotic event, it is possible that uc.48+ siRNA treatment has
beneficial effects on RAW264.7 macrophages under diabetic
conditions. For future studies, the mechanism of apoptosis requires
investigation. Hyperglycemia-induced ROS formation and high
FFA-induced oxidative stress may also contribute to the development
of T2DM (25). The present study
showed that ROS formation was significantly increased when the
RAW264.7 macrophages were exposed to high glucose and high FFAs,
whereas the levels of ROS were sharply decreased when the cells
were treated with uc.48+ siRNA. These findings indicated that
uc.48+ siRNA treatment was involved in the regulation of ROS
formation.
The upregulation of P2X7R is involved in
the pathological processes of diabetes mellitus (15,26). The experiments in the present
study showed that the mRNA and protein expression of
P2X7R in RAW264.7 cells were increased due to high
glucose and high FFA treatment, compared with those in the
controls. This was similar to the results of our preliminary
investigation (6,27), which showed that the
P2X7R may alter the innate immune system contributing to
the development of T2DM. The present study was limited by the fact
that no binding assays were performed to examine the binding of ATP
to P2X7R. Future investigations aim to improve on this
aspect of the pilot study. The present study also revealed that the
upregulated expression of mRNA and protein levels of
P2X7R in RAW264.7 cells due to high glucose and high FFA
treatment were significantly downgraded following uc.48+ siRNA
transfection. Taken together, these results suggested that uc.48+
siRNA treatment may ameliorate the pathophysiological process of
diabetes mellitus through downregulating P2X7R
signaling.
Chronic low-grade inflammation is involved in the
pathogenesis of T2DM (4,28). It has been reported that the
levels of circulating pro-inflammatory cytokines, including IL-1β
and TNF-α, are increased in the pathological process of T2DM
(29,30). IL-1β is essential in orchestrating
effective innate and adaptive immune responses among
pro-inflammatory cytokines (31).
IL-1β enhances the expression of additional pro-inflammatory
cytokines, including IL-6 and TNF-α (32,33). The results of the present study
showed that the levels of pro-inflammatory cytokines (IL-1β and
TNF-α) were higher in theRAW264.7 cells treated with high FFAs and
high glucose, compared with those in the controls, whereas the
concentrations of IL-10, an anti-inflammatory cytokine, in the high
FFAs and high glucose-treated RAW264.7 cells were lower, compared
with those in the controls. When the RAW264.7 macrophages were
treated with uc.48+ siRNA, the levels of IL-1β and TNF-α were
downregulated and that of IL-10 was upregulated. Therefore, uc.48+
siRNA treatment is likely to be involved in the regulation of
cytokine production in monocytes/macrophages in a high FFA and high
glucose state.
The activation of P2X7R may activate the
ERK1/2 signaling pathway to regulate distinct cellular functions
(34,35). To determine the possible mechanism
of uc.48+ siRNA on P2X7R, the ERK pathway was examined
in the present study. It was observed that the phosphorylation of
ERK1/2 was increased on exposure to high FFAs and high glucose,
whereas uc.48+ siRNA treatment reversed the upregulated
phosphorylation of ERK1/2. These data suggested that uc.48+ siRNA
treatment may be associated with intracellular ERK signaling,
relieving P2X7R-mediated inflammatory response in
RAW264.7 cells.
Although the role and possible mechanism of uc.48+
in inflammatory pathological damage due to diabetes remains to be
elucidated, Wu et al focused on the effect of uc.48+ siRNA
in the nervous system (36),
whereas the present study focused on the effect of uc.48+ siRNA in
the mononuclear phagocyte system. Therefore, these two areas of
investigation have different meanings. In addition, although
RAW264.7 cells are not the cell-specific cell type for diabetes,
further investigations are required to validate macrophages as a
cell-type model for diabetes.
In conclusion, the treatment of RAW264.7 cells with
high glucose and FFAs increased the expression of uc.48+, which
evoked P2X7R-mediated immune and inflammatory responses
through several pathways, including cytokine secretion, ROS
formation, and activation of the ERK signaling pathway. uc.48+
siRNA regulated these effects and thus influenced the course and
outcome of immune and inflammatory responses mediated by
P2X7R.
