Introduction
Infection with hepatitis B virus (HBV), which
usually causes chronic hepatitis B (CHB), is a cosmopolitan public
health problem which is highly endemic and associated with a high
mortality rate and heavy societal burden (1,2). A
mortality rate of >1,000/year has been attributed to liver
failure, liver cirrhosis and hepatocellular carcinoma caused by
long-term HBV infection, which accounts for >15% of those
succumbing to chronic diseases globally (2,3).
Although the prevalence of HBV in certain highly endemic regions,
including rural China, has shown improvement, the large absolute
number of HBV-infected susceptible individuals in the population
remains a potential threat (4).
Interferon and nucleoside acid analogs are the major therapeutic
agents that have been used for the treatment of CHB by delaying
disease progression, however, these are limited by severe drug
resistance and relapse following drug withdrawal (2).
The covalently closed circular DNA (cccDNA), which
is formed in the liver cell nucleus following the binding of HBV
with cellular receptors during HBV infection, functions as the main
template for the replication of HBV particles and has been accepted
as the main reason for the difficulty in HBV eradication (5,6).
Due to the persistence of viral cccDNA, which is transcriptionally
active in the liver cell nucleus, lifelong therapy is required
using antiviral drugs, which are currently unable to directly
target the formation of cccDNA (7,8).
The nuclear localization sequence (NLS) of HBV core protein can be
recognized by and is associated with importin, thus having an
important function in transferring HBV relaxed-circular serum DNA
into the hepatocyte nucleus for cccDNA formation (9). Small interfering (si)RNAs targeting
HBV NLS can induce the degradation of mRNAs responsible for HBV
core protein nuclear entry, and lead to the subsequent inhibition
of cccDNA formation and HBV replication (10), demonstrating the potential of HBV
NLS siRNAs in the treatment of HBV infection. However, the
effective inhibition of cccDNA formation by HBV NLS siRNAs depends
on the specific delivery of interfering molecules into the infected
hepatocytes.
The entry of HBV into hepatocytes follows multiple
steps, including the initial attachment of HBV to host cell surface
proteoglycans, early entry following high-affinity binding by
specific receptors, endocytosis-mediated internalization, and
fusion with the cellular membrane (11). Three HBV surface proteins are
important for the entry processes, including the large HBV surface
proteins (LHBs) which contain the preS1, preS2 and S regions, the
middle surface proteins containing the preS2 and S regions, and the
small surface proteins that comprise only the S regions (11). Among these three characteristic
regions in HBV surface proteins, the S and preS1 regions are
reported to be important in HBV infection (11,12). The sodium taurocholate
cotransporting polypeptide (NTCP) has been identified as the
cellular entry receptor for HBV (13), which has attracted increased
attention regarding the molecular mechanisms underlying the entry
steps of HBV infection. NTCP, also known as solute carrier family
10A1 (14), was previously
demonstrated to be a transporter in the hepatocyte basolateral
membrane and function in conjugated bile salt uptake by hepatic
cells (11). Investigations have
shown that NTCP can function as an HBV entry receptor by directly
binding preS1-derived lipopeptide, which was supported by the close
correlation between the expression of NTCP and HBV susceptibility
in multiple cells, and the significantly enhanced HBV
susceptibility conferred by the ectopic expression of NTCP in HepG2
cells (11,13,15,16). These findings suggest that
hepatocytes modified to overexpress NTCP may be useful cellular
models for the development and screening of novel anti-HBV
targeting therapy. The specific interaction between preS1 and NTCP
may be examined for the specific delivery of therapeutic agents
into infected hepatocytes for targeting treatment.
Based on the roles of HBV surface proteins and
cellular receptors in viral infection and disease progression, the
specific delivery of siRNAs by the single chain variable fragment
(scFv), which specifically recognizes HBV surface antigen (HBsAg),
the specific antigen expressed on hepatocytes infected by HBV, has
been shown as a promising approach for inhibiting HBV gene
expression (17,18). The fusion protein containing scFv
and truncated protamine (tP), which binds with DNA with high
affinity (19), can be
internalized by HepG2.2.15 cells and effectively inhibit HBV gene
expression by the siRNAs delivered by the recombinant proteins
(17,18). However, the efficiency of such a
delivery system may be limited by the neutralizing effect of
abundantly existing HBsAg in the serum of patients with CHB.
Therefore, in the present study, the preS1 region of the LHB was
used in place of scFv to construct the recombinant preS1-tP
proteins, which were applied to deliver siRNAs targeting the HBV
NLS to inhibit HBV replication and infection in HepG2.2.15 cells
overexpressing NTCP.
Materials and methods
Cell lines and culture
The HepG2.2.15 cells (cat. no. YB-ATCC-2242) and
293T cells (cat. no. CRL-3216™) were purchased from the Human
American Type Culture Collection (Manassas, VA, USA), and cultured
in DMEM (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany), 2 mM L-glutamine, 100 mg/ml
streptomycin and 100 mg/ml penicillin at 37°C in 5%
CO2.
Generation of HepG2.2.15 cells stably
expressing the NTCP gene
To generate HepG2.2.15 cells stably expressing the
NTCP gene, the human NTCP coding DNA sequences were cloned by
polymerase chain reaction (PCR) using the following specific
primers: NTCP, forward 5′-ATG CTC TAG AGC CAC CAT GGA GGC CCA C-3′
and reverse 5′-ATG GGG ATC CTC AGG CTG TGC AAG GGG AG-3′. The PCR
products were then digested using two restriction endonucleases
EcoRI and XbaI, and then ligated to the LV003 plasmid
(Promega Corporation, Madison, WI, USA) for the construction of
recombinant LV003-NTCP plasmids. Establishment of the HepG2.2.15
cells stably expressing the NTCP gene was performed as previously
described with minor modifications (20). Briefly, the packaging procedure
was performed by the co-infection of 293T cells with pGag/Pol (5
µg), pRev (5 µg) and pVSV-G (5 µg) supplied by
Shanghai GenePharma Co., Ltd. (Shanghai, China) following the
protocol described by the manufacturer. The cell culture medium
containing the recombinant virus was collected and transfected into
HepG2.2.15 cells using lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
instructions. At 3 days post-infection, the HepG2.2.15 cells
showing stable NTCP gene expression were screened using DMEM
containing puromycin, as previously described (21). The expression of NTCP in
HepG2.2.15 cells was finally confirmed using fluorescence
microscopy and reverse transcription-quantitative (RT-q)PCR
analysis 10 days following the screening.
RT-qPCR analysis
The relative mRNA levels of NTCP and associated
genes were analyzed by RT-qPCR analysis in the present study. The
cells were washed with PBS three times, and total RNA samples from
the cultured cells were extracted using TRIzol reagent (cat. no.
15596018) in strict accordance with the manufacturer's protocol
(Thermo Fisher Scientific, Inc.). The cDNA samples were then
synthesized using random primers and the Prime Script RT Master Mix
kit (cat. no. RR036a; Takara Bio, Inc., Otsu, Japan) according to
the manufacturer's protocol. PCR was carried out in a 20 µl
reaction mixture containing 7.5 µl GoTaq® Green
Master Mix (Promega Corporation), 1.5 µl of cDNA, 1
µl of each primer, 9.0 µl nuclease-free water with
the following conditions, 95°C for 120 sec, followed by 40 cycles
at 60°C for 30 sec. The primer sequences were as follows, NTCP:
Forward, 5′-AGG GAG GAG GTG GCA ATC AAG AGT GG-3′, reverse, 5′-CCG
GCT GAA GAA CAT TGA GGC ACT GG-3′; HBV: Forward, 5′-CCG TCT GTG CCT
TCT CAT CTG C-3′, reverse, 5′-ACC AAT TTA TGC CTA CAG CCT C-3′;
GAPDH: Forward, 5′-GAG TCA ACG GAT TTG GTC GT-3′, reverse, 5′-CCC
AGT AGC AGT TCA GGT GG-3′. The gene expression level was finally
determined by RT-qPCR analysis using the SYBR Select Master Mix kit
(cat. no. 4472908; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol. GAPDH was used as the internal control for
gene expression level quantitation. At least three biological
repeats and three technical replicates were performed for the
statistical comparison of gene expression. Data were quantified
using the ΔΔCq method normalizing to GAPDH as reference
genes, and further normalized to the lowest-expressing sample for
comparison (22).
Suppression of HBV by NLS siRNA
The knockdown of the gene expression of NTCP in
HepG2.2.15 cells stably expressing the NTCP gene was performed
using Lipofectamine™ RNAiMAX reagent supplied Invitrogen; Thermo
Fisher Scientific, Inc. (cat. no. 13778-075) following the
manufacturer's protocol. RNAi duplex (~6 pmol) was diluted in 100
µl DMEM without serum and gently mixed, which was added to 1
µl Lipofectamine™ RNAiMAX, mixed gently, and then incubated
at room temperature for 15 min. The HepG2.2.15 cells stably
expressing NTCP were cultured in 24-well plates at ~70% confluence,
and then diluted with DMEM to 1×105 cells/ml. The cells
(~0.5 ml) were then mixed with the previously prepared RNAi duplex
and Lipofectamine™ RNAiMAX mixture, mixed gently and incubated for
72 h at 37°C with 5% CO2. The knockdown of the gene
expression of NTCP was confirmed by RT-qPCR, followed by the
analysis of HBsAg and HBeAg antigens through an enzyme-linked
immunosorbent assay (ELISA). For the successful knockdown of gene
expression, at least three biological and technical replicates were
performed. The siRNA sequences used for suppressing the gene
expression if NTCP are listed in Table I.
 | Table IsiRNA sequences for sodium
taurocholate cotransporting polypeptide knockdown. |
Table I
siRNA sequences for sodium
taurocholate cotransporting polypeptide knockdown.
| siRNA | Sense (5′-3′) | Antisense
(5′-3′) |
|---|
| siRNA-1 |
AAGAUCUCAAUCUCGGGAAUC |
GAUUCCCGAGAUUGAGAUCUU |
| siRNA-2 C |
AGGUCCCCUAGAAGAAGAAC |
GUUCUUCUUCUAGGGGACCUG |
| siRNA-3 |
AACACUUCCGGAAACUACUGU |
ACAGUAGUUUCCGGAAGUGUU |
ELISA
The expression levels of HBsAg (cat. no. KA0286) and
HBeAg (cat. no. KA0290) antigens in the HepG2.2.15 cells stably
expressing NTCP were determined by the ELISA method using the
commercially available kits supplied by Novus Biologicals, Ltd.
(Cambridge, UK) following the manufacturer's protocol. The
quantification of antigen levels was performed by measuring the
absorbance at 405 nm using a microtiter plate spectrophotometer. At
least three biological and technical replicates were performed for
accurate quantification by statistical analysis.
Expression and purification of
recombinant preS1-tP protein in Escherichia coli
The recombinant preS1-tP proteins were expressed and
purified in BL-21 strains of E. coli (American Type Culture
Collection, Manassas, VA, USA) using regular protein expression and
purification protocols. Briefly, the preS1 region of the HBV
surface protein was cloned using the PCR method. The PCR products
of preS1 and tp DNA fragments were ligated into the p-GEX-6P-1
plasmids, which were used for the transformation of E. coli
BL21 strains. Monoclones with the recombinant plasmid were then
screened by culture in LB medium containing 34 µg/ml
chloromycetin and 50 µg/ml ampicillin. The large-scale
expression and purification of recombinant proteins were performed
by overnight induction with 0.5 mM isopropyl β-D-thiogalactoside
(IPTG) at 30°C with agitation. The E. coli cells were
collected by centrifugation at 8 x g over night at 37°C and lysed
by sonication. The supernatant was then purified by two rounds of
affinity chromatography, and visualized using 12% SDS-PAGE
electrophoresis.
Immunofluorescence
As a final examination of the functioning of
recombinant preS1-tP proteins in delivering HBV NLS siRNAs, the NLS
siRNA fragment was synthesized and labeled with Cy3 fluorescent
(Cy3 siNLS), with a shNLS plasmid expressing red fluorescence
protein also applied. Subsequently, 5 µg recombinant
preS1-tP proteins were mixed with 150 pmol Cy3 siNLS or 2 µg
shNLS in 0.2 M NaCl for 30 min, and incubated with the HepG2.2.15
cells for 48 h. The entry of recombinant protein with glutathione
S-transferase (GST) and His tags into hepatocytes was first
determined by immunofluorescence using anti-His antibodies (cat.
no. ab1187; Abcam, Cambridge, UK). The HepG2.2.15 cells stably
expressing NTCP were grown on plates, and covered with 2–3 mm 4%
formaldehyde prepared with PBS solution for 15 min at room
temperature. Following washing for 5 min with PBS solution three
times, the cell plates were then blocked with 5% lipid-free milk
solution for 1 h at room temperature, incubated with anti-His
antibodies (diluted at 1:1,000) overnight 4°C, rinsed three times
with PBS for 5 min each, incubated with fluorochrome-conjugated
secondary antibody (1:2,000; cat. no. ab205718; Abcam) for 2 h in
the dark at room temperature, and rinsed again three times with PBS
for 5 min each. Prolong® Gold Antifade reagent with DAPI
was then added (cat. no. 8961; Cell Signaling Technology, Inc.,
Danvers, MA, USA) overnight at room temperature. The existence of
recombinant proteins in hepatocytes were finally determined using
fluorescence microscopy.