Funding
This study was supported by grants from the National
Natural Science Foundation of China (grant no. 81660144).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YN and HW made substantial contributions to the
conception and design of the study. YN, HW, FW, MJ and QL made
substantial contributions to the acquisition of data. HW drafted
the manuscript; YN critically revised the manuscript for important
intellectual content. All authors read and approved the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the local Ethics
Committee of from the First Affiliated Hospital of Nanchang
University (approval no. 2018 16), and written informed consent was
obtained from each individual.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
T2DM
|
type 2 diabetes mellitus
|
|
FFAs
|
high plasma free fatty acids
|
|
P2X7R
|
P2X7 receptor
|
|
PMA
|
phorbol myristate acetate
|
|
RT-PCR
|
reverse transcription-polymerase chain
reaction
|
|
siRNA
|
small inference RNA
|
|
control-siRNA
|
negative control scrambled siRNA
|
|
HG+HF
|
high glucose combined with high
FFAs
|
|
ROS
|
reactive oxygen species
|
|
DCFH-DA
|
2′,7′-dichlorofluorescin diacetate
|
|
FITC
|
fluorescein isothiocyanate
|
|
DAPI
|
4′,6-diamidino-2-phenylindole
|
Acknowledgments
The authors would like to thank Professor Shangdong
Liang (Department of Physiology, Medical School of Nanchang
University, Nanchang, China) for guidance us during the present
study.
References
|
1
|
Czyżewska-Majchrzak Ł, Grzelak T,
Kramkowska M, Czyżewska K and Witmanowski H: The use of
low-carbohydrate diet in type 2 diabetes-benefits and risks. Ann
Agr Env Med. 21:320–326. 2014. View Article : Google Scholar
|
|
2
|
Zonszein J, Lombardero M, Ismail-Beigi F,
Palumbo P, Foucher S, Groenewoud Y, Cushing G, Wajchenberg B and
Genuth S; Bari 2D Study Group: Triglyceride high-density
lipoprotein ratios predict glycemia-lowering in response to insulin
sensitizing drugs in type 2 diabetes: A post hoc analysis of the
BARI 2D. J Diabetes Res. 2015:1298912015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Martins AR, Nachbar RT, Gorjao R, Vinolo
MA, Festuccia WT, Lambertucci RH, Cury-Boaventura MF, Silveira LR,
Curi R and Hirabara SM: Mechanisms underlying skeletal muscle
insulin resistance induced by fatty acids: Importance of the
mitochondrial function. Lipids Health Dis. 11:302012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Esser N, Legrand-Poels S, Piette J, Scheen
AJ and Paquot N: Inflammation as a link between obesity, metabolic
syndrome and type 2 diabetes. Diabetes Res Clin Pract. 105:141–150.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jansen HJ, Stienstra R, van Diepen JA,
Hijmans A, van der Laak JA, Vervoort GM and Tack CJ: Start of
insulin therapy in patients with type 2 diabetes mellitus promotes
the influx of macrophages into subcutaneous adipose tissue.
Diabetologia. 56:2573–2581. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu H, Nie Y, Xiong H, Liu S, Li G, Huang
A, Guo L, Wang S, Xue Y, Wu B, et al: P2X7 receptor expression in
peripheral blood monocytes is correlated with plasma C-reactive
protein and cytokine levels in patients with type 2 diabetes
mellitus: A preliminary report. Inflammation. 38:2076–2081. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Solini A, Chiozzi P, Morelli A, Adinolfi
E, Rizzo R, Baricordi OR and Di Virgilio F: Enhanced P2X7 activity
in human fibroblasts from diabetic patients: A possible
pathogenetic mechanism for vascular damage in diabetes. Arterioscl
Throm Vas. 24:1240–1245. 2004. View Article : Google Scholar
|
|
8
|
Stolz M, Klapperstuck M, Kendzierski T,
Detro-Dassen S, Panning A, Schmalzing G and Markwardt F:
Homodimeric anoctamin-1, but not homodimeric anoctamin-6, is
activated by calcium increases mediated by the P2Y1 and P2X7
receptors. Pflug Arch. 467:2121–2140. 2015. View Article : Google Scholar
|
|
9
|
Huang SW, Walker C, Pennock J, Else K,
Muller W, Daniels MJ, Pellegrini C, Brough D, Lopez-Castejon G and
Cruickshank SM: P2X7 receptor-dependent tuning of gut epithelial
responses to infection. Immunol Cell Biol. 95:178–188. 2017.