Western blot analysis
The HepG2.2.15 cells stably expressing NTCP were
washed three times with PBS, collected by centrifugation at 150 x g
for 5 min at 37°C, and then used for total protein extraction.
Protein concentrations were determined using the Bradford method.
The cells were lysed by incubating with RIPA buffer containing 1X
Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific, Inc.).
Following boiling for 5 min at 100°C in sample buffer (300 mM
Tris-HCl, 1% SDS, 3.5 M β-mercaptoethanol, 5 mM PMSF and 6%
glycerol and 0.05% bromophenol blue), the protein samples (40
µg) were separated by 10% SDS-PAGE, transferred onto PVDF
membranes, blocked with 5% lipid-free milk, and incubated with
anti-GST antibodies (1:1,000; cat. no. ab19256; Abcam) and
anti-GAPDH (1:2,000; ab9485; Abcam) overnight at 4°C and secondary
antibodies (1:2,000; cat. no. ab205718; Abcam) for 2 h at room
temperature in TBST buffer successively. The protein levels were
finally visualized by adding enhanced chemiluminescence solution
(Amersham; GE Healthcare Life Sciences, Chalfont, UK).
Statistical analysis
The SPSS 18.0 software package (SPSS, Inc., Chicago,
IL, USA) was used for statistical analysis in the present study.
The significance of differences between groups was statistically
analyzed using Student's t-test. At least three biological repeats
were used for statistical evaluation. P<0.05 was considered to
indicate a statistically significant difference.
Results
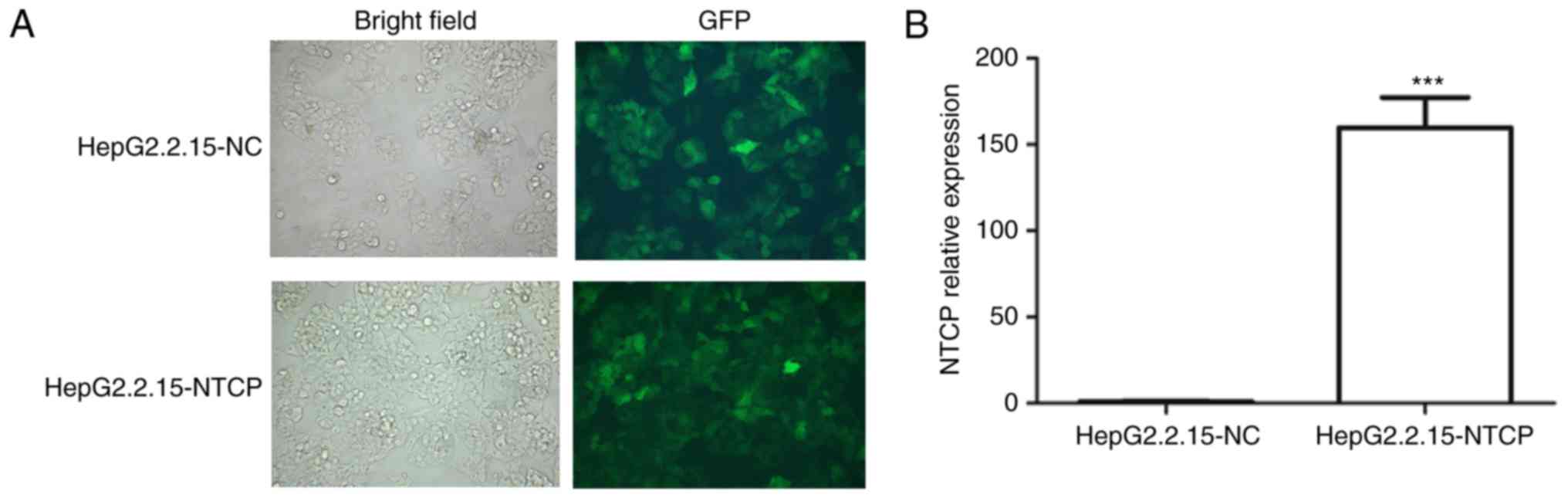
Establishment of HepG2.2.15 cells
overexpressing NTCP
Due to the importance of NTCP protein in the
infection and replication of HBV, hepatocytes overexpressing the
NTCP gene may be applied as an ideal cellular model for the
development and screening of novel anti-HBV therapies. The coding
DNA sequences of the human NTCP gene were amplified by PCR, and
ligated onto the LV003 plasmid. The recombinant LV003-NTCP plasmid
was then used to co-infect 293 cells with pGag/Pol, pRev and pVSV-G
plasmids for packaging into the lentivirus. Finally, the virus
collected from the cell culture medium was used to infect
HepG2.2.15 cells for the screening of cells stably expressing the
NTCP gene (Fig. 1A). For further
confirmation, the gene expression level of NTCP in HepG2.2.15 cells
was verified using the RT-qPCR method, which also showed markedly
elevated gene expression of NTCP in the HepG2.2.15 cells infected
with the virus (Fig. 1B). These
results showed the successful establishment of HepG2.2.15 cells
overexpressing the human NTCP gene, which were used for the
development of a novel siRNA therapy for HBV in the following
experiments.
Suppression of HBV replication and
infection in HepG2.2.15 cells using HBV NLS siRNAs
Previous studies have shown that HBV NLS siRNAs can
effectively suppress HBV infection and replication in hepatocytes
(17). For the development of a
novel HBV NLS siRNA delivery system using recombinant preS1-tP
proteins, three pairs of HBV NLS siRNAs were designed, as described
above. Subsequent to being transfected with these HBV NLS siRNAs,
the total RNA samples of HepG2.2.15 cells stably expressing the
NTCP gene were extracted and used for quantitative analysis of the
mRNA expression of HBV. The results showed that the mRNA levels of
HBV in the HepG2.2.15 cells stabling expressing HBV mRNA were
markedly suppressed by the transfection with all three of the HBV
NLS siRNAs (Fig. 2A). To confirm
the replication of HBV in these HepG2.2.15 cells, the expression
levels of two HBsAgs (HBeAg and HBeAg), were measured using ELISA
methods. The ELISA results also demonstrated that NLS siRNA-2 and
NLS siRNA-3 significantly inhibited the levels of HBsAg in the
HepG2.2.15 cells, and siRNA-3 significantly downregulated the level
of HBeAg in the HepG2.2.15 cells (Fig. 2B and C). Among the three HBV
siRNAs used in this screening, siRNA-3 exerted the most marked
inhibitory effect on the activity of HBV in the HepG2.2.15 cells,
and was used for the subsequent experiments.
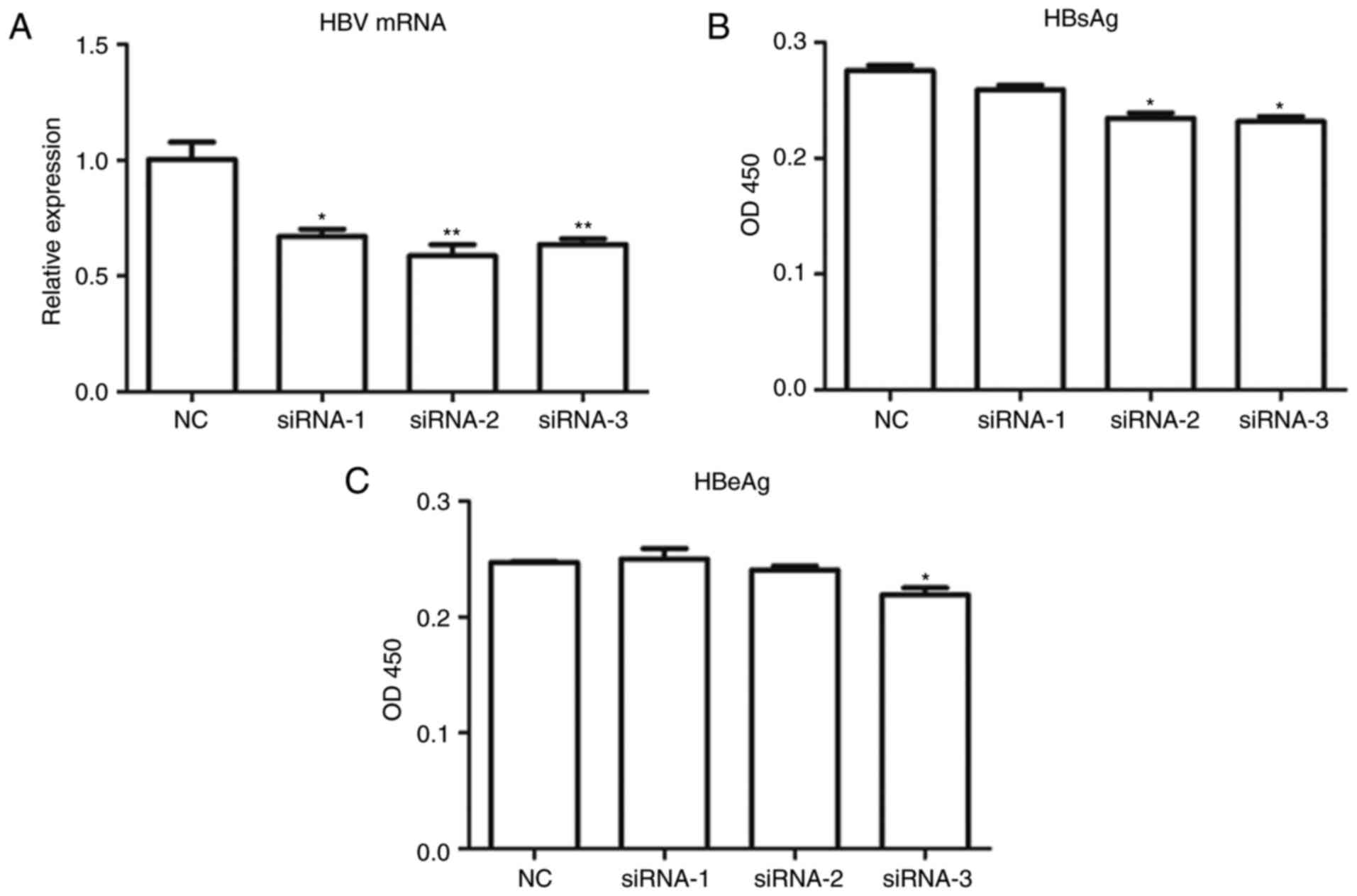 | Figure 2Suppression of HBV infection in
HepG2.2.15 cells using NLS siRNAs. (A) mRNA expression of HBV in
HepG2.2.15 cells stably expressing the NTCP gene by NLS siRNAs,
determined using reverse transcription-quantitative polymerase
chain reaction analysis. Enzyme-linked immunosorbent assay of the
expression of HBV surface antigens (B) HBsAg and (C) HBeAg in
HepG2.2.15 cells stably expressing the NTCP gene. * and
**, compared with the NC group, *P<0.05,
**P<0.01. HBV, hepatitis B virus; siRNAs, small
interfering RNAs; NC, negative control; NTCP, sodium taurocholate
cotransporting polypeptide; NLS, nuclear localization sequence;
HBsAg, Hepatitis B surface antigen; HBeAg, hepatitis B e
antigen. |
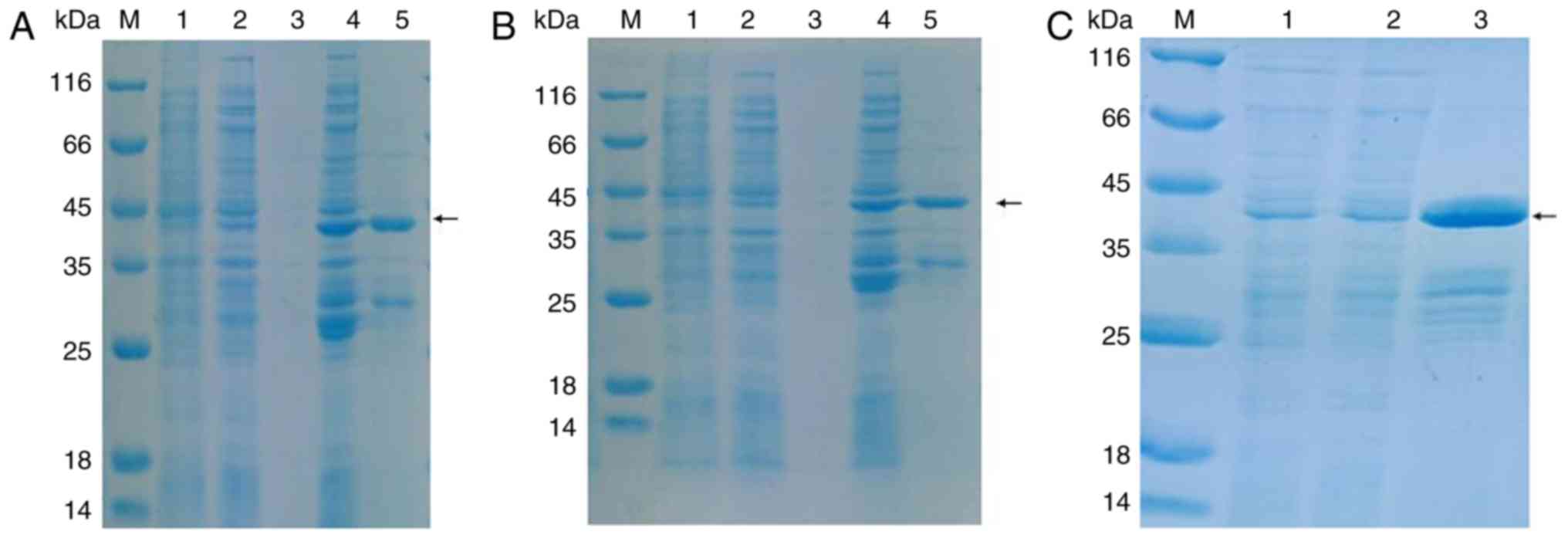
Purification of recombinant preS1-tP
proteins
For the successful delivery of HBV NLS siRNAs into
hepatocytes for the suppression of HBV infection and replication,
the preS1 regions of the LHB gene and tp fragment were cloned
separately, and then ligated to the p-GEX-6P-1 plasmids. The
recombinant preS1-tP proteins tagged with GST were expressed in and
purified from the E. coli BL21 strain. The results of
SDS-PAGE showed that the recombinant preS1-tP proteins were
expressed in E. coli BL21 cells by induction with IPTG at
37°C, but not at 20°C (Fig. 3A
and B). The significant protein band between the 35 and 45 kDa
protein markers indicated the GST-tagged recombinant preS1-tP
protein (Fig. 3A and B). The
recombinant preS1-tP proteins were then expressed and purified on a
large scale by affinity chromatography using glutathione-agarose
and Ni sepharose (Fig. 3C), which
was then applied for the construction of a novel delivery system of
HBV NSL siRNAs.