View Article : Google Scholar
|
|
10
|
Jiang L, Liu W, Zhu A, Zhang J, Zhou J and
Wu C: Transcriptome analysis demonstrate widespread differential
expression of long noncoding RNAs involve in Larimichthys crocea
immune response. Fish Shellfish Immun. 51:1–8. 2016. View Article : Google Scholar
|
|
11
|
Boysen A, Møller-Jensen J, Kallipolitis B,
Valentin-Hansen P and Overgaard M: Translational regulation of gene
expression by an anaerobically induced small non-coding RNA in
Escherichia coli. J Biol Chem. 285:10690–10702. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lutz BM, Bekker A and Tao YX: Noncoding
RNAs: New players in chronic pain. Anesthesiology. 121:409–417.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song G, Shen Y, Ruan Z, Li X, Chen Y, Yuan
W, Ding X, Zhu L and Qian L: LncRNA-uc.167 influences cell
proliferation, apoptosis and differentiation of P19 cells by
regulating Mef2c. Gene. 590:97–108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bejerano G, Pheasant M, Makunin I, Stephen
S, Kent WJ, Mattick JS and Haussler D: Ultraconserved elements in
the human genome. Science. 304:1321–1325. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu C, Wang J, Yuan X, Qian W, Zhang B,
Shi M, Xie J, Shen B, Xu H, Hou Z and Chen H: Long noncoding RNA
uc.345 promotes tumorigenesis of pancreatic cancer by upregulation
of hnRNPL expression. Oncotarget. 7:71556–71566. 2016.PubMed/NCBI
|
|
16
|
Wang S, Xu H, Zou L, Xie J, Wu H, Wu B, Yi
Z, Lv Q, Zhang X, Ying M, et al: LncRNA uc.48+ is involved in
diabetic neuropathic pain mediated by the P2X3 receptor in the
dorsal root ganglia. Purinerg Signal. 12:139–148. 2016. View Article : Google Scholar
|
|
17
|
Lv Q, Xue Y, Li G, Zou L, Zhang X, Ying M,
Wang S, Guo L, Gao Y, Li G, et al: Beneficial effects of evodiamine
on P2X(4)-mediated inflammatory injury of human umbilical vein
endothelial cells due to high glucose. Int Immunopharmacol.
28:1044–1049. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu H, Wu B, Jiang F, Xiong S, Zhang B, Li
G, Liu S, Gao Y, Xu C, Tu G, et al: High fatty acids modulate
P2X(7) expression and IL-6 release via the p38 MAPK pathway in PC12
cells. Brain Res Bull. 94:63–70. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gao Y, Liu H, Deng L, Zhu G, Xu C, Li G,
Liu S, Xie J, Liu J, Kong F, et al: Effect of emodin on neuropathic
pain transmission mediated by P2X2/3 receptor of primary sensory
neurons. Brain Res Bull. 84:406–413. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Suryadevara S, Ueno M, Tello-Montoliu A,
Ferreiro JL, Desai B, Rollini F, Box LC, Zenni M, Guzman LA, Bass
TA and Angiolillo DJ: Effects of pioglitazone on platelet
P2Y12-mediated signalling in clopidogrel-treated patients with type
2 diabetes mellitus. Thromb Haemostasis. 108:930–936. 2012.
View Article : Google Scholar
|
|
21
|
Burnstock G and Novak I: Purinergic
signalling and diabetes. Purinerg Signal. 9:307–324. 2013.