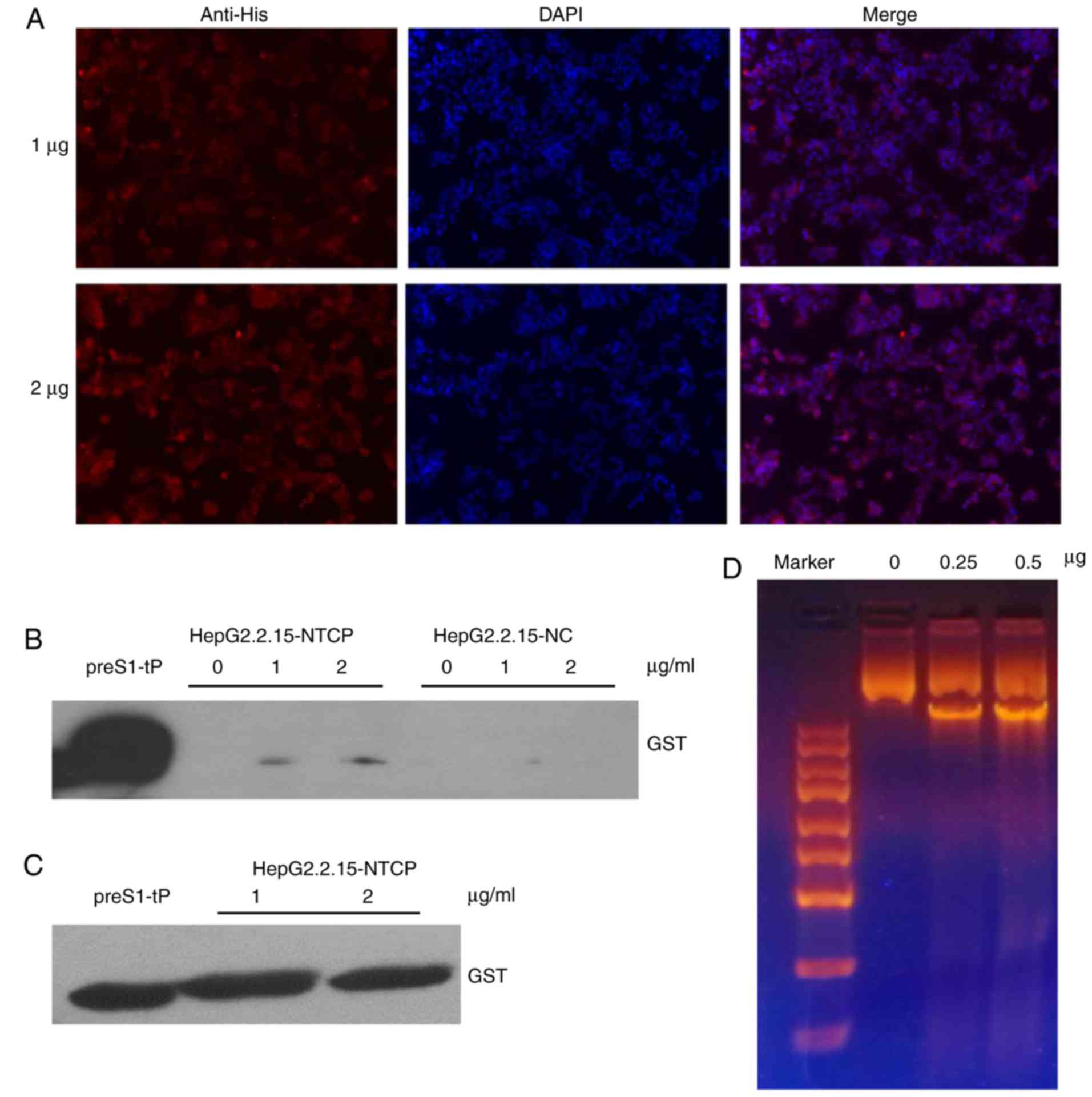
Recombinant preS1-tP protein entry into
HepG2.2.15 cells and DNA binding
For successful establishment of a novel delivery
system of HBV NLS siRNAs, the entry of the carriers, the
recombinant preS1-tP proteins in the present study, into the target
cells and their high-affinity binding with host DNA are key steps.
Therefore, the slides of HepG2.2.15 cells stably expressing the
human NTCP gene were incubated with purified recombinant preS1-tP
proteins overnight. The immunofluorescence assay using anti-His
antibodies showed that the recombinant preS1-tP proteins had
successfully entered into the HepG2.2.15 cells stably expressing
NTCP (Fig. 4A). For further
verification of the recombinant protein entry into the HepG2.2.15
cells, total proteins of the cells incubated with the recombinant
preS1-tP proteins overnight were extracted and subjected to an
immunoblotting assay using anti-GST antibodies. The results of the
western blot analysis also demonstrated entry of the recombinant
proteins into the HepG2.2.15 cells (Fig. 4B and C). However, the results of
the immuno-fluorescence assay showed that the recombinant proteins
were mainly distributed in the cytoplasm of the HepG2.2.15 cells.
As a preliminary assessment of the capacity of recombinant preS1-tP
proteins to bind host cell DNA, 1 µg short hairpin (sh) NLS
plasmids were mixed with 0, 250 or 500 ng recombinant preS1-tP
proteins, and then analyzed by electrophoresis. This also revealed
the binding of recombinant preS1-tP proteins with shNLS plasmids
in vitro (Fig. 4D).
Therefore, the results showed that the recombinant preS1-tP
proteins were able to enter into the HepG2.2.15 cells by simple
co-incubation and bind the DNA in the in vitro assay,
demonstrating the potential of recombinant preS1-tP proteins for
the delivery of NLS siRNA into HepG2.2.15 cells for treatment of
HBV infection.
Inhibition of HBV infection and
production of cccDNA by NLS siRNA delivered by recombinant preS1-tP
proteins in HepG2.2.15 cells
Fluorescence microscopy showed that the Cy3 siNLS
and shNLS had been successfully carried into the HepG2.2.15 cells
by the recombinant preS1-tP proteins (Fig. 5A). To show the effect of siNLS and
shNLS on HBV function in HepG2.2.15 cells, the mRNA levels of HBV
in HepG2.2.15 cells were determined using the RT-qPCR method. It
was observed that the HepG2.2.15 cells incubated with siNLS or
shNLS exhibited significantly repressed mRNA levels of HBV,
compared with the cells incubated with the recombinant preS1-tP
proteins alone (Fig. 5B). In
addition, as demonstrated by ELISA, the levels of HBsAg and HBeAg
were also markedly downregulated in the HepG2.2.15 cells, compared
with the control (Fig. 5C and D).
It was found that the HBV DNA content in HepG2.2.15 cells incubated
with siNLS or shNLS was also markedly decreased, compared with that
in the cells incubated with only the recombinant proteins (Fig. 5E), showing the effect of preS1-tP
protein-delivered NLS siRNAs in suppressing HBV replication.
Finally, the levels of HBV cccDNA in HepG2.2.15 cells showed a
similar decrease in the HepG2.2.15 cells induced by incubation with
preS1-tP proteins and HBV NLS siRNAS (Fig. 5F). Taken together, these results
confirmed that the preS1-tP protein acted as the effective
deliverer of HBV NLS siRNA for the inhibition of HBV infection and
replication in hepatocytes.
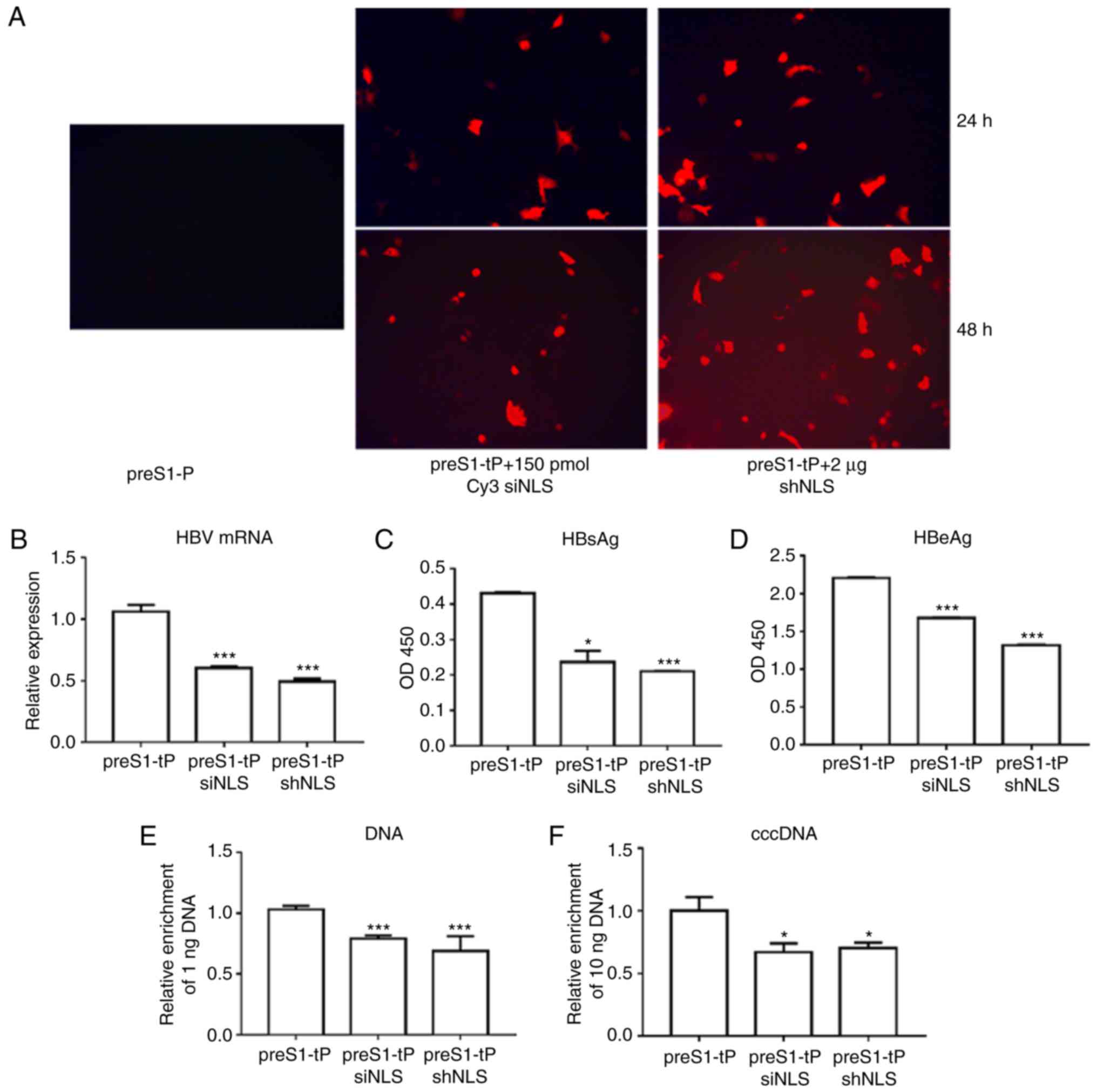 | Figure 5Inhibition of HBV infection by NLS
siRNAs delivered by preS1-tP proteins. (A) Entry of HBV NLS siRNAs
into HepG2.2.15 cells through recombinant preS1-tP proteins by
immunofluorescence (x200). Levels of (B) HBV mRNA, (C) HBsAg, (D)
HBeAg, (E) HBV DNA and (F) cccDNA in HepG2.2.15 cells co-incubated
with recombinant preS1-tP proteins and NLS siRNAs.