View Article : Google Scholar
|
|
22
|
Jaé N and Dimmeler S: Long noncoding RNAs
in diabetic retinopathy. Circ Res. 116:1104–1106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jiang GJ, Zhang T, An T, Zhao DD, Yang XY,
Zhang DW, Zhang Y, Mu QQ, Yu N, Ma XS and Gao SH: Differential
expression of long noncoding RNAs between sperm samples from
diabetic and non-diabetic mice. PloS One. 11:e01540282016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gao W, Wang ZM, Zhu M, Lian XQ, Zhao H,
Zhao D, Yang ZJ, Lu X and Wang LS: Altered long noncoding RNA
expression profiles in the myocardium of rats with ischemic heart
failure. J Cardiovasc Med (Hagerstown). 16:473–479. 2015.
View Article : Google Scholar
|
|
25
|
Younce CW, Wang K and Kolattukudy PE:
Hyperglycaemiainduced cardiomyocyte death is mediated via MCP-1
production and induction of a novel zinc-finger protein MCPIP.
Cardiovasc Res. 87:665–674. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Glas R, Sauter NS, Schulthess FT, Shu L,
Oberholzer J and Maedler K: Purinergic P2X7 receptors regulate
secretion of interleukin-1 receptor antagonist and beta cell
function and survival. Diabetologia. 52:1579–1588. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu S, Zou L, Xie J, Xie W, Wen S, Xie Q,
Gao Y, Li G, Zhang C, Xu C, et al: LncRNA NONRATT021972 siRNA
regulates neuropathic pain behaviors in type 2 diabetic rats
through the P2X7 receptor in dorsal root ganglia. Mol Brain.
9:442016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fernández-Real JM and Pickup JC: Innate
immunity, insulin resistance and type 2 diabetes. Diabetologia.
55:273–278. 2012. View Article : Google Scholar
|
|
29
|
Mirza S, Hossain M, Mathews C, Martinez P,
Pino P, Gay JL, Rentfro A, McCormick JB and Fisher-Hoch SP: Type
2-diabetes is associated with elevated levels of TNF-alpha, IL-6
and adiponectin and low levels of leptin in a population of Mexican
Americans: A cross-sectional study. Cytokine. 57:136–142. 2012.
View Article : Google Scholar
|
|
30
|
Saraswathi V, Ramnanan CJ, Wilks AW,
Desouza CV, Eller AA, Murali G, Ramalingam R, Milne GL, Coate KC
and Edgerton DS: Impact of hematopoietic cyclooxygenase-1
deficiency on obesity-linked adipose tissue inflammation and
metabolic disorders in mice. Metabolism. 62:1673–1685. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dinarello CA: Immunological and
inflammatory functions of the interleukin-1 family. Annu Rev
Immunol. 27:519–550. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ciraci C, Janczy JR, Sutterwala FS and
Cassel SL: Control of innate and adaptive immunity by the
inflammasome. Microbes Infect. 14:1263–1270. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dinarello CA: Blocking interleukin-1β in
acute and chronic autoinflammatory diseases. J Intern Med.
269:16–28. 2011. View Article : Google Scholar :
|
|
34
|
Ponnusamy M, Liu N, Gong R, Yan H and
Zhuang S: ERK pathway mediates P2X7 expression and cell death in
renal interstitial fibroblasts exposed to necrotic renal epithelial
cells. Am J Physiol Renal Physiol. 301:F650–F659. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zanin RF, Bergamin LS, Morrone FB,
Coutinho-Silva R, de Souza Wyse AT and Battastini AM: Pathological
concentrations of homocysteine increases IL-1β production in
macrophages in a P2X7, NF-κB, and erk-dependent manner. Purinerg
Signal. 11:463–470. 2015. View Article : Google Scholar
|
|
36
|
Wu B, Zhang C, Zou L, Ma Y, Huang K, Lv Q,
Zhang X, Wang S, Xue Y, Yi Z, et al: LncRNA uc.48+ siRNA improved
diabetic sympathetic neuropathy in type 2 diabetic rats mediated by
P2X7, receptor in SCG. Auton Neurosci. 197:14–18. 2016. View Article : Google Scholar : PubMed/NCBI
|