*,** and ***, compared with the
preS1-tP group, *P<0.05, **P<0.01,
***P<0.001. HBV, hepatitis B virus; NLS, nuclear
localization sequences; cccDNA, covalently closed circular DNA;
HBsAg, hepatitis B surface antigen; HBeAg, hepatitis B e antigen;
siRNA, small interfering RNA; shRNA, short hairpin RNA; tP,
truncated protamine. |
Discussion
Although the wide application of anti-HBV vaccines
has significantly decreased the incidence of HBV infection, there
were still >350,000,000 patients with CHB globally in 2016
(23). The treatment and
eradication of HBV infections have been limited by the
unavailability of effective therapeutic drugs and methods to
completely remove the cccDNA in the infected hepatocytes. Due to
the significance of the human NTCP gene during HBV infection and
entry, the establishment of hepatocytes with stable NTCP gene
expression has been widely applied as a cellular model for the
investigation of CHB pathology, and for the screening of
therapeutic agents and methods for the treatment of HBV (11, 20, 24–26). Guided by the same rationale, the
present study used HepG2.2.15 cells expressing the NTCP gene to
develop a novel method for delivering HBV NLS siRNAs into
HepG2.2.15 cells. Previous studies have shown that the delivery of
siRNA molecules through s fusion protein containing scFv and tP may
be a novel method for the inhibition of HBV infection and
replication, considering the improved specificity and promoted
affinity (17,18,27,28). However, it is possible that the
high level of existing HBsAg in patients with CHB may be a concern
for the application of recombinant scFv-tP protein-mediated siRNA
delivery in clinics, due to the interaction of asFV with the HBsAg.
To avoid such a disadvantage in siRNA-mediated HBV treatment, the
present study presented a novel anti-HBV siRNA delivery system
using recombinant preS1-tP proteins, in which the preS1 region of
the HBV surface protein was applied to replace the previous
fragment of antibodies against HBsAg.
Based on HepG2.2.15 cells stably expressing the
human NTCP gene and the characterized HBV NLS siRNAs demonstrating
inhibitory effects on the mRNA expression of HBV and production of
two specific antigens in hepatocytes, the present study continued
with the novel delivery method by expressing and purifying the
recombinant preS1-tP proteins in E. coli. The in
vitro co-incubation assay showed that the recombinant preS1-tP
proteins efficiently entered into the HepG2.2.15 cells, and showed
binding of DNA molecules with a high affinity. Finally, it was
shown that the recombinant preS1-tP proteins successfully carried
the HBV NLS siRNAs into the HepG2.2.15 cells stably expressing the
human NTCP gene, which resulted in inhibition of the mRNA
expression of HBV, supported by significantly reduced HBsAg levels
and cccDNA levels in the co-incubated HepG2.2.15 cells. Taken
together, these experimental evidence demonstrated the potential of
this recombinant preS1-tP protein-mediated siRNA delivery strategy
in the clinical treatment of HBV and CHB.
Inspired by the rapid progress of siRNA-based
anti-HBV therapies (23,29), the establishment of this novel
delivery system with improved efficiency may also be applied for
the treatment of HBV with a focus on the siRNA-mediated inhibition
of other associated genes in addition to HBV NLS. Beneficially, a
single siRNA is capable of silencing multiple HBV transcripts
responsible for synthesis of the core protein, surface protein and
polymerase, due to the compact structure of the HBV genome and the
overlapping of HBV transcripts (23). For example, Arrowhead ARC520, the
first siRNA designed for HBV infection and CHB treatment to have
entered clinical development, was shown to reduce the HBV DNA titer
and suppress the expression of HBsAg, HBcAg and HBeAg by targeting
the HBV genome sequence upstream to the core promoter site
(30,31). However, the effect of ARC520 on
inducing a reduction in HBsAg in HBeAg-negative humans and animals
was relatively weak, compared with the HBeAg-positive counterparts.
For the development of more efficient and effective siRNAs for the
treatment of HBV, a number of novel siRNA programmes have been
examined, including ARC-521, TKM-HBV, ALN-HBV and ISIS-HBV, and
even locked nucleic acid (LNA) technology, which silences HBV
transcripts using single-stranded oligodeoxyri-bonucleotides
targeting mRNA molecules derived from HBV cccDNA (23). The novel siRNA-delivering system
developed in the present study, using the recombinant preS1-tP
proteins as carriers, may also be promising if applied for the
treatment of HBV and CHB using these siRNAs, including ARC520 and
LNA technology.
As a preliminary innovation in siRNA delivery, the
technology demonstrated in the present study is far from clinical
application. For example, the specificity of the recombinant
preS1-tP proteins in delivering inhibitory siRNAs into HBV-positive
liver cells requires carefully assessment using both in
vitro and in vivo models prior to its further
development for clinical experiments. The present study was limited
as the efficiency of such a siRNA-delivering system has not been
verified in an animal model. Further confirmation using HBV and CHB
animal models may produce more convincing data and a basis for its
application in the clinical treatment of patients with CHB, with a
focus required on the delivery specificity and safety.
In conclusion, the present study established
HepG2.2.15 cells stably expressing the human NTCP gene as a
cellular model for the development of a novel siRNA-delivering
system for anti-HBV treatment. The recombinant preS1-tP proteins
demonstrated capacity for carrying the HBV NLS siRNAs into the
HepG2.2.15 cells, and subsequently resulting in the significant
suppression of HBV replication, cccDNA formation and HBV-specific
antigen expression. These results showed a promising and improved
technique for the application of siRNA-based anti-CHB therapy,
which may also inspire future innovation in the development of
novel methods for HBV eradication and the treatment of CHB.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 81401706), the Chinese
Foundation for Hepatitis Prevention and Control - Tianqing Liver
Disease Research Fund Subject (grant no. TQGB20150138) and the
Science Research Project of Henan Province (grant no.
142300410372).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
YK conceived and designed this project, and provided
financial support. YZ performed the experiments, analyzed the data
and drafted the manuscript. ZL and JS participated in study design,
study implementation and manuscript revision. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Carey WD: The prevalence and natural
history of hepatitis B in the 21st century. Cleve Clin J Med.
76(Suppl 3): S2–S5. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lai M and Liaw YF: Chronic hepatitis B:
Past, present, and future. Clin Liver Dis. 14:531–546. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lavanchy D: Hepatitis B virus
epidemiology, disease burden, treatment, and current and emerging
prevention and control measures. J Viral Hepat. 11:97–107. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu J, Zhang S, Wang Q, Shen H, Zhang M,
Zhang Y, Yan D and Liu M: Seroepidemiology of hepatitis B virus
infection in 2 million men aged 21-49 years in rural China: A
population-based, cross-sectional study. Lancet Infect Dis.
16:80–86. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lutgehetmann M, Volz T, Köpke A, Broja T,
Tigges E, Lohse AW, Fuchs E, Murray JM, Petersen J and Dandri M: In
vivo proliferation of hepadnavirus-infected hepatocytes induces
loss of covalently closed circular DNA in mice. Hepatology.
52:16–24. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Spangenberg HC, Thimme R and Blum HE:
Tracking cccDNA in chronic HBV infection. Hepatology. 39:1736–1738.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Levrero M, Testoni B and Zoulim F: HBV
cure: Why, how, when? Curr Opin Virol. 18:135–143. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Daoud FS: Branchial cyst: An often
forgotten diagnosis. Asian J Surg. 28:174–178. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kann M, Bischof A and Gerlich WH: In vitro
model for the nuclear transport of the hepadnavirus genome. J
Virol. 71:1310–1316. 1997.PubMed/NCBI
|
|
10
|
Li GQ, Gu HX, Li D and Xu WZ: Inhibition
of Hepatitis B virus cccDNA replication by siRNA. Biochem Biophys
Res Commun. 355:404–408. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Watashi K, Urban S, Li W and Wakita T:
NTCP and Beyond: Opening the door to unveil hepatitis B virus
entry. Int J Mol Sci. 15:2892–2905. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abou-Jaoudé G and Sureau C: Entry of
hepatitis delta virus requires the conserved cysteine residues of
the hepatitis B virus envelope protein antigenic loop and is
blocked by inhibitors of thiol-disulfide exchange. J Virol.
81:13057–13066. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z,
Huang Y, Qi Y, Peng B, Wang H, et al: Sodium taurocholate
cotransporting polypeptide is a functional receptor for human
hepatitis B and D virus. Elife. 1:e000492012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Claro da Silva T, Polli JE and Swaan PW:
The solute carrier family 10 (SLC10): Beyond bile acid transport.
Mol Aspects Med. 34:252–269. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kullak-Ublick GA, Beuers U and Paumgartner
G: Molecular and functional characterization of bile acid transport
in human hepatoblastoma HepG2 cells. Hepatology. 23:1053–1060.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kotani N, Maeda K, Debori Y, Camus S, Li
R, Chesne C and Sugiyama Y: Expression and transport function of
drug uptake transporters in differentiated HepaRG cells. Mol Pharm.
9:3434–3441. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wen WH, Liu JY, Qin WJ, Zhao J, Wang T,
Jia LT, Meng YL, Gao H, Xue CF, Jin BQ, et al: Targeted inhibition
of HBV gene expression by single-chain antibody mediated small
interfering RNA delivery. Hepatology. 46:84–94. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wen WH, Qin WJ, Gao H, Zhao J, Jia LT,
Liao QH, Meng YL, Jin BQ, Yao LB, Chen SY, et al: An hepatitis B
virus surface antigen specific single chain of variable fragment
derived from a natural immune antigen binding fragment phage
display library is specifically internalized by HepG2.2.15 cells. J
Viral Hepat. 14:512–519. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Manasseh D: Protamine binding location on
DNA. 1995.
|
|
20
|
Ko C, Park WJ, Park S, Kim S, Windisch MP
and Ryu WS: The FDA-approved drug irbesartan inhibits HBV-infection
in HepG2 cells stably expressing sodium taurocholate
co-transporting polypeptide. Antivir Ther. 20:835–842. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mcfarland AP, Horner SM, Jarret A, Joslyn
RC, Bindewald E, Shapiro BA, Delker DA, Hagedorn CH, Carrington M,
Gale M Jr, et al: The favorable IFNL3 genotype escapes mRNA decay
mediated by AU-rich elements and hepatitis C virus-induced
microRNAs. Nat Immunol. 15:72–79. 2014. View Article : Google Scholar
|
|
22
|
Urban TJ, Thompson AJ, Bradrick SS, Fellay
J, Schuppan D, Cronin KD, Hong L, McKenzie A, Patel K, Shianna KV,
et al: IL28B genotype is associated with differential expression of
intrahepatic interferon-stimulated genes in patients with chronic
hepatitis C. Hepatology. 52:1888–1896. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gane EJ: Future anti-HBV strategies. Liver
Int. 37(Suppl 1): S40–S44. 2017. View Article : Google Scholar
|
|
24
|
Veloso Alves Pereira I, Buchmann B,
Sandmann L, Sprinzl K, Schlaphoff V, Döhner K, Vondran F, Sarrazin
C, Manns MP, Pinto Marques Souza de Oliveira C, et al: Primary
biliary acids inhibit hepatitis D virus (HDV) entry into human
hepatoma cells expressing the sodium-taurocholate cotransporting
polypeptide (NTCP). PLoS One. 10:e01171522015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen ZW, Luo MY, Hu HH, Zhou H, Jiang HD,
Yu LS and Zeng S: Screening and verifying potential NTCP inhibitors
from herbal medicinal ingredients using the LLC-PK1 cell model
stably expressing human NTCP. Chin J Nat Med. 14:549–560.
2016.PubMed/NCBI
|
|
26
|
Donkers J, Zehnder B, van Westen GJP,
Kwakkenbos MJ, IJzerman AP, Oude Elferink RPJ, Beuers U, Urban S
and van de Graaf SFJ: Reduced hepatitis B and D viral entry using
clinically applied drugs as novel inhibitors of the bile acid
transporter NTCP. Sci Rep. 7:153072017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen YM, Yin B, Guo XC, Wang W, Zheng Q,
Wang F, Sun D, Li D, Ren G and Yin J: Selection of
affinity-improved neutralizing human scFv against HBV PreS1 from
CDR3 VH/VL mutant library. Biologicals. 44:271–275. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang H, Yang Y, Wang W, Guan B, Xun M,
Zhang H, Wang ZL and Zhao Y: Construction and verification of
anti-MM scFv-tP fusion protein expression vector. Nan Fang Yi Ke Da
Xue Xue Bao. 1149–1155. 2017.In Chinese.
|
|
29
|
Marimani M, Hean J, Bloom K, Ely A and
Arbuthnot P: Recent advances in developing nucleic acid-based HBV
therapy. Future Microbiol. 8:1489–1504. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu Z, Chavez D, Guerra B, Littlejohn M,
Peterson R, Locarnini S, Gish R, Anzalone C, Kanner S, Goetzmann J,
et al: Treatment of chronically HBV-infected chimpanzees with RNA
interference therapeutic ARC-520 LED to potent reduction of viral
MRNA, DNA and proteins without observed drug resistance. J Hepatol.
64:S398. 2016. View Article : Google Scholar
|
|
31
|
Yuen MF, Chan HLY, Liu K, Given BD,
Schluep T, Hamilton J, Lai CL, Locarnini SA, Lau JYN, Ferrari C and
Gish RG: Differential reductions in viral antigens expressed from
CCCDNAVS integrated DNA in treatment Naïve HBEAG positive and
negative patients with chronic HBV after RNA interference therapy
with ARC-520. J Hepatol. 64:S390–S391. 2016. View Article : Google Scholar
|